Hox genes are a specific subgroup of homeobox containing genes that are characterized by their structural and functional homology to homeotic genes of the fruit fly Drosophila (1). These genes play important roles in pattern formation along the anterior-posterior axis in the developing embryo in both invertebrates and vertebrates (2). The proteins encoded by Hox genes contain the DNA-binding homeodomain (3), and are therefore believed to act as transcription factors that regulate the expression of other genes during morphogenesis (4, 5). More than 100 of other genes have been identified in mammals that encode homeodomain proteins (6, 7) of which the Hox genes are the best characterized. A growing number of animals with genetically induced aberrant Hox gene expression is providing insight into the crucial function of Hox genes in axial patterning; it is also becoming clear that Hox genes are important in the development of internal organs and the differentiation of tissues.
The development of the lung
starts out from protrusions of the primitive gut, the lung buds (8). Elongation and branching then leads to the formation of the trachea and bronchi and progressive branching elaborates the distal alveoli (9). The proximal-distal orientation of these distinct structures suggests that they could be specified by mechanisms paralleling those that govern anterior-posterior (rostral-caudal) axial specification. The inner surface of the lung is derived from endodermal cells that differentiate into epithelial cells under the influence of the surrounding mesenchymal cells which are of mesodermal origin (reviewed in (10). It is believed that interactions with the mesenchyme are important for epithelial cell differentiation and function. In short, lung development is characterized by two phases, the early specification of the proximal-distal axis and later events of tissue differentiation. As I will present here, there is increasing evidence that Hox genes are involved both in regional specification and cell differentiation in lung development.
The genomic organization of vertebrate Hox genes
is characterized by their arrangement in chromosomal clusters (11). Figure 1 shows schematically that highly similar genes (paralogs) occupy corresponding positions on different clusters. Intriguingly, the position of genes within a cluster is correlated to their order of expression along the anterior-posterior (rostral-caudal) axis (12). In addition, the genes located towards the 5′ end of the clusters have been shown to be expressed in developing limbs in anterior-posterior (thumb-pinkie) and proximal-distal direction (13). The overlapping but distinct expression patterns have been taken to suggest that a combinatorial code of Hox gene expression specifies the subsequent development of a given body region (14).
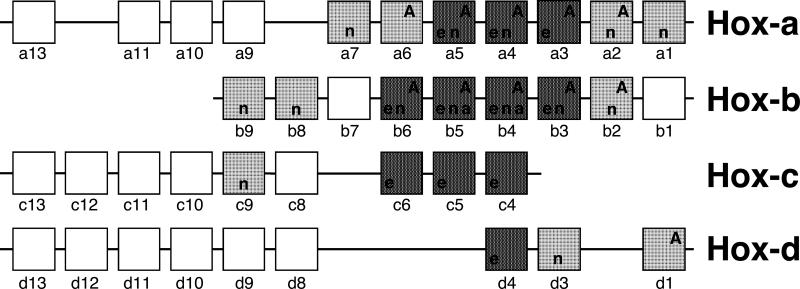
Figure. Chromosomal organization of Hox genes and their expression in the lung.
Hox genes are organized in four clusters on different chromosomes, the Hox-a cluster resides on chromosome #6 (#7 in human), Hox-b on #11 (#17), Hox-c on #12 (#15), and Hox-d on #2 in human and mouse. The order of genes within a cluster is depicted in 5′ to 3′ orientation and paralogous genes are aligned vertically. Expression of Hox genes in the lung has been documented in human adult lung (capital letters), and rat and mouse lung at various embryonic stages (e), newborn (n) and adult (a) (small letters for rodents); data based on Northern or RT-PCR analysis only are represented by light shading, in situ hybridization or immunohistochemistry data are shaded darker. Open boxes represent genes that are not expressed in the developing lung in the mouse (such as Hoxb-1, b-7 and c-8) or have not been examined yet (for review, see 54, 55). The references are given in the text. During development, Hox genes are expressed sequentially in restricted domains along the anterior-posterior axis that reflect their respective position within a cluster (colinearity of position in cluster and expression domain): Hox genes located towards the 3′ end of the clusters are expressed more anteriorly while those located at the 5′ end are expressed in the posterior.
The function of Hox genes in regional specification
is most convincingly demonstrated by the appearance of malformations in Hox-mutant mice and in transgenic mice with aberrant Hox gene expression (reviewed in (15). The targeted inactivation of Hox genes has been shown to affect skeletal development of the limbs as well as the axial skeleton. Loss-of-function of Hox genes typically leads to homeotic transformation of skeletal elements, for example, changes in appearance and identity of specific vertebrae without a change in the overall number of units (16-25). In the developing limbs, disruption of Hox genes induces skeletal abnormalities that indicative of altered patterns of growth (20, 26-29). Gain of Hox gene expression in transgenic mice also leads to defects in the axial skeleton (30-33) and the developing limbs (34). Taken together, these results have established Hox genes as key players in regional specification and pattern formation in mammalian embryonic development. The combinatorial action of Hox genes for a given body region is demonstrated by compounded malformations in those mice that carry multiple mutations in overlapping Hox genes (23, 28, 29, 35, 36).
Hox gene expression in the lung
About half of the 38 known Hox genes have been shown to be expressed in adult human (37, 38) and mouse (39, 40) lung tissue and lungs from newborn rats (41). Information on embryonic expression is only available for the following murine Hox genes: Hoxa- 2 (42), a-3 (43), a-4 (44, 45), a-5 (46), b-2 (47, 48), b-3 (49), b-4 (40), b-5 (50), b-6 (48), c-4 (51), c-5 (51), c-6 (52, 53), and d-4 (45) (reviewed in (54, 55). Figure 1 summarizes the available data for lung expression. Predominantly genes from the 3′ halves of the Hox-clusters a and b have been reported to be expressed in the lung. This is consistent with the expression domains of these genes in cervical and thoracic regions and suggests that Hox genes could play a role in lung development. However, the variations in experimental systems and in developmental stages analyzed make direct comparisons difficult. More importantly, they do not provide insight into temporal, spatial or cell-type specificity of expression. It is in this regard that the study by Bogue et al. of Hox gene expression in the developing lung deserves attention (56). The leading question for these studies is: Does the expression pattern of Hox genes in the lung represent a specific role in regional tissue patterning similar to known processes of positional specification along the anterior-posterior axis?
Bogue et al. focused on four genes that are sequentially located on the Hox-b cluster, Hoxb-2 through b-5. They show that at day 9.5 of development, Hoxb-2 through Hoxb-5 are expressed in the branchial arches and developing foregut in a manner that appears to be collinear with gene order on the chromosome. This is, Hoxb-2 was expressed in more rostral regions that develop into the pharynx whereas Hoxb-3, b-4, and b-5 were localized progressively in more caudal regions. This order of expression conforms well to the relative gene order and confirms the colinearity rule, i.e., genes located more 3′ in a cluster are expressed more anteriorly (rostrally). In the region of the prospective lung bud, Hoxb-2 is only weakly expressed while Hoxb-3, b-4, and b-5 produce stronger hybridization. These results indicate that the pattern of Hox gene expression at beginning of lung development is consistent with a role in regional specification. In particular, Hoxb-3, b-4, and b-5 (and possibly b-6, see Fig.1) could be involved in elaborating the proximal-distal orientation in lung development. Consistent with this interpretation, Hoxb-3 and b-4 are expressed in both proximal and distal lung whereas Hoxb-5 is found restricted to distal lung at 10.5 days of development. However, Bogue et al. also found Hoxb-2 expressed in distal lung, an apparent violation of the collinearity rule. One explanation for this finding is that the posterior limits in the spinal cord and axial mesoderm are not very well defined. Alternatively, it is possible that Hox gene expression is in the lung becomes uncoupled from spatial restriction as the lung differentiates separate from the main body axis. At later stages, Hoxb-3 and Hoxb-4 continue to be expressed in both proximal and distal lung whereas Hoxb-2 and Hoxb-5 are restricted to distal regions (see also (57). These patterns indicate that the expression of Hox genes in the lung is temporally and spatially regulated in a fashion that is collinear with gene order at early stages of lung development. In addition, the possibility exists that Hox gene expression at late stages follows different rules. Data regarding differences in expression in medial-lateral or dorsal-ventral direction are not available. The presence of distinct expression patterns at later stages of development may also suggest that Hox genes could be involved in specifying regional differences between proximal and distal lung development. Future studies which include genes of the Hox-a cluster will clarify this issue.
The regulation of Hox gene expression in the lung
has not been extensively studied to date. The early expression of Hox-b genes in collinear fashion suggests that anterior (rostral) boundaries of expression could be regulated by the same mechanisms found in axial mesoderm. However, the apparent loss of collinearity of Hox gene expression at later stages of lung development seem to indicate that the expression of Hox genes is regulated differently. This would predict that different regulatory elements direct the expression of Hox genes in axial mesoderm as compared to visceral mesoderm. Indeed, distinct regulatory elements have been described for the developing kidney for Hoxb-7 (58, 59) and Hoxb-6 (60) suggesting that certain aspects of Hox gene expression in the visceral mesoderm are controlled through independent mechanisms. Regulatory elements from the Hoxb-3 gene that control expression in the spinal cord and mesoderm are not sufficient to restore expression in the lung underscoring the requirement for additional elements that control Hoxb-3 expression in lung (49). These results imply that the subdivision of mesoderm into axial and visceral mesoderm is associated with the use of alternative enhancers within the Hox-regulatory regions. Identification of these regulatory elements will allow a more precise study of regional specification of the visceral mesoderm and lung mesoderm in particular.
Regional specification within the developing lung
Proximal and distal lung mesenchyme have long been known to differ in their ability to support branching morphogenesis in vitro (61, 62). The distinct patterns of expression that Bogue et al. report are consistent with a role of Hox genes in specification or differentiation of proximal versus distal development of the lung. Interestingly, elements in the 5′ upstream region of the Hoxa-4 gene direct LacZ transgene expression with prominent localization only in the distal region of the lung at 12.5 days of development (63). Thus, it is likely that distinct regulatory elements control the expression of Hox genes in specific regions in the lung, and further studies are warranted to isolate these sequences. The identification of enhancers for correct spatial and temporal regulation of Hox gene expression in the lung will then allow a molecular dissection of the components that control lung development.
Hox genes in lung cell differentiation
Interactions of the mesenchyme and epithelial cells are believed to be critically important for differentiation of epithelial cell types in the lung (64, 65). Bogue et al. confirm earlier findings (57, 66) that the expression of Hox genes is restricted to the mesenchymal cells. These results suggest a function for Hox genes in the mesenchyme and possibly in mesenchymal-epithelial cell interactions. Such a tissue-specific role for Hox genes in cell differentiation pathways has been previously proposed (67) and is being addressed through studies of the role of Hox genes in lung-specific gene expression.
Recently, the Hoxb-3 has been proposed to be an upstream regulator of thyroid transcription factor-1 (TTF-1) (68), a divergent homeodomain transcription factor that is expressed in the developing thyroid and lung and restricted areas of the brain (69). DNA-binding sites for Hoxb-3 in the TTF-1 gene promoter have been identified. More importantly, co-transfection experiments in vitro demonstrate that TTF-1 upstream sequences are specifically activated by Hoxb-3 (68). These results are relevant for lung development because TTF-1 is expressed in the lung and has been shown to regulate the promoters of the lung-specific genes encoding the surfactant proteins SP-A, SP-B, SP-C, and Clara Cell Secretory Protein (CCSP) and of the lung-specific gene CC10 gene (reviewed in (70). Moreover, inhibition of TTF-1 during lung morphogenesis in vitro disrupts the normal differentiation of epithelial cells (71). Taken together, these data open the possibility that Hoxb-3 could be involved in lung-specific gene regulation through TTF-1 as the intermediate. Seemingly conflicting with this hypothesis is the fact that TTF-1 is strongly expressed in epithelial cells but not mesenchyme at day 12.5 of development (69). Conversely, Hox genes are only expressed in mesenchymal cells at this stage (56, 72, 73). However, a potential overlap of Hox gene and TTF-1 gene expression at earlier or later stages of development can not be excluded and warrants further studies. In addition, functional experiments are necessary to explore their possible relationship in vivo. Virtually nothing is known about the role of Hox genes in the adult lung. Tissue-specific gene deletion and expression strategies in genetically manipulated mice will allow a molecular dissection of transcriptional regulation in the lung in the near future.
Lung development in Hox-mutants
Hox genes of the paralogous groups 2 through 6 have been detected in embryonic lung development with expression of groups 3 through 6 in the lung proper. Mutations in the more anteriorly expressed Hox gene Hoxa-1 have an indirect affect on the lung: Hoxa-1 mutants fail to initiate breathing presumably due to abnormal projections of cranial nerves IX (glossopharyngeal) and X (vagus) (74). Hoxa-3 mutants die shortly after birth, presumably due to pulmonary failure (75). They exhibit smaller trachea and bronchi. In addition, the cells of the tracheal lining are disorganized and have lost their columnar appearance (76). Together with the Hoxa-3 dependent defect in differentiation of endodermal cells in the thyroid, this may indicate a role of Hoxa-3 for endodermally derived cells including lung epithelial cells. In addition, Hoxa-3 deficient mice appear to have a dysfunction in the musculature and/or innervation controlling epiglottis and esophagus culminating in accumulation of air in the stomach and intestine (75). In this respect, both structural defects in the lung epithelium and in surrounding tissue could contribute to the breathing problems. In summary, the loss of Hoxa-3 function clearly affects the development of the lung and surrounding tissues. It is interesting to note that the tracheal region affected in Hoxa-3 mutants coincides with the domains for Hoxb-3 and b-4 expression suggesting a possible overlap of functional domains. Since Hoxa-3 expression in the developing lung has not been studied in detail, it is currently not known if the lung defects are caused by a direct effect in epithelial cells or through the mesenchyme. Mutants of the paralogous gene Hoxd-3 are able to breathe but die within the first week, probably of accidental cervical dislocation due to malformations in cervical vertebrae (77). The skeletal and thyroid phenotypes of the Hoxa-3 deficiency are exacerbated by the lack of Hoxd-3 supporting a synergistic role for both genes in the same body region (35) although data specifically pertaining to the lung are presently unavailable. Collectively, these results demonstrate that lung function and morphogenesis can be affected by alterations in Hox genes of paralogous group 3. It will be particularly interesting to see if mutations in Hoxb-3 affect lung morphology and TTF-1 mediated gene expression in the lung. Furthermore, superimposition of two or more Hox-3 mutations would be expected to result in very severe abnormalities of the lung.
Mice homozygous for mutations in Hoxa-4 (21), b-4 (19), d-4 (23), a-6 (22) and b-6 (36 and Kappen, unpublished) are viable and fertile while 50 % of the homozygotes carrying mutations in either Hoxa-5 (18) or Hoxb-4 (19) die prematurely. The cause of death has not been determined but does not seem to involve gross tissue abnormalities. These results do not identify any one Hox gene of group 4 through 6 as singularly critical for lung function. However, it is possible that more subtle or regionally restricted alterations of the lungs are present that are not detrimental to survival and thus have escaped notice. Furthermore, the expression of multiple Hox genes in specific regions of the lung may provide a functional redundancy that would only be revealed in compound mutants. For example, mice deficient for both Hoxa-4 and b-4 and those double mutant for Hoxb-4 and d-4 die shortly after birth, most likely from causes other than their skeletal defects (78). Future analyses of the lungs in double and triple mutants for paralogous Hox genes and compound Hox-mutants for overlapping Hox genes are expected to yield important insights into the relevance of the highly specific expression of Hox genes for lung development, differentiation and function.
Models for Hox gene function in the lung
Based upon interpretations of the phenotypes in Hox-transgenic and Hox-mutant mice, two hypotheses with respect to the role of Hox genes in morphogenesis have been proposed: First, Hox genes play a role in positional specification by imposing a so-called ‘Hox-code’ (79) to specific groups of cells which in turn engage in appropriate cell differentiation pathways. This model and its modifications (such as the posterior prevalence hypothesis (80) are supported by the finding of homeotic transformations in the axial skeleton of Hox-mutants and in transgenic mice. The lung phenotype in Hoxa-3 mutants, interpreted according to this model, implies that Hoxa-3 is important for proper specification of tissues in the tracheal region. This region presumably coincides with the most anterior domain of Hoxa-3 expression in the lung; more distal regions are specified by a more ‘posterior’ code of Hox gene expression (assuming that posterior specification overrides anterior codes (80). This hypothesis would also imply that the epithelial disorganization and histologically evident alterations in cell shape are the result of altered differentiation pathways. This question can experimentally be addressed through the use of cell-type and differentiation stage specific markers. Alternatively, it has been suggested that Hox genes control proliferation rates (81) and thereby affect the presence and shape of a subset of structures in their expression domains. This model best explains the malformation of the hindbrain in Hoxa-1 mutants and the thyroid defects in Hoxa-3 mutants as well as various limb defects in mutants of posteriorly expressed Hox genes. According to this model, the reduced size of trachea and bronchi in Hoxa-3 mutants could be the result of decreased cell proliferation. This, in turn, would lead to altered differentiation of epithelial cells by limiting some necessary component. Further studies are required to distinguish these possibilities. Even though the two major hypotheses for Hox gene function not mutually exclusive (82), a role of Hox genes in cell differentiation as opposed to cell proliferation would imply different downstream targets of Hox gene regulation and ultimately different molecular events.
With respect to lung development, these models allow specific predictions: 1) The Hox genes that can directly contribute to lung development are those located in paralogous groups 3 through 6. It is likely that they regulate lung-specific processes, probably through lung-specific transcription factors. 2) Hox genes of paralogous groups 3 - 6 may control the development of restricted regions in the lung in a proximal-distal progression. Temporal differences in expression could influence the relative contribution of paralogous and neighboring genes. 3) The overlapping Hox gene expression patterns and lack of overt lung phenotypes in some of the mutants indicate a certain degree of functional redundancy in lung development. It is possible that several Hox genes may act in combination to specify particular regions of the lung. Further analyses of Hox genes that affect the same structures in double and triple mutants will help to distinguish these possibilities. 4) Alterations in Hox gene mutants (and Hox-transgenic mice) are expected to primarily affect mesodermal derivatives; effects on epithelial cells may be indirectly mediated through mesenchymal-epithelial interactions. These possibilities can be experimentally resolved through analysis of lung-specific markers and gene regulation in Hox gene mutants. 5) Expression of Hox genes in the adult lung may be relevant to tissue regeneration processes as well as pathological lung cell differentiation and/or proliferation. 6) Identifying the cellular and molecular basis for the morphological alterations in Hox gene mutants and transgenic mice will help in characterizing downstream events in transcriptional regulation. In this way, Hox genes provide a paradigm to bridge the gap between regional specification in development and tissue and cell differentiation in organogenesis.
Hox genes in lung cancer
The capacity of Hox genes to control of cell proliferation has been most convincingly supported by evidence that Hox genes can function as oncogenes. Transfection of NIH3T3 cells with Hox genes, results in a transformed cell phenotype in vitro and in tumor formation upon transplantation into nude mice in vivo (83, 84). Inappropriate expression of the HOXA9 (85), Hoxb-4 (86), and Hoxb-8 (87, 88) genes in the hematopoietic system in vivo leads to the development of leukemias presumably through increased progenitor cell proliferation (reviewed in (89). These results demonstrate that Hox genes control cell proliferation and the emergence of the tumorigenic cell phenotype. Correlative evidence suggests that Hox genes could also be involved in tumorigenesis in the lung. Tiberio et al. conducted complete surveys for expression of the 38 Hox genes in Small Cell Lung Cancers (SCLC) that had been xenografted to nude mice (38). They found that, on average, about 20-25 Hox genes are expressed in these cancers. This number is at least two-fold higher than the number of Hox genes detected in normal adult human lung suggesting that activation of Hox genes might be involved in lung tumorigenesis. Interestingly, the genes not normally expressed in the lung but found in the cancers are located 5′ in the clusters. These 5′ genes have been implicated in regulating cell proliferation in the developing limb, and both HOXA9 (85) and Hoxb-8 (90, 91) are involved in leukemic translocations consistent with deregulated cell proliferation in cancer. Tiberio et al. also attempt to correlate Hox gene expression with clinical features of the tumors, such as histological appearance and malignancy. They show a trend towards a decrease in the number of expressed Hox genes with an increase in malignancy. Although based on a limited number of samples, these results suggest that tumor progression could be associated with a loss of Hox gene expression (38). In summary, the available evidence suggests that Hox genes may be involved in lung cancer in two ways: activation of Hox genes as a step associated with tumorigenesis, and loss of Hox gene expression associated with progression to the metastatic state. Since surveys of tumors are complicated by variable degrees of cellular heterogeneity in the tumor and by chromosomal abnormalities and chromosome losses, more extensive studies will be required to clarify the role of Hox genes in the formation of lung tumors and their progression.
Concluding remarks
The relevance of Hox genes for lung development is becoming increasingly clear. Expression studies have documented temporally dynamic and regionally highly specific patterns of Hox gene expression in the lung. In analogy to their role in patterning other structures, such as the axial skeleton, these findings suggest a role of Hox-transcription factors in regional specification and/or tissue-specific cell differentiation in lung development. Putatively, Hox genes may also be involved in pathological lung cell differentiation. The presence of lung abnormalities in Hoxa-3 mutant mice provides functional evidence for a critical role in lung development. The in vivo analysis of Hox gene function with emphasis on the lung has just begun and is expected to yield insight into the as yet undefined molecular mechanisms that govern regional specification and cell differentiation in the developing lung.
Acknowledgments
I thank Dr. John A. McDonald for the invitation to write this review, and him and Drs. James J. Lee (both Mayo Clinic Scottsdale) and J. Michael Salbaum (The Neurosciences Institute, San Diego) for discussion and critical reading of the manuscript. Work on Hox genes in my laboratory is funded by the March of Dimes Foundation for Birth Defects and Mayo Foundation.
References
- 1.Scott MP, Tamkun JW, Hartzell GW. The structure and function of the homeodomain. Biochim. Biophys. Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 2.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 3.Gehring WJ. The homeobox in perspective. Trends Biochem. Sci. 1992;17:277–280. doi: 10.1016/0968-0004(92)90434-b. [DOI] [PubMed] [Google Scholar]
- 4.Andrew DJ, Scott MP. Downstream of the homeotic genes. New Biologist. 1992;4:5–15. [PubMed] [Google Scholar]
- 5.Edelman GM, Jones FS. Outside and downstream of the homeobox. J.Biol.Chem. 1993;268:20683–20686. [PubMed] [Google Scholar]
- 6.Kappen C, Schughart K, Ruddle FH. Early evolutionary origin of major homeodomain sequence classes. Genomics. 1993;18:54–70. doi: 10.1006/geno.1993.1426. [DOI] [PubMed] [Google Scholar]
- 7.Stein S, Fritsch R, Lemaire L, Kessel M. Checklist - Vertebrate Homeobox Genes. Mech.Develop. 1996;55:91–108. doi: 10.1016/0925-4773(95)00494-7. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman MH. The Atlas of Mouse Development. Academic Press; San Diego: 1992. [Google Scholar]
- 9.Burri PH. Fetal and postnatal development of the lung. Ann. Rev. Physi. 1984;46:617–628. doi: 10.1146/annurev.ph.46.030184.003153. [DOI] [PubMed] [Google Scholar]
- 10.Cardoso WV. Transcription factors and pattern formation in the developing lung. Am. J. Physiol. (Lung Cell Mol. Physiol.) 1995;269:L429–L442. doi: 10.1152/ajplung.1995.269.4.L429. [DOI] [PubMed] [Google Scholar]
- 11.Kappen C, Ruddle FH. Evolution of a regulatory gene family: HOM/HOX genes. Curr. Op. Genet. Develop. 1993;3:931–938. doi: 10.1016/0959-437x(93)90016-i. [DOI] [PubMed] [Google Scholar]
- 12.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 13.Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruss P, Kessel M. Axial specification in higher vertebrates. Curr. Op. Genet. Develop. 1991;1:204–210. doi: 10.1016/s0959-437x(05)80071-1. [DOI] [PubMed] [Google Scholar]
- 15.Krumlauf R. Hox Genes in Vertebrate Development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 16.LeMouellic H, Lallemand Y, Brulet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992;69:251–264. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- 17.Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1332. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 18.Jeanotte L, Lemieux M, Charron J, Poirier F, Robertson EJ. Specification of axial identity in the mouse: role of the Hoxa-5 (Hox1.3) gene. Genes Develop. 1993;7:2085–2096. doi: 10.1101/gad.7.11.2085. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A. Hoxb-4 (hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993;73:279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- 20.Davis AP, Capecchi MR. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development. 1994;120:2187–2198. doi: 10.1242/dev.120.8.2187. [DOI] [PubMed] [Google Scholar]
- 21.Horan GS, Wu K, Wolgemuth DJ, Behringer RR. Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc. Natl. Acad. Sci. USA. 1994;91:12644–12648. doi: 10.1073/pnas.91.26.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostic D, Capecchi MR. Targeted disruptions of the murine hoxa-4 and hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech. Develop. 1994;46:231–247. doi: 10.1016/0925-4773(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 23.Horan GSB, Kovacs EN, Behringer RR, Featherstone MS. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton - evidence for unique and redundant function. Develop. Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 24.Rijli FM, Matyas R, Pellegrini M, Dierich A, Gruss P, Dolle P, Chambon P. Cryptochordism and homeotic transformations of spinal nerves and vertebrae in Hoxa-10 mutant mice. Proc. Natl. Acad. Sci. USA. 1995;92:8185–8189. doi: 10.1073/pnas.92.18.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Small KM, Potter SS. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Develop. 1995;7:2318–2328. doi: 10.1101/gad.7.12a.2318. [DOI] [PubMed] [Google Scholar]
- 26.Davis AP, Witte DP, Hsiehli HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking Hoxa-11 and Hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 27.Davis AP, Capecchi MR. A mutational analysis of the 5' Hoxd genes -dissection of genetic interactions during limb development in the mouse. Development. 1996;122:1175–1185. doi: 10.1242/dev.122.4.1175. [DOI] [PubMed] [Google Scholar]
- 28.Favier B, Rijli FM, Fromental-Ramain C, Fraulob V, Chambon P, Dolle P. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development. 1996;122:449–460. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- 29.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Dolle P, Chambon P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 1996;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 30.Kessel M, Balling R, Gruss P. Variations of cervical vertebrae after expression of a Hox -1.1 transgene in mice. Cell. 1990;61:301. doi: 10.1016/0092-8674(90)90810-2. [DOI] [PubMed] [Google Scholar]
- 31.Jegalian BG, DeRobertis EM. Homeotic transformations in the mouse induced by overexpression of a human Hox3.3 transgene. Cell. 1992;71:901–910. doi: 10.1016/0092-8674(92)90387-r. [DOI] [PubMed] [Google Scholar]
- 32.Pollock RA, Jay G, Bieberich CJ. Altering the boundaries of Hox3.1 expression: evidence for antipodal gene regulation. Cell. 1992;71:911–923. doi: 10.1016/0092-8674(92)90388-s. [DOI] [PubMed] [Google Scholar]
- 33.Pollock RA, Sreenath T, Ngo L, Bieberich CJ. Gain of function mutations for paralogous Hox genes: implications for the evolution of Hox gene function. Proc. Natl. Acad. Sci. USA. 1995;92:4492–4496. doi: 10.1073/pnas.92.10.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charite J, de Graaff W, Shen SB, Deschamps J. Ectopic expression of hoxb-8 causes duplication of the zpa in the forelimb and homeotic transformation of axial structures. Cell. 1994;78:589–601. doi: 10.1016/0092-8674(94)90524-x. [DOI] [PubMed] [Google Scholar]
- 35.Condie BG, Capecchi MR. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature. 1994;370:304–307. doi: 10.1038/370304a0. [DOI] [PubMed] [Google Scholar]
- 36.Rancourt DE, Tsuzuki T, Capecchi MR. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Develop. 1995;9:108–122. doi: 10.1101/gad.9.1.108. [DOI] [PubMed] [Google Scholar]
- 37.Bernacki SH, Nervi C, Vollberg TM, Jetten AM. Homeobox 1.3 expression: induction by retinoic acid in human bronchial fibroblasts. Am. J. Resp. Cell Mol. Biol. 1992;7:3–9. doi: 10.1165/ajrcmb/7.1.3. [DOI] [PubMed] [Google Scholar]
- 38.Tiberio C, Barba P, Magli MC, Arvelo F, Le Chevalier T, Poupon MF, Cillo C. HOX gene expression in human small-cell lung cancers xenografted into nude mice. Int. J. Cancer. 1994;58:608–615. doi: 10.1002/ijc.2910580426. [DOI] [PubMed] [Google Scholar]
- 39.Jackson IJ, Schofield P, Hogan B. A mouse homeo box gene is expressed during embryogenesis and in adult kidney. Nature. 1985;317:745–748. doi: 10.1038/317745a0. [DOI] [PubMed] [Google Scholar]
- 40.Graham A, Papalopulu N, Lorimer J, McVey JH, Tuddenham EGD, Krumlauf R. Characterization of a murine homeo box gene, Hox-2.6, related to the Drosophila deformed gene. Genes Develop. 1988;2:1424–1438. doi: 10.1101/gad.2.11.1424. [DOI] [PubMed] [Google Scholar]
- 41.Bogue CW, Lou LJ, Vasavade H, Wilson CM, Jacobs HC. Expression of Hoxb genes in the developing mouse foregut and lung. Am. J. Respir. Cell Mol. Biol. 1996 doi: 10.1165/ajrcmb.15.2.8703472. this issue. [DOI] [PubMed] [Google Scholar]
- 42.Tan DP, Ferrante J, Nazarali A, Shao X, Kozak CA, Guo V, Nirenberg M. Murine Hox-1.11 homeobox gene structure and expression. Proc. Natl. Acad. Sci . USA. 1992;89:6280–6284. doi: 10.1073/pnas.89.14.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaunt SJ, Sharpe PT, Duboule D. Spatially restricted domains of homeo gene transcripts in mouse embryos: relation to a segmented body plan. Development. 1988;104(Suppl.):169. [Google Scholar]
- 44.Galliot B, Dollé P, Vigneron M, Featherstone MS, Baron A, Duboule D. The mouse Hox-1.4 gene: primary structure, evidence for promoter activity and expression during development. Development. 1989;107:343. doi: 10.1242/dev.107.2.343. [DOI] [PubMed] [Google Scholar]
- 45.Gaunt SJ, Krumlauf R, Duboule D. Mouse homeo-genes within a subfamily, Hox-1.4, -2.6 and -5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development. 1989;107:131–141. doi: 10.1242/dev.107.1.131. [DOI] [PubMed] [Google Scholar]
- 46.Dony C, Gruss P. Specific expression of the Hox 1.3 homeo box gene in murine embryonic structures originating from or induced by the mesoderm. EMBO J. 1987;6:2965–2975. doi: 10.1002/j.1460-2075.1987.tb02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt P, Wilkinson D, Krumlauf R. Patterning the vertebrate head: murine Hox 2 genes mark distinct subpopulations of premigratory and migrating cranial neural crest. Development. 1991;112:43–50. doi: 10.1242/dev.112.1.43. [DOI] [PubMed] [Google Scholar]
- 48.Conlon RA, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 49.Sham MH, Hunt P, Nonchev S, Papalopulu N, Graham A, Boncinelli E, Krumlauf R. Analysis of the murine Hox-2.7 gene: conserved alternative transcripts with differential distributions in the nervous system and the potential for shared regulatory regions. EMBO J. 1992;11:1825–1836. doi: 10.1002/j.1460-2075.1992.tb05234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graham A, Holland PW, Lumsden A, Krumlauf R, Hogan BL. Expression of the homeobox genes Hox 2.1 and 2.6 during mouse development. Curr. Top. Microbiol. Immunol . 1988 doi: 10.1007/978-3-642-50059-6_14. [DOI] [PubMed] [Google Scholar]
- 51.Geada A, Gaunt S, Azzawi M, Shimeld S, Pearce J, Sharpe P. Sequence and embryonic expression of the murine Hox-3.5 gene. Development. 1992;116:497–506. doi: 10.1242/dev.116.2.497. [DOI] [PubMed] [Google Scholar]
- 52.Oliver G, Wright CV, Hardwicke J, De Robertis EM. Differential antero-posterior expression of two proteins encoded by a homeobox gene in Xenopus and mouse embryos. EMBO J. 1988;7:3199–3209. doi: 10.1002/j.1460-2075.1988.tb03187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimeld SM, Gaunt SJ, Coletta PL, Geada AMC, Sharpe PT. Spatial localisation of transcripts of the Hox-C6 gene. J. Anat. 1993;183:515–523. [PMC free article] [PubMed] [Google Scholar]
- 54.Shashikant CS, Utset MF, Violette SM, Wise TL, Einat P, Einat M, Pendleton JW, Schughart K, Ruddle FH. Homeobox genes in mouse development. Crit. Rev. Eukar. Gene Expr. 1991;1:207–245. [PubMed] [Google Scholar]
- 55.Deschamps J, Meijlink F. Mammalian homeobox genes in normal development and neoplasia. Crit. Rev. Oncogen. 1992;3:117–173. [PubMed] [Google Scholar]
- 56.Bogue CW, Gross I, Vasavada H, Dynia DW, Wilson CM, Jacobs HC. Identification of hox genes in newborn lung and effects of gestational age and retinoic acid on their expression. Am. J. Physiol. (Lung Cell Mol. Physiol.) 1994;266:L488–L454. doi: 10.1152/ajplung.1994.266.4.L448. [DOI] [PubMed] [Google Scholar]
- 57.Holland PW, Hogan BL. Spatially restricted patterns of expression of the homeobox-containing gene Hox 2.1. during mouse embryogenesis. Development. 1988;102:159–174. doi: 10.1242/dev.102.1.159. [DOI] [PubMed] [Google Scholar]
- 58.Kress C, Vogels R, De Graaff W, Bonnerot C, Meijlink F, Nicolas JF, Deschamps J. Hox-2.3 upstream sequences mediate lacZ expression in intermediate mesoderm derivatives of transgenic mice. Development. 1990;109:775–86. doi: 10.1242/dev.109.4.775. [DOI] [PubMed] [Google Scholar]
- 59.Vogels R, Charite J, De Graaff W, Deschamps J. Proximal cis-acting elements cooperate to set Hoxb-7 (Hox-2.3) expression boundaries in transgenic mice. Development. 1993;118:71–82. doi: 10.1242/dev.118.1.71. [DOI] [PubMed] [Google Scholar]
- 60.Eid R, Koseki H, Schughart K. Analysis of LacZ reporter genes in transgenic embryos suggests the presence of several cis-acting regulatory elements in the murine Hoxb-6 gene. Develop. Dyn. 1993;196:205–216. doi: 10.1002/aja.1001960307. [DOI] [PubMed] [Google Scholar]
- 61.Alescio T, Cassini A. Induction in vitro of tracheal buds by pulmonary mesenchyme grafted on tracheal epithelium. J. Exp. Zool. 1962;150:83–94. doi: 10.1002/jez.1401500202. [DOI] [PubMed] [Google Scholar]
- 62.Alescio T, Piperno EC. A quantitative assessment of mesenchymal contribution to epithelial growth rate in mouse embryonic lung developing in vitro. J. Embryol. Exp. Morphol. 1967;17:213–227. [PubMed] [Google Scholar]
- 63.Behringer RR, Crotty DA, Tennyson VM, Brinster RL, Palmiter RD, Wolgemuth DJ. Sequences 5′ of the homeobox of the hox-1.4 gene direct tissue-specific expression of lacz during mouse development. Development. 1993;117:823–833. doi: 10.1242/dev.117.3.823. [DOI] [PubMed] [Google Scholar]
- 64.Spooner BS, Wessels NK. Mammalian lung development: intereactions in primordium formation and bronchial morphogenesis. J. Exp. Zool. 1970;175:455–454. doi: 10.1002/jez.1401750404. [DOI] [PubMed] [Google Scholar]
- 65.Wessels NW. Mammalian lung development: interactions in formation and morphogenesis of tracheal buds. J. Exp. Zool. 1970;175:455–466. doi: 10.1002/jez.1401750405. [DOI] [PubMed] [Google Scholar]
- 66.Wall NA, Jones CM, Hogan BL, Wright CV. Expression and modification of Hox 2.1 protein in mouse embryos. Mech. Develop. 1992;37:111–20. doi: 10.1016/0925-4773(92)90073-s. [DOI] [PubMed] [Google Scholar]
- 67.Holland PWH, Hogan BLM. Expression of homeo box genes during mouse development: a review. Genes Develop. 1988;2:773–782. doi: 10.1101/gad.2.7.773. [DOI] [PubMed] [Google Scholar]
- 68.Guazzi S, Lonigro R, Pintonello L, Boncinelli E, Di Lauro R, Mavilio F. The thyroid transcription factor-1 gene is a candidate target for regulation by Hox proteins. EMBO J. 1994;13:3339–47. doi: 10.1002/j.1460-2075.1994.tb06636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lazarro D, Price M, DeFelice M, DiLauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–11-4. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 70.Whitsett JA, Korfhagen TR. Regulation of Gene Transcription in Respiratory Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 1996;14:118–120. doi: 10.1165/ajrcmb.14.2.8630260. [DOI] [PubMed] [Google Scholar]
- 71.Minoo P, Hamdan H, Bu D, Warburton D, Stepanik P, Delemos R. Ttf-1 regulates lung epithelial morphogenesis. Develop. Biol. 1995;172:694–698. doi: 10.1006/dbio.1995.8080. [DOI] [PubMed] [Google Scholar]
- 72.Krumlauf R, Holland PW, McVey JH, Hogan BL. Developmental and spatial patterns of expression of the mouse homeobox gene, Hox 2.1. Development. 1987;99:603–17. doi: 10.1242/dev.99.4.603. [DOI] [PubMed] [Google Scholar]
- 73.Kuratani SC, Wall NA. Expression of Hox 2.1 protein in restricted populations of neural crest cells and pharyngeal ectoderm. Develop. Dyn. 1992;195:15–28. doi: 10.1002/aja.1001950103. [DOI] [PubMed] [Google Scholar]
- 74.Chisaka O, Musci TS, Capecchi MR. Developmental defects in the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992;355:516–20. doi: 10.1038/355516a0. [DOI] [PubMed] [Google Scholar]
- 75.Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991;350:473–479. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 76.Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121:1989–2003. doi: 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- 77.Condie BG, Capecchi MR. Mice homozygous for a targeted disruption of Hoxd-3 (Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and the axis. Development. 1993;119:579–595. doi: 10.1242/dev.119.3.579. [DOI] [PubMed] [Google Scholar]
- 78.Horan GSB, Ramirez-Solis R, Featherstone MS, Wolgemuth DJ, Bradley A, Behringer RR. Compound mutants for the paralogous hoxa4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Develop. 1995;9:1667–1677. doi: 10.1101/gad.9.13.1667. [DOI] [PubMed] [Google Scholar]
- 79.Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concommittant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- 80.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 81.Duboule D. Temporal colinearity and the phylogenetic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development - Suppl. 1994;120:135–142. [PubMed] [Google Scholar]
- 82.Duboule D. Vertebrate Hox-genes and proliferation: an alternative pathway to homeosis? Curr. Op. Genet. Develop. 1995;5:525–528. doi: 10.1016/0959-437x(95)90058-o. [DOI] [PubMed] [Google Scholar]
- 83.Aberdam D, Negreanu V, Sachs L, Blatt C. The Oncogenic Potential of an Activated Hox-2.4 Homeobox Gene in Mouse Fibroblasts. Mol. Cell. Biol. 1991;11:554–557. doi: 10.1128/mcb.11.1.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maulbecker CC, Gruss P. The oncogenic potential of deregulated homeobox genes. Cell Growth Diff. 1993;4:431–41. [PubMed] [Google Scholar]
- 85.Borrow J, Shearman AM, Stanton VP, Becher R, Collins T, Williams AJ, Dube I, Katz F, Kwong YL, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman DE. The t(7;11)(p15;p15) translocation in acute myeloid leukemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HoxA9. Nature genetics. 1996;12:159–166. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 86.Sauvageau G, Thorsteins-Dottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Develop. 1995;9:1753–65. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 87.Perkins A, Konsuwan K, Visvader J, Adams JM, Cory S. Homeobox gene expression plus autocrine growth factor production elicits myeloid leukemia. Proc. Natl. Acad. Sci. USA. 1990;87:8398. doi: 10.1073/pnas.87.21.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blatt C, Lotem J, Sachs L. Inhibition of specific pathways of myeloid cell differentiation by an activated Hox-2.4 homeobox gene. Cell Growth Diff. 1992;3:671–676. [PubMed] [Google Scholar]
- 89.Kehrl JH. Homeobox genes in hematopoiesis. Crit. Rev. Onc.-Hematol. 1994;16:145–156. doi: 10.1016/1040-8428(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 90.Blatt C, Aberdam D, Schwartz R, Sachs L. DNA rearrangement of a homeobox gene in myeloid leukaemic cells. EMBO J. 1988;7:4283–4290. doi: 10.1002/j.1460-2075.1988.tb03326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kongsuwan K, Webb E, Housiaux P, Adams JM. Expression of multiple homeobox genes within diverse mammalian haemopoietic lineages. EMBO J. 1988;7:2131–2138. doi: 10.1002/j.1460-2075.1988.tb03052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]