Abstract
Low dietary folate intake is associated with an increased risk for colon cancer; however, relevant genetic animal models are lacking. We therefore investigated the effect of targeted ablation of two folate transport genes, folate binding protein 1 (Folbp1) and reduced folate carrier 1 (RFC1), on folate homeostasis to elucidate the molecular mechanisms of folate action on colonocyte cell proliferation, gene expression, and colon carcinogenesis. Targeted deletion of Folbp1 (Folbp1+/− and Folbp1−/−) significantly reduced (P < 0.05) colonic Folbp1 mRNA, colonic mucosa, and plasma folate concentration. In contrast, subtle changes in folate homeostasis resulted from targeted deletion of RFC1 (RFC1+/−). These animals had reduced (P < 0.05) colonic RFC1 mRNA and exhibited a 2-fold reduction in the plasma S-adenosylmethionine/S-adenosylhomocysteine. Folbp1+/− and Folbp1−/− mice had larger crypts expressed as greater (P < 0.05) numbers of cells per crypt column relative to Folbp1+/+ mice. Colonic cell proliferation was increased in RFC1+/− mice relative to RFC1+/+ mice. Microarray analysis of colonic mucosa showed distinct changes in gene expression specific to Folbp1 or RFC1 ablation. The effect of folate transporter gene ablation on colon carcinogenesis was evaluated 8 and 38 weeks post-azoxymethane injection in wild-type and heterozygous mice. Relative to RFC1+/+ mice, RFC1+/− mice developed increased (P < 0.05) numbers of aberrant crypt foci at 8 weeks. At 38 weeks, RFC1+/− mice developed local inflammatory lesions with or without epithelial dysplasia as well as adenocarcinomas, which were larger relative to RFC1+/+ mice. In contrast, Folbp1+/− mice developed 4-fold (P < 0.05) more lesions relative to Folbp1+/+ mice. In conclusion, Folbp1 and RFC1 genetically modified mice exhibit distinct changes in colonocyte phenotype and therefore have utility as models to examine the role of folate homeostasis in colon cancer development.
Introduction
Colon cancer is a major health concern in the United States and is the second leading cause of death from cancer (1). Interestingly, an insufficient supply of methyl group donors from folate has been linked to the promotion of colorectal tumorigenesis (2–8). The implications of low folate status are four-fold and include (a) alterations in global methylation (9–11), (b) impaired DNA repair (12–14), (c) enhanced cell proliferation (15, 16), and (d) impaired DNA synthesis (17, 18). There is cogent clinical and epidemiologic evidence linking low folate status to increased proliferation of colonocytes and an elevated risk for developing colon cancer in humans (4, 7, 8, 19) and in experimental animal models (2, 3, 6, 20, 21). Conversely, folate supplementation is protective. Patients at risk for colon cancer supplemented with folate were shown to have decreased colonic mucosal cell proliferation (22). In addition, rats supplemented with folate were protected against the development of macroscopic colonic tumors induced by the colon-specific carcinogen, dimethylhydrazine (2). However, the precise mechanisms by which aberrant folate status perturbs crypt cytokinetics and enhances colon cancer risk remain to be determined.
Dietary intervention has been a primary means to alter folate status in rodent animal models of colon cancer (2, 3, 6, 20, 21). Of concern, these models may be prone to moderate short-term compensatory up-regulation of DNA methyltransferase activity associated with dietary induced folate deficiency (5, 6). In addition, diet-induced folate deficiency observed in rodent models is often very extreme and may not resemble the subclinical folate deficient status found in 30% of the U.S. population (23) nor the modestly reduced folate status found in some colon cancer patients (24).
At present, the molecular basis of folate deficiency–mediated colon carcinogenesis is not well understood. We therefore engineered mice with targeted ablation of either the reduced folate carrier 1 (RFC1) or folate binding protein 1 (Folbp1) gene to elucidate the molecular and cellular mechanisms of folate action on colonocyte cell proliferation, gene expression, and colon carcinogenesis. RFC1 and Folbp1 are well-characterized folate transporters (25–27). RFC1 is a facilitative anion exchanger that mediates folate delivery into a variety of cells, including colonocytes (27), and has a high affinity for reduced folates, such as the primary physiologic substrate, 5-methyltetrahydrofolate (27). In comparison, folate receptors (Folbp1 and Folbp2 in mice and α and β in humans) are coupled to the plasma membrane via a glycosylphosphatidylinositol linkage and transport folic acid and 5-methyltetrahydrofolate with high affinity (28). Folbp1 is highly expressed in the kidney proximal tubule (28) but not in the small intestine (29, 30) and is found in low levels in the colon (31).
Using these mouse models, we were able to show that inactivation of cellular folate transport mechanisms (RFC1 and Folbp1) perturbs folate status, alters colonic cytokinetics, and increases the development of preneoplastic biomarkers of colon cancer. The results of this study highlight the utility of these mouse models to examine the relationship between compromised folate homeostasis and colon cancer development.
Materials and Methods
Animals and Diet
Genetically modified C57BL/6J × 129/Sv Folbp1 and SWV/Fnn RFC1 male mice, 80 to 100 days old, were housed in a temperature and humidity controlled animal facility with a 12-hour light/dark cycle and initially fed defined diets (Harlan Teklad, Indianapolis, IN) containing 2.7 mg folate/kg. Animals enrolled into the carcinogen phase of the study were maintained on a semipurified defined diet (Harlan Teklad) containing 2.0 mg folate/kg, 44.57 (g/100 g diet) corn starch, 15.0 casein, 15.5 sucrose, 15.0 corn oil (Degussa Bio Actives, Champaign, IL), 5.0 fiber (cellulose), 3.5 AIN-93 mineral mix, 1.0 AIN-93-VX mineral mix, 0.25 choline bitartrate, and 0.18 L-cysteine. All procedures were done in accordance with the animal experimentation guidelines of Texas A&M University.
Folbp1 and RFC1 Mice
The generation of Folbp1 knockout mice has been described previously (9, 32). Folbp1−/− mice are embryonic lethal but may be rescued by supplementation of dams with folate 2 weeks before mating and throughout pregnancy, and pups do not require further folate supplementation (9, 32). Therefore, these mice have limited utility and were only used for basic characterization purposes in this study. The generation of RFC1 knockout mice is described herein. RFC1−/− mice are also embryonic lethal; however, supplementation of dams with various folate sources does not yield viable offspring (data not shown); thus, no data are presented for RFC1−/− mice in this study. In a comparable RFC1 knockout mouse model, Zhao et al. (33) also reported that folate rescue of RFC1−/− mice is not possible.
Cloning of RFC1
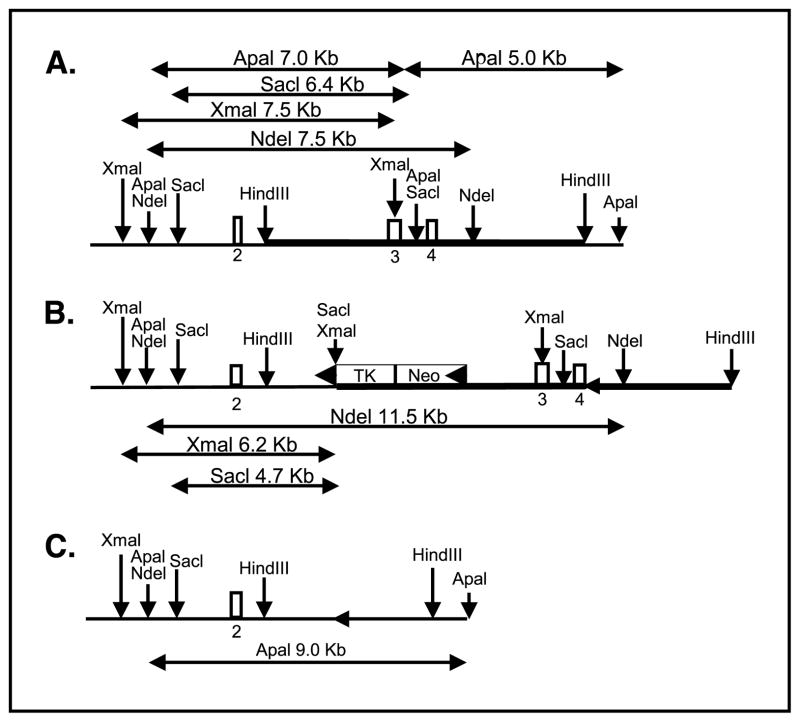
A bacteriophage Pl library (Genome Systems, St. Louis, MO) prepared from the inbred mouse strain 129/Sv genomic DNA was screened by PCR with primers specific for exon 3 of mouse RFC1 (TGCGATACAAGCCAGTCTTGG and GCACCAGGGAGAATATGTAG-GAGG). We identified one positive clone containing an 80-kb insert. Digestion of the clone with HindIII and probing with exon 3 identified a 7.5-kb fragment containing exons 3 and 4 (Fig. 1). This fragment was subcloned into pBluescript (Stratagene, La Jolla, CA) and used to develop a CreloxP-based conditional knockout targeting construct. The RFC1 clone was restriction mapped and a loxP-flanked neo-TK was introduced into the unique AocI site. An additional loxP site was introduced into the NsiI site located downstream from exon 4 by partial digestion, thus also ensuring that the NsiI site was destroyed. The targeting construct (Fig. 1) was linearized with HindIII and introduced into the E14 ES cell line.
Figure 1.
Targeted inactivation of the murine RFC1. A, endogenous genomic loci. B, targeted RFC1 loci before Cre-mediated excision. Thick line, targeting construct; arrowhead, location of loxP sites. C, targeted locus following Cre-mediated excision.
Electroporation and Validation
The ES cell line, E14TG2a, was used and cells were maintained in DMEM supplemented with 15% fetal bovine serum, 0.1 mmol/L 2-mercaptoethanol, and 2 mmol/L glutamine. ES cells were cocultured with embryonic fibroblast feeders as described previously (34). Cells were electroporated essentially as described by Reid et al. (35) using linearized DNA at a final concentration of 2 to 5 nmol/L. Electroporated cells were plated at a density of 1 × 106 to 2 × 106 cells per 10 cm plate. Twenty-four hours after electroporation, cells were placed in 150 μg/mL G418. After 7 to 10 days, G418-resistant colonies were selected for expansion and analysis. From 1,350 colonies resistant to G418, 162 were analyzed by Southern blot and 4 were found to be targeted. Targeted colonies were identified by genomic Southern blotting. As shown in Fig. 1 for the initial targeting event, digestion with SacI, XmaI, or NdeI and probing with exon 2 can differentiate between the endogenous allele and the targeted allele. Once a targeted colony was identified, it was expanded and electroporated with 3 nmol/L Cre-expressing plasmid (36). Following electroporation, cells were placed in ganciclovir to select for loss of the neo-TK insert. Of 60 clones analyzed, 33 had lost the neo-TK but not exons 3 and 4, whereas 27 clones had lost both the neo-TK insert and exons 3 to 4. The latter colonies were further expanded and injected into C57BL/6 blastoacysts, and chimeric progeny were bred to C57BL/6 to ascertain germ line transmission of the mutant allele. The resulting lines were maintained on a SWV/Fnn background. As shown in Fig. 1C, the deletion event resulted in the complete removal of exons 3 and 4 of the RFC1 gene. This deleted allele was differentiated from the endogenous allele using an ApaI digest and probing with exon 2.
Folate, S-Adenosylmethionine, and S-Adenosylhomocysteine Assays
The total folate levels in plasma were determined using the Lactobacillus casei turbidimetric assay (37) with slight modification as described previously (38). Plasma concentrations of S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) were determined by high-pressure liquid chromatography using electrochemical detection as described previously (39). Tissue folate levels were determined using scraped colonic mucosa as described previously (40). Tissue SAM and SAH were also quantified (41).
Measurement of Colonic Cell Cytokinetics
In vivo cell proliferation was determined by immunohistochemical detection. Mice were injected i.p. with bromodeoxyuridine (BrdUrd, 50 mg/kg body weight). Mice were terminated 1 hour after injection. Colons were excised, slit open longitudinally, and rinsed thoroughly in cold PBS. Colons were then fixed in paraformaldehyde and embedded in paraffin as Swiss rolls (42, 43). The incorporation of BrdUrd into DNA of actively dividing cells was determined using a commercially available kit (Zymed, South San Francisco, CA). Crypt size and proliferative activity in distal sections were determined as described previously (44).
Quantitative Real-time PCR
Expression of Folbp1 and RFC1 in knockout mice was determined using mRNA isolated from colonic mucosa and whole kidneys. In addition, selected genes implicated in colon cancer development and regulated by CpG promoter methylation were quantified by real-time PCR. These genes included E-cadherin (Cdh1), caudal type homeobox 1 (Cdx1), decorin (Dcn), estrogen receptor-1α (Esr1), insulin-like growth factor-II (Igf2), N-myc downstream regulated 2 (Ndr2), phosphoinositide 3′-kinase catalytic polypeptide (Pik3cg), and prostaglandin-endoperoxide synthase 2 (Ptgs2). Isolation and analysis of RNA was done as described previously by Davidson et al. (45). RNA was isolated using the Totally RNA extraction kit (Ambion, Austin, TX). Total RNA was quantified with the Agilent 2100 Bioanalyzer. Real-time PCR was done using the ABI 7700 (Applied Biosystems, Foster City, CA) and Taqman Probes (Assayson-Demand, Applied Biosystems).
CodeLink Microarray Analysis
Colonic mucosa from ~100-day-old adult mice was processed in strict accordance to the CodeLink Gene Expression Assay manual (Amersham, Piscataway, NJ) and analyzed using the Mouse UniSet 10K Expression Bioarray. Each array contained a broad range of genes (~10,000) derived from publicly available, well-annotated mRNA sequences. Microarray analyses were done on colonic mucosa comparing gene expression in Folbp1+/+ versus Folbp1+/− and in RFC1+/+ versus RFC1+/− mice (n = 3 for each genotype). For local background subtraction, median intensity values from all background pixels of a particular gene were subtracted from the median signal for that gene; effective signals <0 were increased to 1 using a floor function to avoid downstream “division by 0” or “logs of 0” errors. Global median scaling factors were computed for each array separately. Every effective gene signal was divided by its corresponding scaling factor for normalization. Standard ratios were calculated as the median signal for the heterozygous group divided by the median signal for the wild-type group. The ratios were logged (base 2) to transform the data into an equidistant domain. Average intensities were calculated as Ia = sqrt(I1 × I2) (46). Probability estimates for each gene were determined by Student’s t test, where the resulting P was solely used for relative ranking purposes. The data sets were filtered based on the following criteria: (a) genes with P > 0.05 were excluded, (b) genes with an average intensity below the median expression intensity of all genes on the same array were excluded, and (c) genes with >2-fold differential expression were excluded.
Carcinogen Treatment, Aberrant Crypt, and Histologic Analyses
Adult male mice, ~100 days old, were injected with the carcinogen, azoxymethane. Mice received an initial s.c. injection of azoxymethane at 10 mg/kg body weight followed by a second injection of 5 mg/kg body weight 7 days later. Eight weeks after the second azoxymethane injection, mice were terminated for aberrant crypt foci (ACF) analysis. The colon was excised and flushed with cold PBS, inflating it to twice its normal diameter. Fecal pellets were worked up and down to break the circular muscle fibers and to remove mucin adhering to the epithelium. The colon was slit longitudinally and fixed flat between two pieces of Whatman No. 1 filter paper and placed under a glass plate in 4% paraformaldehyde for 4 hours. The fixed tissue was then washed in 50% ethanol four times followed by three washes with 70% ethanol. Each wash was 20 minutes in duration. Fixed colons were stained with 0.2% methylene blue in PBS for 5 minutes and then placed on a light microscope equipped with a clear grid and visualized under low magnification (47). ACFs were scored for total number and multiplicity (number of crypts per focus) as described previously (48).
Thirty-eight weeks after the last azoxymethane injection, mice were terminated and colons were excised, slit longitudinally, and flushed with PBS. Lesions were mapped, excised, and fixed in 4% paraformaldehyde as described above. H&E-stained sections were subsequently viewed and blindly scored by a board-certified pathologist.
Statistical Analysis
Data were analyzed using the least significant difference test on multiple means, and the Student’s t test (one-tailed) was used in analyses with two means. Data are expressed as mean ± SE.
Results
Expression of RFC1 and Folbp1 in the Colon and Kidney
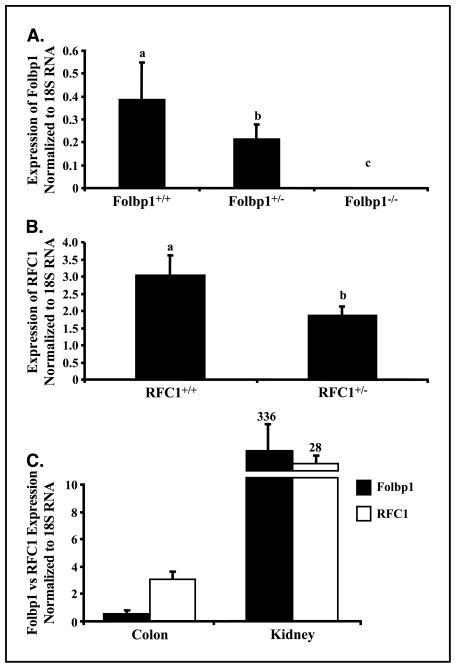
Data presented in Fig. 2 show that allelic ablation of Folbp1 (Fig. 2A) and RFC1 (Fig. 2B) result in the expected reduction in the level of mRNA for each respective gene in colonic mucosa. RFC1 expression in the colon of wild-type mice was substantially greater than that of Folbp1, consistent with the concept that folate absorption in the colon is primarily mediated by RFC1 (27). In contrast, Folbp1 was highly expressed in the kidney relative to RFC1. This is consistent with the reported role of Folbp1 in the reabsorption of folate (49).
Figure 2.
Expression of Folbp1 and RFC1 in knockout mice. Expression was determined by quantitative real-time PCR as described in Materials and Methods. A, Folbp1 mRNA expression in the colon in Folbp knockout mice. B, RFC1 mRNA expression in the colon in RFC1 knockout mice. C, relative expression of Folbp1 and RFC1 in colonic mucosa and kidney in wild-type animals. Columns, mean (n = 5–8) normalized to 18S rRNA expression; bars, SE.
Perturbation of Folate, SAM, and SAH Status in RFC1 and Folbp1 Mice
The effect of reduced expression of RFC1 and Folbp1 on colonic tissue and plasma folate status is summarized in Table 1. It should be noted that observed changes in measures between Folbp1 and RFC1 mice are likely attributable to strain differences. Therefore, direct comparisons between these two genetic mouse models cannot be made. Tissue and plasma folate levels in RFC1+/− mice were not altered by RFC1 gene ablation. In contrast, tissue and plasma folate levels in Folbp1+/− and Folbp1−/− mice decreased in a predictable manner, consistent with a reduction in Folbp1 expression. These data indicate that Folbp1 gene ablation is effective in reducing circulating and tissue folate stores.
Table 1.
Plasma and tissue folate, SAH, and SAM in Folbp and RFC1 knockout mice
| Folbp1+/+ | Folbp1+/− | Folbp1−/− | RFC1+/+ | RFC1+/− | |
|---|---|---|---|---|---|
| Plasma | |||||
| Folate (ng/mL) | 30.4 ± 2.0a | 17.0 ± 2.8b | 10.4 ± 0.5c | 21.6 ± 7.6 | 28.6 ± 2.4 |
| SAH (nmol/L) | 41.4 ± 8.8 | 51.4 ± 6.9 | 32.6 ± 3.1 | 23.7 ± 4.2 | 41.5 ± 9.7 |
| SAM (nmol/L) | 49.5 ± 9.4 | 79.7 ± 12.7 | 42.5 ± 1.6 | 124.1 ± 9.5 | 101.0 ± 5.5 |
| SAM/SAH | 1.4 ± 0.4 | 1.6 ± 0.2 | 1.3 ± 0.1 | 5.8 ± 1.3x | 2.7 ± 0.5y |
| Colonic mucosa | |||||
| Folate (ng/mg protein) | 19.2 ± 2.2a | 13.1 ± 0.8b | 10.5 ± 0.4c | 11.9 ± 2.2 | 10.0 ± 1.8 |
| SAH (pmol/mg tissue) | 3.8 ± 0.8 | 4.4 ± 0.6 | 5.7 ± 1.0 | 8.8 ± 0.6 | 9.6 ± 0.6 |
| SAM (pmol/mg tissue) | 14.3 ± 3.1 | 21.5 ± 2.2 | 21.6 ± 1.7 | 17.1 ± 1.6 | 19.0 ± 1.4 |
| SAM/SAH | 5.3 ± 2.1 | 5.2 ± 0.7 | 4.4 ± 0.6 | 1.9 ± 0.1 | 2.1 ± 0.2 |
NOTE: Scraped colonic mucosa and blood plasma from mice fed chow diets containing 2.7 mg folate/kg were analyzed for folate content using a standard microbiological bioassay as described in Materials and Methods. Plasma SAH and SAM were determined by HPLC as described in Materials and Methods. Mean ± SE (n = 2–7). Mouse models with different letters are significantly different (P < 0.05) within a row.
We also evaluated additional key folate metabolic intermediates in colonic mucosa (i.e., SAM and SAH) given that folate can directly affect cellular SAM status, which is the primary methyl donor for DNA methylation (21, 50). The concentration of colonic SAM and SAH (pmol/mg tissue) was not altered in either RFC1+/−, Folbp1+/−, or Folbp1 −/− mice (Table 1). In addition to absolute levels of SAM and SAH, the relative ratios of SAM and SAH in plasma may predict cellular methylation potential (39). Although there was no significant effect of RFC1 and Folbp1 ablation on plasma SAM and SAH levels, these values did approach significance. When expressed as the ratio of SAM/SAH, the ratio was significantly (P < 0.05) reduced in RFC1+/− mice. These results suggest that one carbon metabolism may be adversely affected by RFC1 gene ablation in RFC1+/− mice.
Reduced Expression of RFC1 and Folbp1 Adversely Affects Colonic Cytokinetics
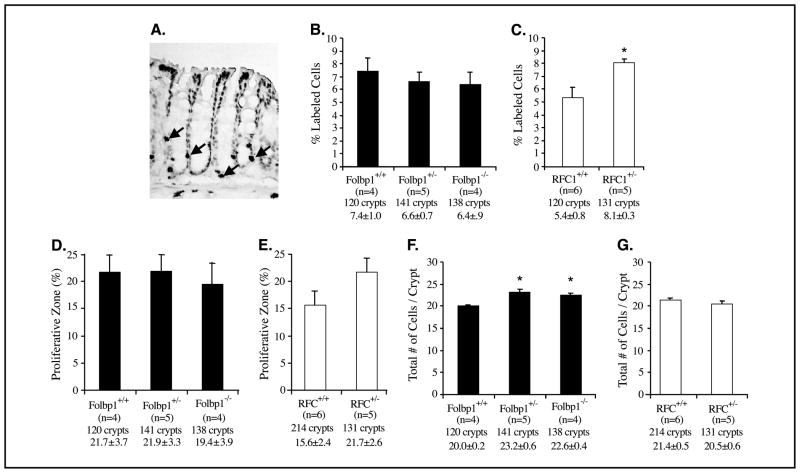
Because reduced folate status can affect cell proliferation, we also examined the effect of RFC1 and Folbp1 targeted deletion on colonocyte proliferation by assessing BrdUrd incorporation into dividing cells. The labeling index (% labeled cells divided by total cells) was significantly (P < 0.05) increased in RFC1+/− but not in the Folbp1 mice (Fig. 3B and C). The size of the proliferative zone, measured as the position of the highest labeled cell in the crypt column, was not different among the Folbp1 genotypes but was enlarged in RFC1+/− mice although not significantly (P > 0.05; Fig. 3D and E). In contrast, a significantly (P < 0.05) greater number of colonic cells per crypt column were found in Folbp1 but not RFC1 heterozygous mutants (Fig. 3F and G). These results indicate that reduced RFC1 expression stimulates hyperproliferation within the colonic crypt, whereas reduced Folbp1 expression disrupts homeostatic mechanisms regulating cell numbers in the colonic epithelium.
Figure 3.
Effect of targeted inactivation of Folbp1 and RFC1 on colonic cell proliferation. A, representative photomicrographs (×200) of colonic crypts stained for BrdUrd. Arrows, cycling cells stained with BrdUrd. Labeling index or percentage of colonic cells proliferating [Folbp mice (B) and RFC1 mice (C)] is percentage of BrdUrd-stained cells relative to the total number of cells per crypt column. Proliferative zone data [Folbp mice (D) and RFC1 mice (E)] are the measure of the highest labeled cell within the crypt divided by the total number of cells within a crypt (×100). This provides an indication of the size of the proliferative compartment within the crypt column. Absolute number of cells in a crypt column [Folbp mice (F) and RFC1 mice (G)] is the overall size of the crypt. Comparisons are not made between Folbp and RFC1 genotypes because these animals are on different genetic backgrounds. Below each column, genotype, number of animals, total number of crypts scored, and mean ± SE. *, P < 0.05, relative to wild-type animals.
Microarray Analysis and Real-time PCR Validation
To further elucidate the effect of folate transport gene ablation on adaptive responses in the colon, microarray analyses were done on colonic mucosa isolated from “baseline” (noninjected) mice. In total, seven genes were differentially expressed (more than 2-fold difference) between RFC1+/− and RFC1+/+ mice (Table 2). In contrast, a total of 115 genes having a minimum 2-fold change in gene expression were differentially expressed in Folbp1+/− and Folbp1+/+ mice (Table 3). Only genes with annotations are reported in Table 3. The complete microarray data set is available online.9
Table 2.
Differentially expressed genes in RFC1+/+ and RFC1+/− mice
| Accession no. | Symbol | Gene description | Fold change, heterozygous/wild-type | P |
|---|---|---|---|---|
| Transcription/translation | ||||
| NM_011543 | Skp1a | S-phase kinase-associated protein 1A | 0.44 | 0.04 |
| NM_009099 | Trim30 | Tripartite motif protein 30 | 0.36 | 0.01 |
| Immune response | ||||
| NM_010724 | Psmb8 | Proteosome (prosome, macropain) subunit, β type 8 (large multifunctional protease 7) | 0.43 | 0.05 |
| NM_010738 | Ly6a | Lymphocyte antigen 6 complex, locus A | 0.49 | 0.04 |
| G-protein signaling | ||||
| NM_008330 | Olfr56 | Olfactory receptor 56 | 0.49 | 0.01 |
| NM_007588 | Calcr | Calcitonin receptor | 2.85 | 0.04 |
| Others | ||||
| NM_013864 | Ndr2 | N-myc downstream regulated 2 (cell differentiation) | 2.17 | 0.04 |
NOTE: Genes were selected based on a significant (P < 0.05) and a differential fold change in expression >2 or <0.5. From each genotype, colonic mucosa from three noninjected (100-day-old baseline) mice was collected and processed for RNA isolation. Fold change refers to the normalized signal intensity of a given gene in RFC1+/− (heterozygous) relative to RFC1+/+ (wild-type) mice.
Table 3.
Differentially expressed genes in Folbp1+/+ and Folbp1+/− mice
| Accession no. | Symbol | Gene description | Fold change, heterozygous/wild-type | P |
|---|---|---|---|---|
| Apoptosis | ||||
| NM_010062 | DNase2a | DNase IIα | 0.49 | 0.01 |
| NM_011050 | Pdcd4 | Programmed cell death 4 | 0.39 | 0.01 |
| NM_007465 | Birc3 | Baculoviral IAP repeat-containing 3 | 0.35 | 0.02 |
| Cell adhesion | ||||
| NM_009675 | Aoc3 | Amine oxidase, copper-containing 3 | 2.11 | 0.05 |
| NM_008483 | Lamb2 | Laminin, β2 | 0.43 | 0.03 |
| Cell cycle/cell proliferation | ||||
| NM_013525 | Gas5 | Growth arrest–specific 5 | 2.35 | 0.005 |
| AF236887 | Atr | Ataxia telangiectasia and Rad3 related | 0.42 | 0.03 |
| NM_011369 | Shcbp1 | Shc SH2-domain binding protein 1 | 0.39 | 0.02 |
| NM_010784 | Mdk | Midkine | 0.38 | 0.03 |
| Nucleotide metabolism and transport | ||||
| AB020203 | Ak3l | Adenylate kinase 3α like | 2.66 | 0.01 |
| NM_053103 | Lysal2 | Lysosomal apyrase-like 2 | 2.00 | 0.01 |
| NM_007398 | Ada | Adenosine deaminase | 0.45 | 0.05 |
| NM_016690 | Hnrpdl | Heterogeneous nuclear ribonucleoprotein D like | 0.44 | 0.04 |
| DNA binding | ||||
| NM_009561 | Zfp61 | Zinc finger protein 61 | 0.48 | 0.04 |
| NM_020618 | Smarce1 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily E, member 1 | 0.45 | 0.04 |
| NM_015781 | Nap1l1 | Nucleosome assembly protein 1 like 1 | 0.42 | 0.01 |
| NM_021790 | Solt | SoxLZ/Sox6 leucine zipper binding protein in testis | 0.37 | 0.04 |
| RNA binding | ||||
| NM_024199 | Cstf1 | Cleavage stimulation factor, 3′ pre-RNA, subunit 1 | 2.05 | 0.01 |
| AK008240 | Snrpf | Small nuclear ribonucleoprotein polypeptide | 0.45 | 0.04 |
| Transcription/translation | ||||
| AK011545 | Basp1 | Neuronal axonal membrane protein (Nap-22) | 0.31 | 0.03 |
| NM_009385 | Titf1 | Thyroid transcription factor 1 | 2.95 | 0.02 |
| NM_011297 | Rps24 | Ribosomal protein S24 | 0.45 | 0.03 |
| D83146 | Six5 | Sine oculis-related homeobox 5 homologue (Drosophila) | 0.43 | 0.02 |
| AK019500 | Syncrip | Synaptotagmin binding, cytoplasmic RNA interacting protein | 0.39 | 0.01 |
| S66855 | Hoxb9 | Homeobox B9 | 0.31 | 0.05 |
| Eicosanoid | ||||
| NM_010160 | Cugbp2 | CUG triplet repeat, RNA binding protein 2 | 0.23 | 0.02 |
| AF233645 | Cyp4f15 | Cytochrome P450, family 4, subfamily F, polypeptide 15 | 0.13 | 0.05 |
| Glycosylation | ||||
| NM_009178 | Siat4c | Sialyltransferase 4C (β-galactoside α-2,3-sialytransferase) | 3.74 | 0.02 |
| NM_009176 | Siat6 | Sialyltransferase 6 (N-acetyllacosaminide α-2,3-sialyltransferase) | 2.71 | 0.01 |
| NM_028189 | B3gnt3 | UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase 3 | 2.54 | 0.005 |
| AK006263 | Siat10 | Sialyltransferase 10 (α-2,3-sialyltransferase VI) | 2.29 | 0.05 |
| G-protein related | ||||
| AK008273 | Arhgdib | Rho, GDP dissociation inhibitor (GDI) β | 0.32 | 0.01 |
| NM_010336 | Edg2 | Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 | 0.32 | 0.002 |
| NM_008376 | Imap38 | Immunity-associated protein | 0.28 | 0.0004 |
| Immune function | ||||
| NM_009399 | Tnfrsf11a | Tumor necrosis factor receptor superfamily, member 11a | 4.39 | 0.02 |
| NM_008532 | Tacstd1 | Tumor-associated calcium signal transducer 1 | 3.48 | 0.01 |
| NM_007657 | Cd9 | CD9 antigen | 3.39 | 0.01 |
| AJ006130 | Ik | IK cytokine | 0.48 | 0.003 |
| NM_011157 | Prg | Proteoglycan, secretory granule | 0.46 | 0.003 |
| NM_008510 | Xcl1 | Chemokine (C motif) ligand 1 | 0.41 | 0.01 |
| NM_017372 | Lyzs | Lysozyme | 0.30 | 0.03 |
| NM_019391 | Lsp1 | Lymphocyte-specific 1 | 0.25 | 0.03 |
| NM_013590 | Lzp-s | P lysozyme structural | 0.25 | 0.04 |
| NM_013706 | Cd52 | CD52 antigen | 0.25 | 0.01 |
| Kinase/phosphatase | ||||
| NM_026268 | Dusp6 | Dual-specificity phosphatase 6 | 2.47 | 0.05 |
| NM_007394 | Acvr1 | Activin A receptor, type 1 | 2.19 | 0.04 |
| NM_054097 | Pip5k2c | Phosphatidylinositol-4-phosphate 5-kinase type II | 2.06 | 0.002 |
| NM_008846 | Pip5k1a | Phosphatidylinositol-4-phosphate 5-kinase type Iα | 2.03 | 0.02 |
| AK011110 | Riok3 | RIO kinase 3 (yeast) | 0.48 | 0.01 |
| AK007215 | Wdfy1 | WD40 and FYVE domain containing 1 | 0.40 | 0.01 |
| Lipid related | ||||
| NM_011868 | Peci | Peroxisomal 3, 2-enoyl-CoA isomerase | 2.79 | 0.02 |
| NM_008094 | Gba | Glucosidase, β acid | 2.49 | 0.01 |
| NM_010941 | Nsdhl | NAD(P)-dependent steroid dehydrogenase-like | 2.28 | 0.05 |
| NM_009415 | Tpi | Triosephosphate isomerase | 2.13 | 0.02 |
| NM_008149 | Gpam | Glycerol-3-phosphate acyltransferase, mitochondrial | 0.46 | 0.01 |
| NM_020573 | Osbpl1a | Oxysterol binding protein-like 1A | 0.30 | 0.04 |
| Protein degradation | ||||
| AF303829 | Ube2v1 | Ubiquitin-conjugating enzyme E2 variant 1 | 2.15 | 0.03 |
| NM_019562 | Uchl5 | Ubiquitin carboxyl-terminal esterase L5 | 0.48 | 0.02 |
| AF098949 | Coq7 | Demethyl-Q7 | 0.40 | 0.001 |
| NM_020593 | Fbxo3 | F-box only protein 3 | 0.35 | 0.003 |
| Amino acid transport/metabolism | ||||
| NM_008577 | Slc3a2 | Solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 2.70 | 0.003 |
| U68526 | Bcat2 | Branched chain aminotransferase 2, mitochondrial | 2.04 | 0.04 |
| NM_025314 | Hars2 | Histidyl tRNA synthetase 2 | 0.44 | 0.01 |
| Transport | ||||
| U33012 | Aqp4 | Aquaporin 4 | 6.25 | 0.04 |
| NM_017474 | Clca3 | Chloride channel calcium activated 3 | 6.22 | 0.001 |
| NM_022411 | Slc13a2 | Solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 | 2.38 | 0.02 |
| NM_017391 | Mrps6 | Mitochondrial ribosomal protein S6 | 2.00 | 0.02 |
| NM_015751 | Abce1 | ATP-binding cassette, subfamily E (OABP), member 1 | 0.50 | 0.02 |
| AK005223 | Ap1s2 | Adaptor-related protein complex 1, 2 subunit | 0.24 | 0.04 |
| NM_021301 | Slc15a2 | Solute carrier family 15 (H+/peptide transporter), member 2 | 0.14 | 0.01 |
| Metabolism | ||||
| NM_007409 | Adh1 | Alcohol dehydrogenase 1 (class I) | 2.80 | 0.05 |
| X03796 | Aldo3 | Aldolase 3, C isoform | 2.28 | 0.02 |
| U27014 | Sdh1 | Sorbitol dehydrogenase 1 | 2.05 | 0.01 |
| NM_023113 | Aspa | Aspartoacylase (aminoacylase) 2 | 0.41 | 0.03 |
| Other | ||||
| NM_053108 | Glrx1 | Glutaredoxin 1 (thioltransferase) (protein metabolism) | 2.43 | 0.05 |
| NM_021895 | Actn4 | Actinin α4 (actin and calcium binding) | 2.43 | 0.001 |
| NM_011503 | Stxbp2 | Syntaxin binding protein 2 (protein trafficking) | 2.36 | 0.03 |
| NM_011222 | Pvt1 | Plasmacytoma variant translocation 1 (B-cell malignancy) | 2.35 | 0.02 |
| NM_019649 | Clptm1 | Cleft lip and palate associated transmembrane protein 1 (cleft lip) | 2.27 | 0.03 |
| AA409743 | Tagln2 | Transgelin 2 (muscle development) | 2.09 | 0.01 |
| NM_007509 | Atp6v1b2 | ATPase, H+ transporting, V1 subunit B, isoform 2 (ATPase) | 2.08 | 0.04 |
| NM_009304 | Syngr2 | Synaptogyrin 2 (synaptic protein) | 2.05 | 0.05 |
| NM_011072 | Pfn1 | Profilin 1 (actin binding protein) | 2.03 | 0.001 |
| NM_016902 | Nphp1 | Nephronophthisis 1 (juvenile) homologue (human) (kidney disease) | 0.50 | 0.02 |
| NM_009311 | Tac1 | Tachykinin 1 (neural signaling) | 0.50 | 0.03 |
| NM_009441 | Ttc3 | Tetratricopeptide repeat domain 3 (pathogenesis) | 0.48 | 0.04 |
| NM_009804 | Cat | Catalase (antioxidant) | 0.46 | 0.02 |
| AK011899 | Tbce | Tubulin-specific chaperone (tubulin folding) | 0.44 | 0.05 |
| NM_008017 | Smc2l1 | SMC2 structural maintenance of chromosomes 2-like 1 (yeast) (chromosomal protein) | 0.43 | 0.04 |
| NM_011169 | Prlr | Prolactin receptor (receptor) | 0.40 | 0.003 |
| NM_026352 | Ppid | Peptidylprolyl isomerase D (cyclophilin D) (protein folding) | 0.39 | 0.01 |
| D00208 | S100a4 | S100 calcium binding protein A4 (calcium binding) | 0.39 | 0.003 |
| NM_019939 | Mpp6 | Membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (cell polarity) | 0.27 | 0.04 |
NOTE: Genes were selected based on a significant (P < 0.05) and a minimum 2-fold change in expression. From each genotype, colonic mucosa from three noninjected (100-day-old baseline) mice was collected and processed for RNA isolation. Fold change is the normalized signal intensity of a given gene in Folbp1+/− (heterozygous) relative to Folbp1+/+ (wild-type) mice.
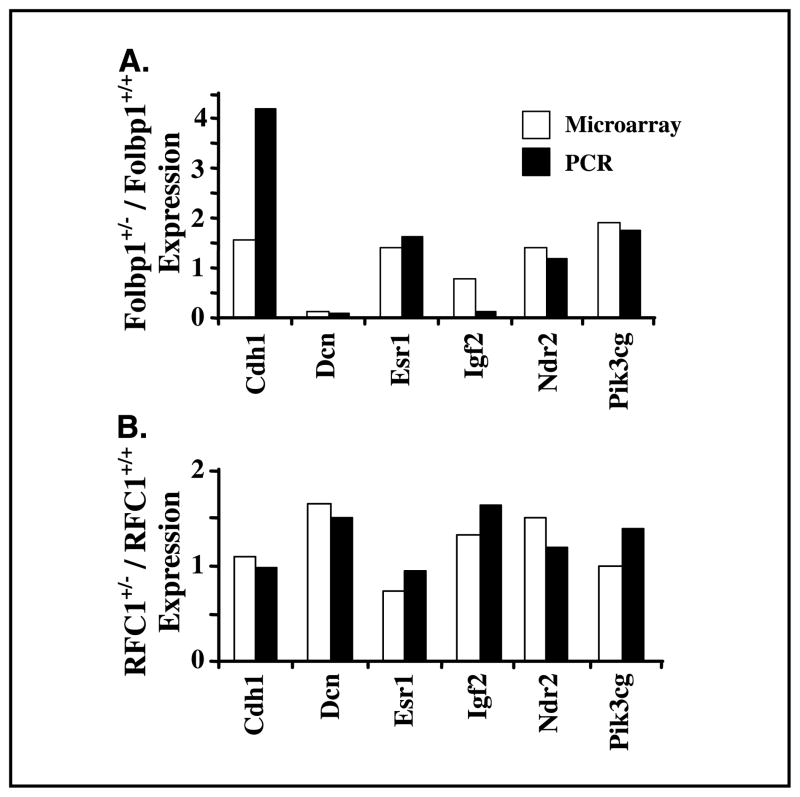
To validate expression patterns, real-time PCR analyses were done on preselected colon cancer–related genes, which included Cdh1, Dcn, Esr1, Igf2, Ndr2, and Pik3cg. There was good correlation between PCR and microarray data, which are described in Fig. 4.
Figure 4.
Correlation of gene expression by microarray and real-time PCR. RNA was isolated from scraped colonic mucosa. Real-time PCR was done using the ABI 7700 and Taqman Probes. Columns, level of gene expression in colonic mucosa from heterozygous mice relative to wild-type, noninjected mice at baseline by microarray (n = 3) and real-time PCR (n = 4–5). A subset of genes listed in Table 1 were also present on the CodeLink arrays. These genes include Cdh1, Dcn, Esr1, Igf2, Ndr2, and Pik3cg.
Carcinogen Increases the Development of Aberrant Crypt Foci in RFC1+/− Mice and Grossly Visible Dysplastic Masses in Folbp1+/− Mice
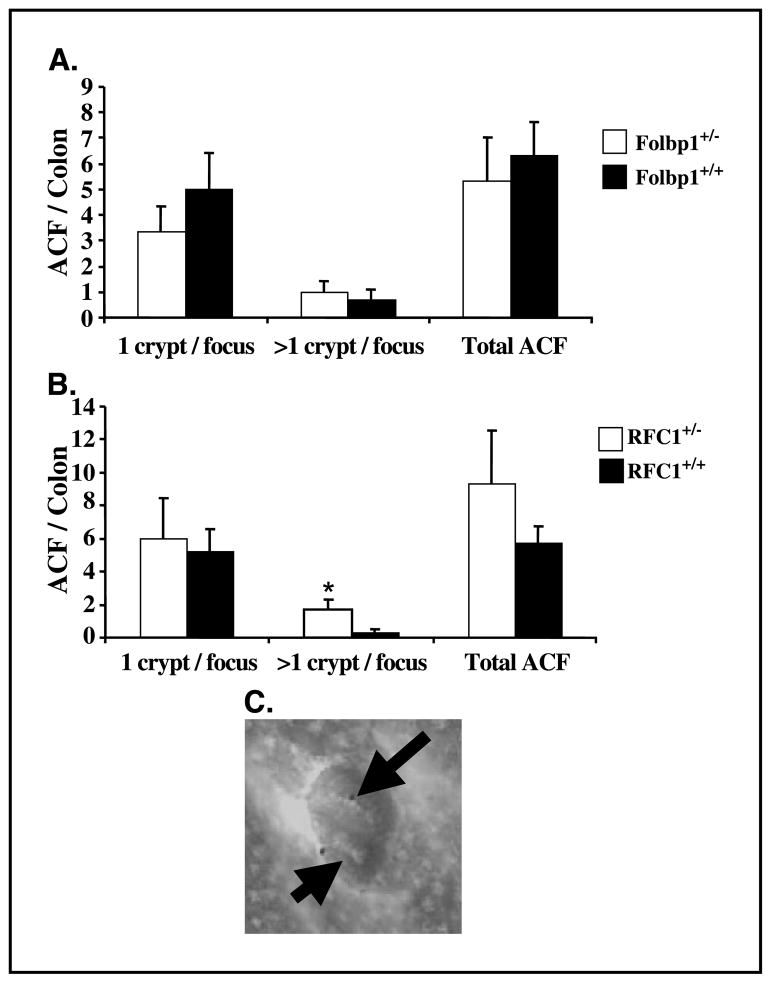
Increased cell proliferation is a significant risk factor for the development of colon cancer (51). Therefore, we investigated whether RFC1+/− and Folbp1+/− mice have increased susceptibility to colon carcinogenesis arising from azoxymethane exposure. Azoxymethane has been extensively characterized with respect to its ability to induce ACF formation, which represent well-established preneoplastic colonic lesions in both rodents and humans (52, 53). ACF are readily recognized by their enlarged size, pericryptal area, and thickened epithelial lining (52, 53). The number of ACF is believed to be predictive of tumor development (48, 54). At 8 weeks post-azoxymethane injection, there was no difference in the total number of ACF present in the colonic epithelium of RFC1+/− or Folbp1+/− mice relative to wild-type animals. However, when ACF multiplicity was assessed, there were significantly (P < 0.05) more aberrant crypts per focus in RFC1 +/− compared with RFC1+/+ mice (Fig. 5).
Figure 5.
Effect of RFC1 and Folbp1 gene ablation on ACF development. A, no difference in ACF development was observed in Folbp1 mice. B, RFC1+/− mice had significantly elevated numbers of ACF with >1 crypt per focus (n = 6–8). C, representative ACF characterized by an enlarged lumen and perinuclear region. Arrows, individual crypts.
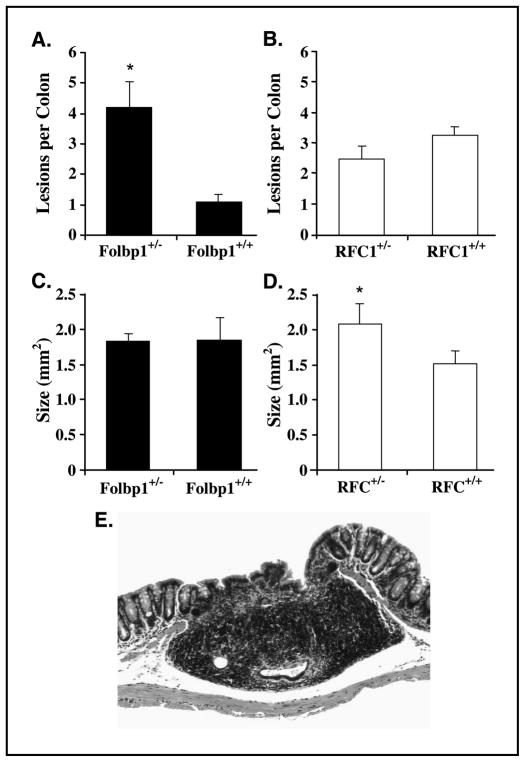
At 38 weeks post-azoxymethane injection, mice were terminated and colons were examined for evidence of tumor development. Grossly visible masses were detected in both Folbp1 and RFC1 mice. These masses were determined to be a combination of focal inflammatory lesions with or without epithelial dysplasia as well as adenocarcinomas. Specifically, there were ~4-fold more lesions in Folbp1+/− mice relative to Folbp1+/+ mice (Fig. 6). Although not significant, 50% (5 of 10) of Folbp1+/− mice developed adenocarcinomas relative to 27% (3 of 11) of Folbp1+/+ mice. In addition, no significant difference in absolute numbers or incidence of adenocarcinoma in RFC1 mice was observed. Interestingly, lesion size was significantly (P < 0.05) larger in RFC1+/− relative to RFC1+/+ mice (Fig. 6). Folate status at weeks 8 and 38 was assessed by measuring plasma folate levels and was unchanged throughout (data not shown). Therefore, azoxymethane exposure does not influence folate status.
Figure 6.
Development of dysplastic lesions in Folbp1 and RFC1 mice. Mice treated with azoxymethane were terminated 38 weeks after the last azoxymethane injection. Colons were removed, cleaned, and opened longitudinally. Grossly visible colonic lesions were mapped and enumerated before fixation in 4% paraformaldehyde. Slides were cut and evaluated in a blinded manner. Variable degrees of focal inflammatory lesions with or without epithelial dysplasia as well as adenocarcinomas were detected. A and B, Folbp1+/− mice developed significantly (P < 0.05) greater numbers of lesions relative to Folbp1+/+ mice. C and D, RFC1+/− mice developed significantly (P < 0.05) larger lesions relative to RFC1+/+ mice. E, representative section of an adenocarcinoma stained with H&E. Magnification, ×200.
Expression of Genes Involved in Colon Cancer Development Was Affected by Folate Transport Gene Ablation and Carcinogen Exposure
The expression of colon cancer–related genes known to contain CpG island promoter regions was evaluated by real-time PCR at baseline and at 8 and 38 weeks after the last azoxymethane injection in Folbp1 and RFC1 wild-type and heterozygous mice (Table 4). Over time, Cdh1, Dcn, Esr1, Igf2, Pik3cg, and Ptgs2 were more highly expressed in Folbp1+/− mice relative to Folbp1+/+ mice. In contrast, Cdh1, Cdx1, Igf2, and Ptgs2 were down-regulated in RFC1+/− mice relative to RFC1+/+ mice. These data suggest that partial ablation of Folbp1 or RFC1 increases colon cancer risk via distinct molecular pathways.
Table 4.
Expression of methylation-regulated genes involved in colon cancer at baseline and at 8 and 38 weeks post-azoxymethane injection
| Folbp1+/− (heterozygous)/Folbp1+/+ (wild-type)
|
RFC1+/− (heterozygous)/RFC1+/+ (wild-type)
|
|||||
|---|---|---|---|---|---|---|
| Baseline | 8 wk | 38 wk | Baseline | 8 wk | 38 wk | |
| Cdh1 | 4.2* | 2.0 † | 3.5 † | 1.0 | 1.1 | 0.7* |
| Cdx1 | 1.4 | 0.9 | 1.1 | 0.8 | 0.6 † | 0.6 † |
| Dcn | 0.1 | 0.6 | 3.3 † | 1.5 | 0.6 † | 1.7 |
| Esr1 | 1.6 | 1.9 † | 4.8 † | 0.9 | 0.8 | 0.9 |
| Igf2 | 0.1 | 0.7 | 6.6 † | 1.6 | 0.6 | 0.7 |
| Ndr2 | 1.2 | 0.9 | 1.5 | 1.2 | 1.0 | 1.0 |
| Pik3cg | 1.8 † | 1.0 | 1.5 | 1.4 | 1.0 | 0.8 |
| Ptgs2 | 0.4 | 0.6 | 5.0* | 1.3 | 0.8 | 0.6 |
NOTE: RNA from normal colonic mucosa was isolated at baseline before azoxymethane injection at 8 and 38 weeks post-azoxymethane injection. mRNA was quantified by real-time PCR (n = 4–5). Mean ± SE fold change in gene expression in heterozygous mice relative to homozygous mice.
P < 0.10, significant down-regulation of gene expression for a given gene at a specific time point.
P < 0.05, significant up-regulation of gene expression for a given gene at a specific time point.
Discussion
Epidemiologic and animal studies have shown that low folate status is associated with an increased risk for colon cancer development. Elucidation of molecular and cellular mechanisms by which folate modulates colon carcinogenesis is important for substantiating the chemopreventive health benefits of folate. In this study, we used RFC1 and Folbp1 knockout mice as genetic models to elucidate mechanisms of folate action on colonocyte cell proliferation, gene expression, and colon carcinogenesis. We show for the first time that the reduced expression of folate transporters (RFC1 and Folbp1) differentially perturbs colonocyte folate status, gene expression profiles, and development of preneoplastic biomarkers.
The partial and complete ablation of Folbp1 coincided with marked decreases in both plasma and tissue folate, consistent with its role in the reabsorption of folate by the kidney (49). These data are similar to observations made in rodent studies where animals were fed folate-deficient diets (2, 21). In contrast, reduced expression of RFC1 did not elicit a similar effect. Although RFC1 contributes significantly to folate absorption within the small intestine and colon, passive diffusion and other carriers within the small intestine also regulate folate absorption (49, 55). The contribution of these ancillary mechanisms likely explain why overt systemic folate deficiency was not observed in RFC1+/− mice. In addition, the presence of one allele may be adequate to maintain RFC1 function, albeit in a compromised state.
There is overwhelming evidence linking dietary folate intake to aberrant DNA methylation and colon cancer (10, 56–58). Tissue levels of SAM and SAH are frequently used as indicators of cellular methylation potential (50). Interestingly, the pronounced suppression of plasma and colonic tissue folate concentrations in Folbp1+/− mice was not associated with a perturbation in the absolute levels or the ratio of SAM/SAH. The resistance of SAM and SAH pools in the colon to changes in folate status has been reported previously (21, 59) and may be explained by the induction of homeostatic mechanisms present in the colon. Because RFC1 expression did not affect mucosa or plasma folate concentrations (Table 1), it is not unexpected that tissue SAM and SAH were not altered. Surprisingly, there was a significant reduction in the plasma SAM/SAH ratio in RFC1+/− mice, suggesting that DNA methylation status may be altered. Despite the lack of overt systemic folate deficiency, these data suggest that reduced RFC1 expression can cause a subtle perturbation in folate homeostasis within the colon.
Increased cell proliferation is an important biomarker of preneoplastic events and colon cancer risk (44). Our data show that the perturbed folate status associated with reduced expression of Folbp1 and RFC1 differentially affects specific indices of cell proliferation in the colon. These alterations in cellular proliferation may be indicative of increased susceptibility to the development of colon cancer. We therefore examined the effect of Folbp1 and RFC1 haploinsufficiency on azoxymethane-induced ACF formation. This is a highly relevant biological end point given that ACFs (52, 53), and especially high multiplicity ACF (60–62), are considered to be early preneoplastic lesions in humans and rodents. Interestingly, Folbp1+/− mice, which display overt clinical signs of folate deficiency, did not develop increased numbers of ACF relative to Folbp1+/+ mice. In contrast, RFC1+/− mice, characterized as having a subtle perturbation in folate metabolism, developed significantly greater numbers of ACF classified as >1 aberrant crypt per focus. Our observations made in RFC1+/− mice support the current dogma, which suggests that only a modest reduction in folate status is necessary to enhance colon carcinogenesis (24). This is significant from a public health perspective given that subclinical folate deficiency is found in 30% of the U.S. population (23).
To further evaluate the effects of folate transport gene ablation on colon cancer risk, we also examined mice at 38 weeks post-azoxymethane treatment. Grossly visible colonic lesions were detected in both Folbp1 and RFC1 mice. These lesions were identified histologically as a combination of focal inflammatory lesions with or without epithelial dysplasia as well as adenocarcinomas (63). The ~4-fold increase in the number of lesions in Folbp1+/− mice relative to Folbp1+/+ mice (Fig. 6) suggests that systemic folate deficiency induced by Folbp1 gene ablation elevates the risk for colon cancer. Although this initial finding is noteworthy, the correlation between Folbp1 gene ablation and the formation of these lesions requires additional studies to define molecular mechanisms. With respect to RFC1 mice, although no difference in the absolute numbers of lesions was detected, the average lesion size was significantly (P < 0.05) increased in RFC1+/− relative to RFC1+/+ mice (Fig. 6). These observations combined with ACF data (Fig. 5) suggest an increased risk for colon cancer development in this model. Additional studies are in progress to further evaluate the effects of an expanded azoxymethane injection regimen using greater numbers of mice to conclusively determine whether folate status affects the incidence of tumor development in Folbp1+/− and RFC1+/− mice.
To date, a limited number of studies have examined the effect of folate status on gene expression (58, 64). Therefore, microarray analysis was used to generate a molecular portrait of gene expression profiles from RFC1+/− and Folbp1+/− colonic mucosa at baseline before azoxymethane treatment. Only a limited number of genes were differentially expressed between RFC1+/− and RFC1+/+. This is not surprising given that folate status was only modestly affected by reduced RFC1 expression. Interestingly, Skp1a (Table 2) was significantly down-regulated, and defects in this gene contribute to improper chromosomal separation leading to neoplastic transformation (65). Folate deficiency also causes chromosomal breakage, which may contribute and enhance the effect of Skp1a down-regulation (18). Although it is tempting to speculate that the expression of this gene is directly related to the altered phenotype of RFC1+/− mice, further experimentation is required to clarify the nature of these associations.
In contrast to the RFC1+/− knockout mice, a substantially greater number of genes were differentially up or down-regulated in Folbp1+/− mice compared with Folbp1+/+ controls. It is noteworthy that a large number of immune-related genes were differentially expressed. Similar observations have been reported in the colons of rats fed folate-deficient diets and may indicate altered immune monitoring for abnormal preneoplastic cells (64). It is possible that the complex molecular portrait of gene expression profiles we observed may in part be related to the source of RNA used in this study. Our microarray findings are based on the use of (nontumor) mucosal scrapings containing a heterogeneous population of colonic cells, similar to the procedure used by Crott et al. (64). Therefore, the inclusion of additional cell populations may have influenced our results. These issues highlight the complexity of obtaining an in vivo global view of gene expression. The unique genomic profiles of the RFC1+/− compared with Folbp1+/− mice may help explain the distinct phenotypes of these models.
Folate plays an important role in CpG island methylation, which is involved in the regulation of gene expression. This has been postulated as a mechanism by which folate deficiency causes dysregulation of gene expression via global DNA hypomethylation and, paradoxically, gene-specific hypermethylation (24). Although the methylation status of specific genes was not directly evaluated, in this study, we determined the effects of folate transport gene ablation on the mRNA expression of known colon cancer–related genes, which are regulated by DNA methylation. Cdh1, Cdx1, Dcn, Esr1, Igf2, Ndr2, Pik3cg, and Ptgs2 have been shown previously to be hypermethylated in colon cancer and are therefore presumably silenced by DNA methylation. Interestingly, several of these genes, including Cdx1, Dcn, Esr1, and Igf2, were highly expressed in Folbp1+/− mice by 38 weeks, which may indicate the involvement of a hypomethylation-related mechanism. This is consistent with the apparent systemic and colonic folate-deficient phenotype of Folbp1+/− mice (Table 1), which would be expected to promote global DNA hypomethylation. In comparison, RFC1+/− mice exhibited a contrasting trend with respect to the expression of these genes, suggesting a role for DNA hypermethylation and gene silencing in this model.
The lack of suitable animal models in which to evaluate the effect of folate status on colon carcinogenesis is partly responsible for the paucity of experimental evidence concerning the optimization of dietary intervention strategies for colon cancer prevention. By using a genetically engineered folate transporter knockout model approach, we have differentially manipulated folate status to identify regulatory relationships among folate-responsive genes considered to drive colonic tumor development. We believe that these models will provide valuable insight into the role of folate in cancer biology, which will ultimately lead to the establishment of public health recommendations for cancer prevention.
Acknowledgments
Grant support: American Institute for Cancer Research grant 03A038, NIH grant CA59034, National Institute of Environmental Health Sciences grant P30-ES-09106, Texas A&M Life Sciences Task Force, Natural Sciences and Engineering Research Council of Canada (D.W.L. Ma), and Alberta Heritage for Medical Research Foundation postdoctoral fellowships (D.W.L. Ma).
We thank Drs. Chandrika Piyathilake and Conrad Wagner for their expertise in measuring folate intermediates and the staff at Mayo Clinic for the generation of the RFC1 knockout mouse.
Footnotes
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Kim YI, Salomon RN, Graeme-Cook F, et al. Dietary folate protects against the development of macroscopic colonic neoplasia in a dose responsive manner in rats. Gut. 1996;39:732–40. doi: 10.1136/gut.39.5.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YI, Pogribny IP, Basnakian AG, et al. Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am J Clin Nutr. 1997;65:46–52. doi: 10.1093/ajcn/65.1.46. [DOI] [PubMed] [Google Scholar]
- 4.Biasco G, Zannoni U, Paganelli GM, et al. Folic acid supplementation and cell kinetics of rectal mucosa in patients with ulcerative colitis. Cancer Epidemiol Biomarkers Prev. 1997;6:469–71. [PubMed] [Google Scholar]
- 5.Song J, Medline A, Mason JB, Gallinger S, Kim YI. Effects of dietary folate on intestinal tumori genesis in the apcMin mouse. Cancer Res. 2000;60:5434–40. [PubMed] [Google Scholar]
- 6.Song J, Sohn KJ, Medline A, Ash C, Gallinger S, Kim YI. Chemopreventive effects of dietary folate on intestinal polyps in Apc+/−Msh2−/− mice. Cancer Res. 2000;60:3191–9. [PubMed] [Google Scholar]
- 7.Janne PA, Mayer RJ. Chemoprevention of colorectal cancer. N Engl J Med. 2000;342:1960–8. doi: 10.1056/NEJM200006293422606. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs EJ, Connell CJ, Patel AV, et al. Multivitamin use and colon cancer mortality in the Cancer Prevention Study II cohort (United States) Cancer Causes Control. 2001;12:927–34. doi: 10.1023/a:1013716323466. [DOI] [PubMed] [Google Scholar]
- 9.Finnell RH, Spiegelstein O, Wlodarczyk B, et al. DNA methylation in Folbp1 knockout mice supplemented with folic acid during gestation. J Nutr. 2002;132:2457–61S. doi: 10.1093/jn/132.8.2457S. [DOI] [PubMed] [Google Scholar]
- 10.Friso S, Choi SW, Girelli D, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–11. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibani S, Melnyk S, Pogribny IP, et al. Studies of methionine cycle intermediates (SAM, SAH), DNA methylation and the impact of folate deficiency on tumor numbers in Min mice. Carcinogenesis. 2002;23:61–5. doi: 10.1093/carcin/23.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Choi SW, Kim YI, Weitzel JN, Mason JB. Folate depletion impairs DNA excision repair in the colon of the rat. Gut. 1998;43:93–9. doi: 10.1136/gut.43.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. FASEB J. 1998;12:1491–7. [PubMed] [Google Scholar]
- 14.Duthie SJ, Narayanan S, Blum S, Pirie L, Brand GM. Folate deficiency in vitro induces uracil misincorporation and DNA hypomethylation and inhibits DNA excision repair in immortalized normal human colon epithelial cells. Nutr Cancer. 2000;37:245–51. doi: 10.1207/S15327914NC372_18. [DOI] [PubMed] [Google Scholar]
- 15.Jaszewski R, Khan A, Sarkar FH, et al. Folic acid inhibition of EGFR-mediated proliferation in human colon cancer cell lines. Am J Physiol. 1999;77:C1142–8. doi: 10.1152/ajpcell.1999.277.6.C1142. [DOI] [PubMed] [Google Scholar]
- 16.Akoglu B, Faust D, Milovic V, Stein J. Folate and chemoprevention of colorectal cancer: is 5-methyl-tetrahydrofolate an active antiproliferative agent in folate-treated colon-cancer cells? Nutrition. 2001;17:652–3. doi: 10.1016/s0899-9007(01)00594-9. [DOI] [PubMed] [Google Scholar]
- 17.Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94:3290–5. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi SW, Mason JB. Folate status: effects on pathways of colorectal carcinogenesis. J Nutr. 2002;132:2413–8S. doi: 10.1093/jn/132.8.2413S. [DOI] [PubMed] [Google Scholar]
- 19.Terry P, Jain M, Miller AB, Howe GR, Rohan TE. Dietary intake of folic acid and colorectal cancer risk in a cohort of women. Int J Cancer. 2002;97:864–7. doi: 10.1002/ijc.10138. [DOI] [PubMed] [Google Scholar]
- 20.Nensey YM, Arlow FL, Majumdar AP. Aging. Increased responsiveness of colorectal mucosa to carcinogen stimulation and protective role of folic acid. Dig Dis Sci. 1995;40:396–401. doi: 10.1007/BF02065427. [DOI] [PubMed] [Google Scholar]
- 21.Sohn KJ, Stempak JM, Reid S, Shirwadkar S, Mason JB, Kim YI. The effect of dietary folate on genomic and p53-specific DNA methylation in rat colon. Carcinogenesis. 2003;24:81–90. doi: 10.1093/carcin/24.1.81. [DOI] [PubMed] [Google Scholar]
- 22.Khosraviani K, Weir HP, Hamilton P, Moorehead J, Williamson K. Effect of folate supplementation on mucosal cell proliferation in high risk patients for colon cancer. Gut. 2002;51:195–9. doi: 10.1136/gut.51.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selhub J, Jacques PF, Wilson PW, Rush D, Rosenberg IH. Vitamin status and intake as primary determinants of homocysteinemia in an elderly population. JAMA. 1993;270:2693–8. doi: 10.1001/jama.1993.03510220049033. [DOI] [PubMed] [Google Scholar]
- 24.Kim YI. Folate and carcinogenesis: evidence, mechanisms, and implications. J Nutr Biochem. 1999;10:66–88. doi: 10.1016/s0955-2863(98)00074-6. [DOI] [PubMed] [Google Scholar]
- 25.Sierra EE, Goldman ID. Recent advances in the understanding of the mechanism of membrane transport of folates and antifolates. Semin Oncol. 1999;26:11–23. [PubMed] [Google Scholar]
- 26.Spiegelstein O, Eudy JD, Finnell RH. Identification of two putative novel folate receptor genes in humans and mouse. Gene. 2000;258:117–25. doi: 10.1016/s0378-1119(00)00418-2. [DOI] [PubMed] [Google Scholar]
- 27.Matherly LH. Molecular and cellular biology of the human reduced folate carrier. Prog Nucleic Acid Res Mol Biol. 2001;67:131–62. doi: 10.1016/s0079-6603(01)67027-2. [DOI] [PubMed] [Google Scholar]
- 28.Kamen BA. Folate receptor α. In: Rao MS, editor. Stem cells and CNS development. New Jersey: Humana Press; 2002. pp. 117–35. [Google Scholar]
- 29.Wang Y, Zhao R, Russell RG, Goldman ID. Localization of the murine reduced folate carrier as assessed by immunohistochemical analysis. Biochim Biophys Acta. 2001;1513:49–54. doi: 10.1016/s0005-2736(01)00340-6. [DOI] [PubMed] [Google Scholar]
- 30.Weitman SD, Lark RH, Coney LR, et al. Distribution of the folate receptor GP38 in normal and malignant cell lines and tissues. Cancer Res. 1992;52:3396–401. [PubMed] [Google Scholar]
- 31.Holm J, Hansen SI, Hoier-Madsen M, Sondergaard K, Bzorek M. The high-affinity folate receptor of normal and malignant human colonic mucosa. APMIS. 1994;102:828–36. doi: 10.1111/j.1699-0463.1994.tb05241.x. [DOI] [PubMed] [Google Scholar]
- 32.Piedrahita JA, Oetama B, Bennett GD, et al. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet. 1999;23:228–32. doi: 10.1038/13861. [DOI] [PubMed] [Google Scholar]
- 33.Zhao R, Russell RG, Wang Y, et al. Rescue of embryonic lethality in reduced folate carrier-deficient mice by maternal folic acid supplementation reveals early neonatal failure of hematopoietic organs. J Biol Chem. 2001;276:10224–8. doi: 10.1074/jbc.c000905200. [DOI] [PubMed] [Google Scholar]
- 34.Piedrahita JA, Oetama B, Bennett GD, et al. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet. 1999;23:228–32. doi: 10.1038/13861. [DOI] [PubMed] [Google Scholar]
- 35.Reid LH, Shesely EG, Kim HS, Smithies O. Cotransformation and gene targeting in mouse embryonic stem cells. Mol Cell Biol. 1991;11:2769–77. doi: 10.1128/mcb.11.5.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rucker EB, Piedrahita JA. Cre-mediated recombination at the murine whey acidic protein (mWAP) locus. Mol Reprod Dev. 1997;48:324–31. doi: 10.1002/(SICI)1098-2795(199711)48:3<324::AID-MRD4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 37.Horne DW, Patterson D. Lactobacillus casei microbiological assay of folic acid derivatives in 96-well microtiter plates. Clin Chem. 1988;34:2357–9. [PubMed] [Google Scholar]
- 38.Spiegelstein O, Merriweather MY, Wicker NJ, Finnell RH. Valproate-induced neural tube defects in folate-binding protein-2 (Folbp2) knockout mice. Birth Defects Res Part A Clin Mol Teratol. 2003;67:974–8. doi: 10.1002/bdra.10128. [DOI] [PubMed] [Google Scholar]
- 39.Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: alterations with plasma homocysteine and pyridoxal 5′-phosphate concentrations. Clin Chem. 2000;46:265–72. [PubMed] [Google Scholar]
- 40.Piyathilake CJ, Johanning GL, Macaluso M, White-side MA, Heimburger DC, Grizzle WE. Localized deficiencies of folate and vitamin B12 in lung tissues are associated with global DNA methylation. Nutr Cancer. 2000;37:99–107. doi: 10.1207/S15327914NC3701_13. [DOI] [PubMed] [Google Scholar]
- 41.Hamedani MP, Valko K, Qi X, Welham KJ, Gibbons WA. Two-dimensional high-performance liquid chromatographic method for assaying S-adenosyl-L-methionine and its related metabolites in tissues. J Chromatogr. 1993;619:191–8. doi: 10.1016/0378-4347(93)80108-g. [DOI] [PubMed] [Google Scholar]
- 42.Park CM, Reid PE, Walker DC, MacPherson BR. A simple, practical “Swiss roll” method of preparing tissues for paraffin or methacrylate embedding. J Microsc. 1987;145:115–20. doi: 10.1111/j.1365-2818.1987.tb01321.x. [DOI] [PubMed] [Google Scholar]
- 43.Beckstead JH. A simple technique for preservation of fixation-sensitive antigens in paraffin-embedded tissues. J Histochem Cytochem. 1994;42:1127–34. doi: 10.1177/42.8.8027531. [DOI] [PubMed] [Google Scholar]
- 44.Chang WC, Chapkin RS, Lupton JR. Predictive value of proliferation, differentiation and apoptosis as intermediate markers for colon tumorigenesis. Carcinogenesis. 1997;18:721–30. doi: 10.1093/carcin/18.4.721. [DOI] [PubMed] [Google Scholar]
- 45.Davidson LA, Lupton JR, Miskovsky E, Fields A, Chapkin RS. Quantification of human intestinal gene expression profiles using exfoliated colonocytes: a pilot study. Biomarkers. 2003;8:51–61. doi: 10.1080/1354750021000042268. [DOI] [PubMed] [Google Scholar]
- 46.Yang YH, Buckley MJ, Speed TP. Analysis of cDNA microarray images. Brief Bioinform. 2001;2:341–9. doi: 10.1093/bib/2.4.341. [DOI] [PubMed] [Google Scholar]
- 47.Murray NR, Davidson LA, Chapkin RS, Clay Gustafson W, Schattenberg DG, Fields AP. Overexpression of protein kinase C βII induces colonic hyperproliferation and increased sensitivity to colon carcinogenesis. J Cell Biol. 1999;145:699–711. doi: 10.1083/jcb.145.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLellan EA, Medline A, Bird RP. Dose response and proliferative characteristics of aberrant crypt foci: putative preneoplastic lesions in rat colon. Carcinogenesis. 1991;12:2093–8. doi: 10.1093/carcin/12.11.2093. [DOI] [PubMed] [Google Scholar]
- 49.Matherly LH, Goldman DI. Membrane transport of folates. Vitam Horm. 2003;66:403–56. doi: 10.1016/s0083-6729(03)01012-4. [DOI] [PubMed] [Google Scholar]
- 50.Caudill MA, Wang JC, Melnyk S, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine β-synthase heterozygous mice. J Nutr. 2001;131:2811–8. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 51.Preston-Martin S, Pike MC, Ross RK, Jones PA, Henderson BE. Increased cell division as a cause of human cancer. Cancer Res. 1990;50:7415–21. [PubMed] [Google Scholar]
- 52.McLellan EA, Bird RP. Aberrant crypts: potential preneoplastic lesions in the murine colon. Cancer Res. 1988;48:6187–92. [PubMed] [Google Scholar]
- 53.Takayama T, Katsuki S, Takahashi Y, et al. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med. 1998;339:1277–84. doi: 10.1056/NEJM199810293391803. [DOI] [PubMed] [Google Scholar]
- 54.Pretlow TP, O’Riordan MA, Somich GA, Amini SB, Pretlow TG. Aberrant crypts correlate with tumor incidence in F344 rats treated with azoxymethane and phytate. Carcinogenesis. 1992;13:1509–12. doi: 10.1093/carcin/13.9.1509. [DOI] [PubMed] [Google Scholar]
- 55.Brzezinska A, Winska P, Balinska M. Cellular aspects of folate and antifolate membrane transport. Acta Biochim Pol. 2000;47:735–49. [PubMed] [Google Scholar]
- 56.Hernandez-Blazquez FJ, Habib M, Dumollard JM, et al. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–93. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang JY, Xiao SD. Alteration of DNA methylation in gastrointestinal carcinogenesis. J Gastroenterol Hepatol. 2001;16:960–8. doi: 10.1046/j.1440-1746.2001.02554.x. [DOI] [PubMed] [Google Scholar]
- 58.Jhaveri MS, Wagner C, Trepel JB. Impact of extracellular folate levels on global gene expression. Mol Pharmacol. 2001;60:1288–95. doi: 10.1124/mol.60.6.1288. [DOI] [PubMed] [Google Scholar]
- 59.Davis CD, Uthus EO. Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr. 2003;133:2907–14. doi: 10.1093/jn/133.9.2907. [DOI] [PubMed] [Google Scholar]
- 60.Magnuson BA, Carr I, Bird RP. Ability of aberrant crypt foci characteristics to predict colonic tumor incidence in rats fed cholic acid. Cancer Res. 1993;53:4499–504. [PubMed] [Google Scholar]
- 61.Papanikolaou A, Wang QS, Papanikolaou D, Whiteley HE, Rosenberg DW. Sequential and morphological analyses of aberrant crypt foci formation in mice of differing susceptibility to azoxymethane-induced colon carcinogenesis. Carcinogenesis. 2000;21:1567–72. [PubMed] [Google Scholar]
- 62.Park HS, Goodlad RA, Wright NA. The incidence of aberrant crypt foci and colonic carcinoma in dimethylhydrazine-treated rats varies in a site-specific manner and depends on tumor histology. Cancer Res. 1997;57:4507–10. [PubMed] [Google Scholar]
- 63.Ward JM. Morphogenesis of chemically induced neoplasms of the colon and small intestine in rats. Lab Invest. 1974;30:505–13. [PubMed] [Google Scholar]
- 64.Crott JW, Choi SW, Ordovas JM, Ditelberg JS, Mason JB. Effects of dietary folate and aging on gene expression in the colonic mucosa of rats: implications for carcinogenesis. Carcinogenesis. 2004;25:69–76. doi: 10.1093/carcin/bgg150. [DOI] [PubMed] [Google Scholar]
- 65.Piva R, Liu J, Chiarle R, Podda A, Pagano M, Inghirami G. In vivo interference with Skp1 function leads to genetic instability and neoplastic transformation. Mol Cell Biol. 2002;22:8375–7. doi: 10.1128/MCB.22.23.8375-8387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]