Abstract
Engineered analog-sensitive (AS) protein kinases have emerged as powerful tools for dissecting phospho-signaling pathways, for elucidating the cellular function of individual kinases, and for deciphering unanticipated effects of clinical therapeutics. A crucial and necessary feature of this technology is a bioorthogonal small molecule that is innocuous towards native cellular systems but can potently inhibit the engineered kinase. In order to generalize this method we sought a molecule capable of targeting divergent AS-kinases. Here we employ X-ray crystallography and medicinal chemistry to unravel the mechanism of current inhibitors and use these insights to design the most potent, selective and general AS-kinase inhibitors reported to date. We use large-scale kinase inhibitor profiling to characterize the selectivity of these molecules as well as examine the consequences of potential off-target effects in chemical genetic experiments. The molecules reported here will serve as powerful tools in efforts to extend AS-kinase technology to the entire kinome and the principles discovered may help in the design of other engineered enzyme/ligand pairs.
Eukaryotic protein kinases catalyze the transfer of phosphate from ATP to serine, threonine or tyrosine residues on substrate proteins. The resulting phosphorylation event constitutes a critical mode of post-translational protein modification and a core mechanism for cellular signal transduction. Nearly every physiological process in eukaryotic cells involves phospho-signaling networks and many human diseases occur when these pathways are disregulated1. Potent and selective inhibitors of protein kinases are valuable tools for probing these networks and for elucidating the cellular functions of individual kinases2. However, due to the large number of protein kinases in a cell and their highly homologous active sites, it has proven challenging to find specific inhibitors for many individual kinases3. Our laboratory has developed a chemical genetic technique which enables systematic generation of highly specific inhibitors for individual kinases4. This approach combines the advantages of a small molecule inhibitor (temporal resolution, reversibility, etc.) with the specificity of a genetic manipulation. When a residue at a structurally conserved position in the kinase active site (termed the gatekeeper5) is mutated from the natural bulky amino acid side chain (methionine, leucine, phenylalanine, threonine, etc.) to smaller glycine or alanine, a novel pocket that is not found in wild-type (WT) kinases is created within the ATP-binding site. Such an engineered kinase is termed an analog-sensitive (AS) allele because it can be potently and specifically targeted by inhibitor analogs that contain a bulky substituent complementing the enlarged ATP binding pocket6.
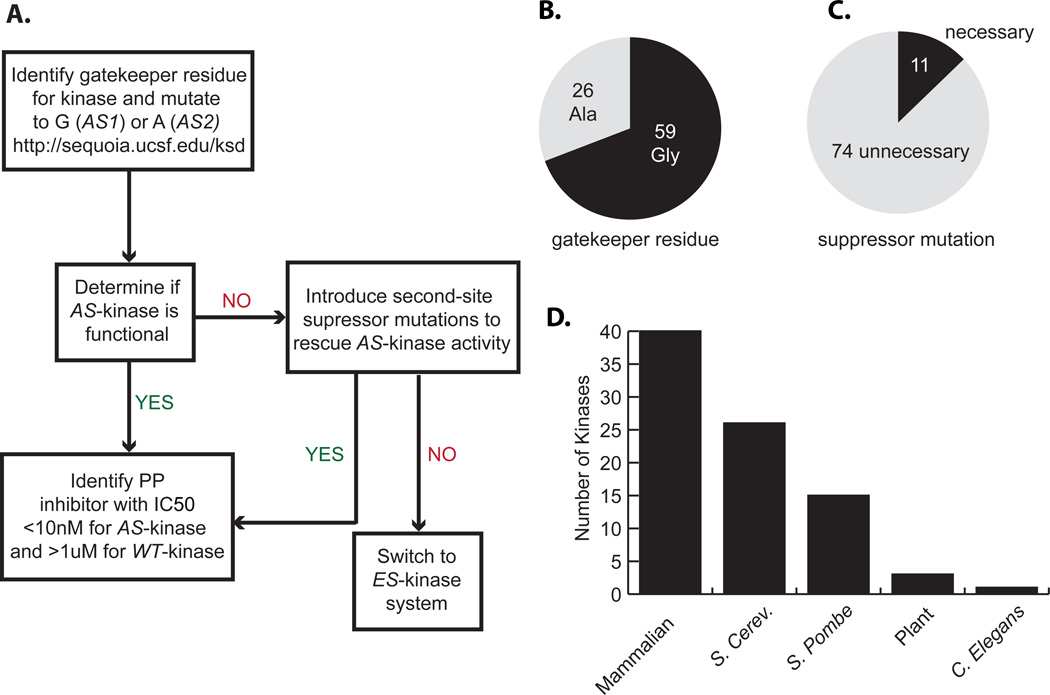
A major goal of our laboratory is to extend AS-kinase technology to the entire kinome; the fact that nearly all kinases contain a bulky gatekeeper residue suggests this undertaking is possible in principle. However, two criteria must be satisfied for a particular kinase-of-interest to be amenable to the approach. First, the kinase must tolerate mutation of the gatekeeper residue to glycine or alanine without severe loss of catalytic activity or cellular function. Second, a potent and bioorthogonal small molecule inhibitor of the AS-kinase must be identified. In the course of our studies we have encountered a number of kinases that do not meet one or both of the criteria; they are either intolerant of the gatekeeper mutation or insensitive to available pyrazolo[3,4-d]pyrimidine (PP) inhibitors. We have developed a systematic workflow to overcome each of these limitations as they present themselves (Fig. 1A). For example, if the kinase does not tolerate a glycine gatekeeper, which provides the maximal expansion of the ATP pocket, the larger amino acid alanine may be used instead (Fig. 1B). We have also demonstrated that the introduction of second-site suppressor mutations can be employed to rescue the activity of weakened AS-kinase alleles7 (Fig. 1C). In extreme cases where second-site suppressor mutations fail to rescue activity a second approach termed an electrophile-sensitive (ES)-kinase may be used8. In ES-kinases the gatekeeper is mutated to cysteine instead of alanine or glycine, thereby retaining a more hydrophobic residue at the gatekeeper position, maintaining the integrity of the hydrophobic spine of the kinase9 and thus enhancing enzyme activity, while sensitizing the ES-kinase to PP-inhibitors bearing an appropriately positioned electrophile. Once an active AS-kinase or ES-kinase allele has been engineered, a potent and selective AS-kinase or ES-kinase inhibitor must be identified from a small library of PP molecules based on the semi-promiscuous tyrosine kinase inhibitor PP1.
Figure 1.
(A) Flow-chart outlines systematic approach for application of AS-kinase technology. (B) The pie chart illustrates the number of reported AS-kinases with alanine, A, or Glycine, G, gatekeeper residues. (C) The number of reported AS-kinases that require suppressor mutations to rescue activity. (D) The bar graph denotes the number of AS-kinases that have been reported in peer-reviewed publications for each class of kinase.
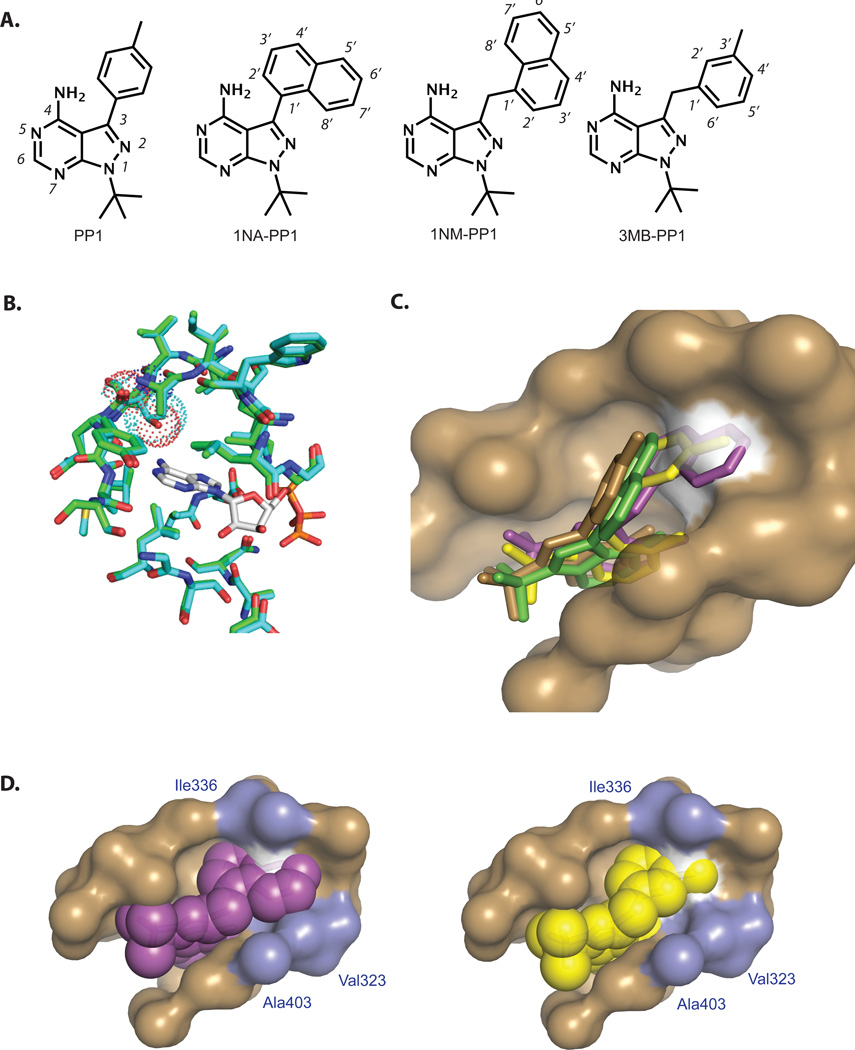
PP1 was initially identified as a potent ATP-competitive inhibitor of Src family kinases10. The co-crystal structure of PP1 bound to a Src family kinase, Hck, showed that the pyrazolopyrimidine core of PP1 mimics the adenine ring of ATP in its binding to the nucleobase pocket while the p-tolyl group at the C3 position projects into a deep hydrophobic pocket situated between the gatekeeper (T338) and the catalytic lysine residue (K295)11. Based on this crystal structure, it was predicted that PP1 analogs with an enlarged C3 substituent would suffer a steric clash with the native gatekeeper residue in WT kinases. However, mutation of the gatekeeper to glycine or alanine should create extra space to accommodate the enlarged C3 substituent. It has been demonstrated that potent and specific inhibitors can be readily identified for various AS kinases by screening a small panel of PP’s with enlarged C3 substituents4.
1NA-PP1 and 1NM-PP1 were initially found to be the most effective inhibitors for AS-kinases and have been used to target numerous AS-kinase alleles from a variety of organisms and kinase families (Fig. 1D, Table S1). In the course of our studies, however, we have encountered a number of AS-kinases that are insensitive to 1NA-PP1 and 1NM-PP1, thereby precluding the application of our chemical genetic technique. Our initial hypothesis was that the PP structure was sub-optimal for some kinases and thus we sought alternative scaffolds that might be complementary or even superior to PP. For example, we used an inhibitor based on the aminoindazole scaffold (a known Akt binder) to target AS-Akt alleles12, 13. However, this molecule and its derivatives were largely inactive towards other AS-kinases (unpublished results). We also attempted to design AS-kinase inhibitors based on purine, pyrimidine, and quinazoline scaffolds14, 15, but these molecules also proved to be largely ineffective against diverse AS-kinases. Since the PP scaffold has so far proven to be the most general for AS-kinases, we embarked on a medicinal chemistry effort to find a new PP analog capable of targeting recalcitrant AS-kinases. The result of this undertaking was the identification of a new molecule 3MB-PP1 that has enhanced potency against a diverse set of AS-kinases, including several that were insensitive to all previously known AS-kinase inhibitors12, 16–18. The focus of this study is to understand the mechanism of efficacy of 3MB-PP1 by analysis of co-crystal structures of Src-AS1 kinase bound to 3MB-PP1, 1NA-PP1, or 1NM-PP1. We then use insights gained from these structures to design next-generation PP inhibitors with increased potency and selectivity for AS-kinases.
Results and Discussion
X-ray Crystal Structures of PP Inhibitors Bound to Src-AS1 Reveal Mechanism of Enhanced Efficacy of 3MB-PP1
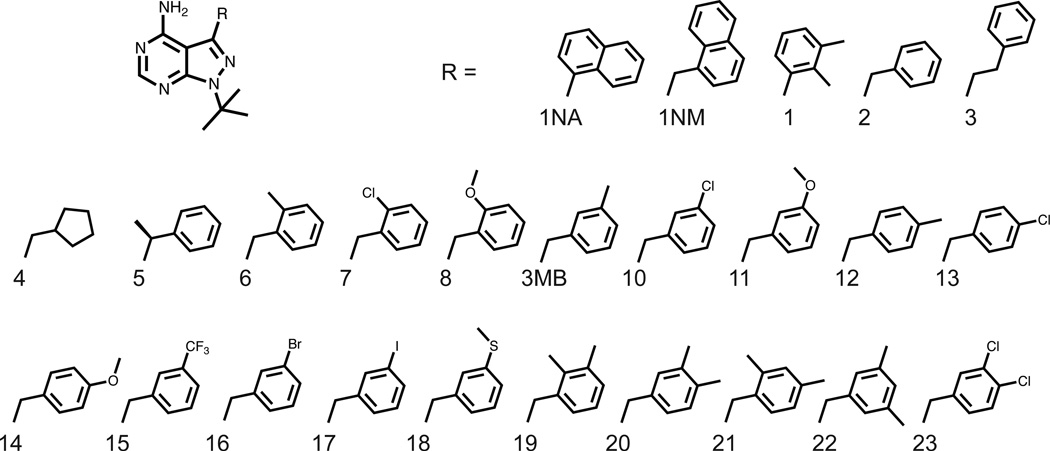
We previously reported that 3MB-PP1 and a closely related analog 16 (Fig. 2A, Fig. 3) are capable of targeting several AS-kinases that are impervious to 1NA-PP1 and 1NM-PP112, 16–18. In order to better understand the enhanced efficacy of 3MB-PP1 and the potential deficiencies of 1NA-PP1 and 1NM-PP1 in targeting divergent AS-kinases, we solved co-crystal structures of Src-AS1 (T338G) in complex with 1NA-PP1, 1NM-PP1 or 3MB-PP1 (Fig. 2C). Comparison of the Src-AS1 structures to the previously solved Src-WT/AMP-PNP complex reveals little movement of protein backbone atoms (with an RMSD of 0.22 A) upon introduction of the gatekeeper mutation T338G, indicating that the mutation does not perturb the overall structure of the Src protein (Fig. 2B). The most notable effect of the T338G mutation in Src is the expanded ATP-binding pocket that we hypothesized would accommodate bulky groups at the C3 position of PP1. Next we created an overlay of the Src-AS1 structures with that of the highly homologous tyrosine kinase Hck bound to PP1 (Fig. 2C). We found that the PP analogs share nearly identical binding orientations and hydrogen bond interactions with the hinge region of the kinase. As predicted, the bulky naphthyl, naphthylmethyl and 3’-methylbenzyl substituents of 1NA-PP1, 1NM-PP1 and 3MB-PP1, respectively, project toward the gatekeeper and occupy the pocket created by the T338G mutation. While PP1 is accommodated in the ATP pocket of Hck, it is clear that all three AS-kinase inhibitors are prevented from binding due to a steric clash with the Thr gatekeeper residue; however, the degree of clash varies amongst the compounds. Specifically, the napthyl ring of 1NA-PP1 (Fig. 2C, green) appears to have minimal overlap with the surface of the gatekeeper side chain, while the clash between the gatekeeper and 1NM-PP1 (Fig. 2C, purple) is maximal and that with 3MB-PP1 (Fig. 2C, yellow) is intermediate. The minimal clash between 1NA-PP1 and a Thr gatekeeper explains why 1NA-PP1 has weak activity towards WT-kinases with small gatekeeper residues but not towards kinases with larger Ile or Phe gatekeeper residues4. In contrast, the C5’ position of 1NM-PP1 projects deep into the gatekeeper side chain surface of Hck, thus explaining the enhanced orthogonality of 1NM-PP1 towards WT kinases relative to 1NA-PP1.
Figure 2.
(A) PP1 is a promiscuous kinase inhibitor that targets kinases with medium size gatekeeper residues (Thr, Val, Ser). 1NA-PP1, 1NM-PP1 and 3MB-PP1 are three of the most commonly used AS-kinase inhibitors. (B) Overlay of Src-WT (teal) and Src-AS1 (green) with the WT-gatekeeper surface illustrated in dots and the Src-WT ligand AMP-PNP (white). (C) Overlay of 1NA-PP1(green), 1NM-PP1(pink) and 3MB-PP1(yellow) with the PP1/Hck-WT structure (brown). The surface of the gatekeeper residue is highlighted in white. (D) Overlay of Hck ATP pocket surface with 3MB-PP1 (right, yellow spheres) and 1NM-PP1 (left, pink spheres). Surface of residues V323, I336 and A403 are highlighted in blue.
Figure 3.
Chemical structures of PP compounds
A key feature of both 1NM-PP1 and 3MB-PP1 is the methylene linker between the aryl ring and the heterocyclic core of the PP scaffold. This linkage imbues the molecules with rotational freedom that allows for optimal orientation of the bulky phenyl and napthyl rings in the gatekeeper pocket, thereby maximizing potential affinity interactions with the enlarged ATP pocket of AS-kinases. Simultaneously, the methylene linker ensures that the bulky aryl groups project more deeply into the gatekeeper pocket, resulting in greater steric clash with the gatekeeper side chain of WT-kinases. The 3’-methyl group of 3MB-PP1 and C5’ of 1NM-PP1 appear to occupy the same position within the ATP binding pocket and provide the most direct steric clash with the gatekeeper side chain. An important difference between 3MB-PP1 and 1NM-PP1, however, is that the additional atoms of the naphthyl ring of 1NM-PP1 (C6’, C7’, and C8’) provide additional clash with the Hck gatekeeper, but also appear to overlap with the surface of other residues in the ATP binding pocket (V323, I336 and A403; Fig. 2D). We hypothesize that subtle differences in the ATP binding sites of divergent kinases might alleviate or escalate this undesired interaction and, in extreme cases, prevent 1NM-PP1 from binding to the AS-kinase despite the enlarged gatekeeper pocket. In fact, it has been demonstrated that some kinases bearing larger amino acids at the position analogous to Ala403 can be further sensitized to PP derivatives by mutation of that position to alanine19.
Structure-guided Inhibitor Design Yields More Potent, Selective and General AS-kinase Inhibitors
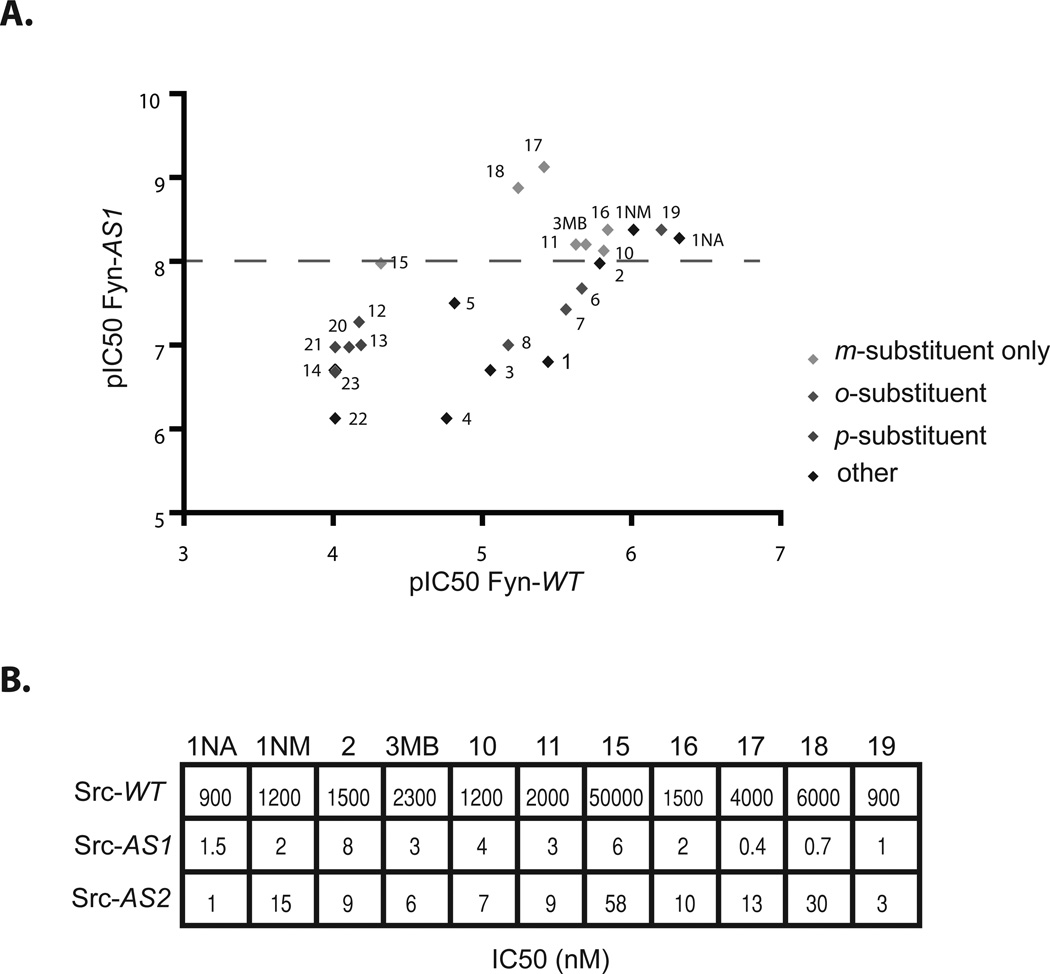
Next we sought to test our hypothesis that the benzyl moiety is the optimal orthogonality element for PP AS-kinase inhibitors as well as to design more potent, selective and general AS-kinase inhibitors using our enhanced understanding of 3MB-PP1 as a guide. Thus, we synthesized a panel of PP derivatives based largely on the benzyl PP scaffold with various substituents designed to project into the gatekeeper pocket, as does the methyl group of 3MB-PP1 (Fig. 3). The PP analogs were synthesized in four steps using a previously described route6 and were analyzed for potency and selectivity towards Fyn-AS1 over Fyn-WT. We chose Fyn, a Src family tyrosine kinase, because it is a potent target of PP15; thus, an inhibitor of AS-kinases that does not target WT-Fyn would ensure that potency was not gained at the expense of selectivity. We plotted the negative logarithm of the IC50 value (pIC50) of each compound for Fyn-AS1 against the pIC50 for Fyn-WT to illustrate the potency and selectivity of the inhibitors (Fig. 4A). Strikingly, all of the new PP’s with pIC50 (Fyn-AS1) ≥ 8 contain the m-substituted benzyl functional group. Increasing the linkage between the aryl group and the heterocyclic core from one methylene to two (3) or removing the methylene group altogether (1) results in a dramatic decrease in activity. Remarkably, all compounds that are mono-substituted at the C3’ position of the benzyl group are quite potent and selective inhibitors of Fyn-AS1. C2’-subsituted benzyl PP inhibitors (6–8) showed decreasing potency with increasing size of the substituent, indicating an unfavorable steric clash that can be minimized with smaller substituents. Molecules with substituents at the C4’ position (12–14) display an even more dramatic decrease in potency towards both Fyn alleles. Taken together, these results confirm that the C3’-substitued benzyl PP structure is an ideal scaffold for AS-kinase inhibitors.
Figure 4.
(A) The pIC50 of each compound for Fyn-AS1 is plotted against the pIC50 for Fyn-WT to illustrate the potency and selectivity of PP inhibitors. The dotted grey line indicates threshold for compounds with suitable potency (pIC50 > 8) against Fyn-AS1. (B) IC50 values for select PP’s against Src-WT, Src-AS1 and Src-AS2. IC50 values were determined from a single dose-response measurement.
Notably, two C3’-benzyl PP’s, 17 and 18, stand out as being the most potent and selective inhibitors of Fyn-AS1; these molecules contain larger, more polarizeable groups relative to 3MB-PP1. We wished to verify that the enhanced efficacy of these compounds was a general phenomenon and not unique to Fyn, so we measured their ability to target Src-WT, Src-AS1 (SrcT338G), and Src-AS2 (SrcT338A). Once again, 17 and 18 have poor (IC50 > 10µM) activity towards the WT-kinase but are the most potent inhibitors for the kinase bearing a glycine gatekeeper. 17 and 18 are less potent towards Src-AS2, suggesting the iodo and thiomethyl substituents may ideally complement the space created by a glycine gatekeeper but are perhaps too large to fit in the smaller ATP pocket created by the alanine gatekeeper. Taken together, our results suggest that the C3’-substituted PP core is the optimal scaffold for AS-kinase inhibitors; however, the substituent that best complements a particular AS-kinase might vary.
Selectivity Profiles of PP Inhibitors
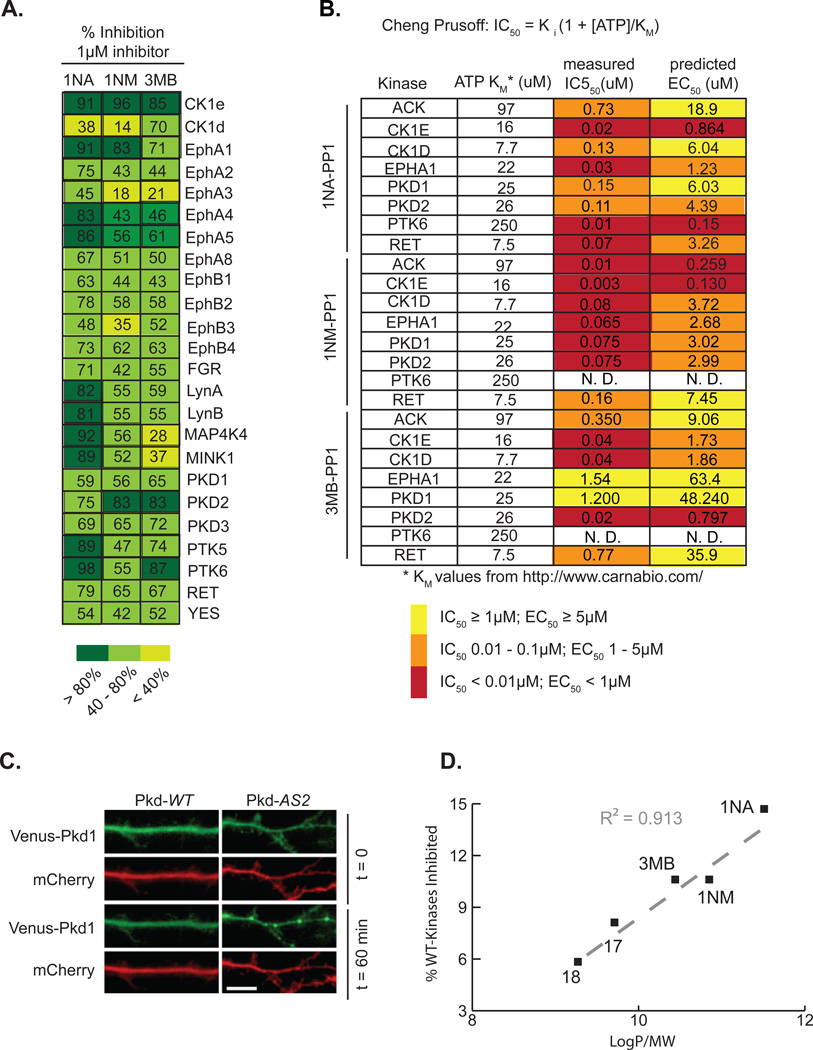
A crucial property that an effective AS-kinase inhibitor must possess is selectivity; it must potently inhibit the activity of an AS-kinase but not affect WT-kinases or other endogenous proteins. We and others have previously reported that 1NA-PP1 and 1NM-PP1 are very selective but imperfect AS-kinase inhibitors, as they weakly target some WT-kinases4, 20. To gain a more comprehensive understanding of the off-target effects of our AS-kinase inhibitors we tested the ability of 1 µM 1NA-PP1, 1NM-PP1, 3MB-PP1, 17 and 18 to inhibit the activity of several hundred WT-kinases in vitro and found that several are indeed sensitive to one or more PP inhibitors (Table S2). This is a very high standard for selectivity as AS-kinase inhibitors typically have IC50 values < 100 nM towards AS-kinases and the cellular concentration of the competitor ligand ATP is relatively high (1–10mM). Nonetheless, we wished to determine if off-target inhibition of WT-kinases by PP compounds is significant in the broad panel of AS-technology. We asked which of the WT-kinases targeted by our PP inhibitors are likely to be inhibited in cells at concentrations typically used in chemical genetic experiments. To this end we first measured IC50 values for 1NA-PP1, 1NM-PP1 and 3MB-PP1 against a representative subset of the PP-sensitive WT-kinases identified in our screen (Fig. 5A and Fig. 5B). We found that kinases inhibited greater than 80% in our screen generally have IC50 values less than 100 nM, while those inhibited between 40–80% have IC50 values ranging between 100 nM and 1.5 µM (Fig. 5B). The correlation between the fractional kinase activity determined in our screen and the measured IC50 values gives us confidence that data from the screen can be used to accurately predict the in vitro potency of our inhibitors against a large portion of the kinome.
Figure 5.
(A) Fraction of activity of wild-type PP-sensitive kinases inhibited by 1uM 1NA-PP1, 1NM-PP1 and 3MB-PP1. (B) IC50 values were measured for 1NA-PP1, 1NM-PP1 and 3MB-PP1 towards eight WT-kinases. The Cheng-Prusoff equation and KM values of each kinase for ATP were used to calculate predicted EC50 values for 1NA-PP1, 1NM-PP1, and 3MB-PP1towards eight WT-kinases. (C) Neurons were transfected with Venus-Pkd1-WT or Venus-Pkd1-AS2, treated with DMSO or 1uM 1NA-PP1, and analyzed for puncta formation. (D) The fraction of WT-kinases inhibited by 40% or more was plotted as a function CLogP value for 1NA-PP1, 1NM-PP1, 3MB-PP1, 17 and 18.
To address whether PP-sensitive WT-kinases are likely to be bona fide targets in cells we calculated the predicted EC50 values for each inhibitor/kinase pair using the Cheng Prusoff equation (Fig. 5B) which relates the measured IC50 value, the KM of the enzyme for ATP, and the cellular concentration of ATP which we assume to be ~5mM21, 22. PP-inhibitors are typically used in mammalian cells at concentrations between 500 nM and 5 µM (Table S1). We calculated that many of the identified PP-sensitive WT-kinases would only be targeted in cells at concentrations greater than 5 µM and we therefore predicted their cellular inhibition should be minimal under typical experimental conditions.
We sought to confirm that weak in vitro inhibition of WT-kinases by PP molecules is not a practical impediment to our technique by demonstrating the analog-sensitive approach can be applied to Protein Kinase D1 (Pkd1), one of the few kinases inhibited by all PP analogs tested. This unusual sensitivity to PP analogs may be due to a lysine residue at position 667 that occurs in all three Pkd family members in place of a conserved hydrophobic amino acid found in nearly all other kinases. K667 is located in the hinge region of the kinase domain and it is possible that this amino acid side chain allows greater flexibility in the inhibitor binding orientation although this remains to be tested. Pkd1 is a widely expressed Ser/Thr protein kinase involved in a number of physiological processes including the trafficking of membrane proteins in neurons23. We transfected HEK 293T cells with Pkd1-AS2 (M665A) or Pkd1-WT and found that a concentration of 1 µM 1NA-PP1 completely abrogates autophosphorylation by the analog-sensitive allele but not the wild type kinase, despite the fact that Pkd1-WT is inhibited by 1NA-PP1 in vitro with an IC50 of 150 nM (Fig. S1). We wished to examine the effects of 1NA-PP1 on a physiological function of Pkd1 and thus we examined the localization of the Pkd1-WT and Pkd1-AS2 in the presence of inhibitor. Kinase-dead alleles of Pkd1 have been shown to constitutively localize to the trans-Golgi network (TGN) and TGN-derived vesicles24. We co-expressed Venus-tagged Pkd1-WT and Pkd1-AS2 in neurons and assessed puncta formation in the presence of 1µM 1NA-PP1and found that Venus-Pkd1-WT neurons were unaffected, while Venus-Pkd1-AS2 recapitulated the kinase-dead phenotype, forming discrete puncta after one hour of inhibitor treatment (Fig. 5C).
The example of Pkd1demonstrates that kinases predicted to be targeted by PP inhibitors in cells with EC50 values greater than several micromolar are unlikely to be inhibited under the typical conditions of chemical genetic experiments. Additionally, 1NA-PP1 and 3MB-PP1 have been used to selectively target AS-alleles of Ret, EphB2, EphB3 and EphB4 in cells and mice despite weak inhibition of their WT-alleles in vitro25, 26. Thus, many of the WT-kinases identified as targets of 1NA-PP1, 1NM-PP1 or 3MB-PP1 in our screen and in the work of others are not likely to be bona fide targets in a cellular context.
As discussed above, weak in vitro inhibition of WT-kinases by PP inhibitors is unlikely to be relevant in cells due to competition with the high concentration of ATP. However, we postulate that such weak inhibition might serve as a proxy measurement for overall promiscuity of molecules towards unrelated proteins in a cell. Specifically, PP inhibitors that bind to fewer WT-kinases are less likely to bind to diverse proteins including metabolic enzymes capable of inactivating drug molecules in vivo. An important physical property that contributes to the promiscuity of small molecule inhibitors is lipophilicity; lipophilic molecules tend to bind non-specifically to hydrophobic regions of proteins such as ligand binding sites27. Furthermore, lipophilic molecules are more likely to have poor bioavailability, poor solubility and to form aggregates that may non-specifically inhibit unrelated enzymes28. We found that the promiscuity of our PP inhibitors was related to both the lipophilicity and the molecular weight of the molecule; larger molecules (greater molecular weight) are less promiscuous while more lipophilic molecules (greater LogP) tend to be more promiscuous. We plotted the fraction of WT-kinases whose activity is inhibited by 40% or more as a function of LogP/MW for 1NA-PP1, 1NM-PP1, 3MB-PP1, 17, and 18, and discovered this yields a linear relationship (Fig. 5D). This observation was surprising as we initially expected 1NM-PP1 to be the most selective inhibitor due to the greater steric clash between the napthyl moiety and the ATP-binding pocket observed in the overlay of the 1NM-PP1/Src-AS1 and the Hck structures. In fact 3MB-PP1 has a selectivity profile that is similar to 1NM-PP1, while 17 and 18 inhibit a much smaller fraction of WT-kinases. We interpret these results to reflect the fact that two mechanisms of selectivity are at work; increasing size of the functional group that projects towards the gatekeeper contributes to selectivity as we originally designed. However, minimizing lipophilicity also contributes by reducing non-specific affinity of the molecules for the hydrophobic ATP binding pocket of kinases. This discovery highlights the importance of a more comprehensive evaluation of drug molecules that incorporates consideration of physical properties such as lipophilicity in addition to the more obvious metrics of on-target potency (IC50, EC50, etc).
Summary
We have reported the mechanism of enhanced efficacy of the AS-kinase inhibitor 3MB-PP1 as well as the design and development of new, more potent and selective AS-kinase inhibitors. We employed X-ray crystallography to decipher the binding modes of 1NA-PP1, 1NM-PP1, and 3MB-PP1 and discovered that the methylene linker of benzyl PP analogs projects the phenyl group towards the gatekeeper, preventing binding to most WT-kinases while providing rotational flexibility that allows optimal orientation to complement the enlarged ATP pocket of AS-kinases. We used insights gained from these structures to design a small panel of benzyl-PP’s and discovered two molecules, 17 and 18 that are the most potent and selective AS-kinase inhibitors yet developed. The increased potency and reduced off-target effects of the new PPs combine to yield a major advantage when targeting an individual AS-kinase in cells or organisms that express hundreds of native kinases. Recent studies have identified 3MB-PP1 and 16 as capable of targeting at least 10 AS-kinase that are resistant to inhibition by any other known PP analog12, 16–18. Importantly, these kinases are from several different organisms and span four of the seven major kinase families (CMGC, STE, AGC, and TK) suggesting the enhanced efficacy of the inhibitors is not unique to the tyrosine kinases described in this report. We also examined the potential limitations presented by off-target inhibition of WT-kinases by PP inhibitors and discovered that many of the WT-kinases inhibited in vitro are unlikely to be targeted under cellular conditions. We have combined 1NA-PP1 and 1NM-PP1 with a panel of 3-substituted benzyl PP molecules to create a repertoire of potent and selective AS-kinase inhibitors that allows us to extend AS-kinase technology to the edges of the kinome and propels us towards the larger goal of understanding the logic of cellular signal transduction.
Online Methods
Chemical synthesis
Materials obtained commercially were reagent grade and were used without further purification. 1H NMR and 13C NMR spectra were recorded on a Varian 400 spectrometer at 400 and 100 MHz, respectively. High-resolution electron impact mass spectra were recorded on a MicoMass VG70E spectrometer at the University of California-San Francisco center for Mass Spectrometry. Reactions were monitored by thin layer chromatography (TLC), using Merck silica gel 60 F254 glass plates (0.25 mm thick). Flash chromatography was conducted with Merck silica gel 60 (230–400 mesh).
Protein crystallization
Src-AS1 co-crystal structure with 1NA-PP1
The structure of 1NA-PP1 in complex with Src-AS1 (residues 83–533, including the SH3, SH2, kinase, and tail regions) was determined essentially as previously described for an N6-Benzyl-ADP complex29. Briefly, the Src-AS1 protein, homogeneously phosphorylated on Tyr 527, was concentrated to 10–15 mg/ml in storage buffer (20 mM HEPES [pH 7.6], 100 mM NaCl, and 10 mM DTT), and combined with a slight molar excess of 1NA-PP1. The complex was crystallized in hanging drops at 22°C by combining 2 μl of the above solution with 1 μl of a well solution (50 mM PIPES [pH 6.5], 12% PEG4000, 10 mM DTT). Crystals were briefly transferred to a buffer containing crystallization solution plus 20% glycerol and frozen at −165°C. Diffraction data were recorded with a Princeton 2k CCD detector on the A1 beamline at CHESS. All diffraction data were integrated and scaled with the programs DENZO and SCALEPACK. The structure was determined by molecular replacement using PDB entry 2SRC as a search model and refined using Refmac. Data collection and refinement statistics are available in Table S3.
Src-AS1 co-crystal structures with 1NM-PP1 and 3MB-PP1
The kinase domain of c-Src-AS1 was expressed and purified as previously described15. Src-drug complexes were formed by mixing 10mM DMSO stocks of the drug compounds into 100 µM Src, 100 mM NaCl, 50 mM Tris [8.0], 2 mM DTT, and 5 % glycerol at room temperature. Crystals were obtained in hanging drops of a 1:1 mixture (protein-drug and mother liquor) by vapor diffusion atop a mother liquor solution of 100 mM MES [6.5], 50 mM NaAc, 12% glycerol, and 6% PEG 4000. Just prior to data collection, crystals were flash frozen in mother liquor containing 25% glycerol. Data for the 1NM-PP1 and 3MB-PP1 structures were collected at the Advanced Light Source, beam lines 8.2.2 and 8.2.1, respectively. Data were processed using HKL2000. Molecular replacement solutions for the 1NM-PP1 and 3MB-PP1 structures were solved using Phaser in the CCP4 program suite with chain A of c-Src (PDB ID: 1YOJ) as the search model. Structures were optimized through iterative rounds of manual fitting in Coot and using maximum likelihood refinement protocols in Refmac. Coordinate and topology files for the ligands 1NM and 3MB were derived in PRODRG (Schuettelkopf and van Aalten, 2004). Data collection and refinement statistics are available in Table S3.
Supplementary Material
Acknowledgement
We thank C. Schartner and B. Aizenstein of Invitrogen Corporation for screening of inhibitors against library of kinases. This study was funded by NIH (EB001987). M.S. Lopez is supported by a National Science Foundation Graduate Research Fellowship award. We also thank D.O. Morgan for careful reading of the manuscript and for many helpful comments.
Footnotes
Associated Content
Supporting Information. Compiled list of each reported analog-sensitive kinase allele and inhibitor, inhibitor profiling data, x-ray crystallography data collection and refinement statistics, western blot demonstrating effect of 1NA-PP1 on Pkd1 autophosphorylation, chemical characterization and kinase assay conditions. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The atomic coordinates and structure factors of Src-AS1 in complex with inhibitors have been deposited in the Protein Data Bank with accession codes 4K11 (Src-AS1/1NA-PP1), 4LGH (Src-AS1/1NM-PP1), and 4LGG (Src-AS1/3MB-PP1).
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T. The protein kinase complement of the human genome. Science. 2002 doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Lampson MA, Kapoor TM. Unraveling cell division mechanisms with small-molecule inhibitors. Nat. Chem. Biol. 2006;2:19–27. doi: 10.1038/nchembio757. [DOI] [PubMed] [Google Scholar]
- 3.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shokat KM, Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Bishop A, Witucki L, Kraybill B, Shimizu E, Tsien J, Ubersax J, Blethrow J, Morgan DO, Shokat KM. Structural basis for selective inhibition of Src family kinases by PP1. Chem. Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 6.Bishop AC, Kung C, Shah K, Witucki L. Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. Journal of the …. 1999 [Google Scholar]
- 7.Zhang C, Kenski DM, Paulson JL, Bonshtien A, Sessa G, Cross JV, Templeton DJ, Shokat KM. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 8.Garske AL, Peters U, Cortesi AT, Perez JL, Shokat KM. Chemical genetic strategy for targeting protein kinases based on covalent complementarity. Proc. Natl. Acad. SciU.SA. 2011;108:15046–15052. doi: 10.1073/pnas.1111239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat. Struct. Mol. Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, Pollok BA, Connelly PA. Discovery of a Novel, Potent, and Src Family-selective Tyrosine Kinase Inhibitor: STUDY OF Lck- AND FynT-DEPENDENT T CELL ACTIVATION. J. Biol. Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 11.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 12.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Inhibitor hijacking of Akt activation. Nat. Chem. Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuzumi T, Ducker GS, Zhang C, Aizenstein B, Hoffman R, Shokat KM. Synthesis and evaluation of indazole based analog sensitive Akt inhibitors. Mol Biosyst. 2010;6:1389–1402. doi: 10.1039/c003917a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Shokat KM. Enhanced selectivity for inhibition of analogsensitive protein kinases through scaffold optimization. Tetrahedron. 2007;63:5832–5838. [Google Scholar]
- 15.Blair JA, Rauh D, Kung C, Yun CH, Fan QW. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nature chemical …. 2007 doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 16.Cipak L, Zhang C, Kovacikova I, Rumpf C, Miadokova E, Shokat KM, Gregan J. Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle. 2011;10:3527–3532. doi: 10.4161/cc.10.20.17792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Au-Yeung BB, Levin SE, Zhang C, Hsu L-Y, Cheng DA, Killeen N, Shokat KM, Weiss A. A genetically selective inhibitor demonstrates a function for the kinase Zap70 in regulatory T cells independent of its catalytic activity. Nat. Immunol. 2010;11:1085–1092. doi: 10.1038/ni.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drubin DG, Weiss EL, Bishop AC, Shokat KM. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- 20.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H, Klevernic I, Arthur JSC, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Cheng YC, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973 doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Ruiloba L, Cabrera-Poch N, Rodriguez-Martinez M, Lopez-Menendez C, Jean-Mairet RM, Higuero AM, Iglesias T. Protein Kinase D Intracellular Localization and Activity Control Kinase D-interacting Substrate of 220- kDa Traffic through a Postsynaptic Density-95/Discs Large/Zonula Occludens-1-binding Motif. J. Biol. Chem. 2006;281:18888–18900. doi: 10.1074/jbc.M603044200. [DOI] [PubMed] [Google Scholar]
- 24.Liljedahl M, Maeda Y, Colanzi A, Ayala I, Van Lint J. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans- Golgi network. Cell. 2001 doi: 10.1016/s0092-8674(01)00228-8. [DOI] [PubMed] [Google Scholar]
- 25.Savitt J, Singh D, Zhang C, Chen L-C, Folmer J, Shokat KM, Wright WW. The in vivo response of stem and other undifferentiated spermatogonia to the reversible inhibition of glial cell line-derived neurotrophic factor signaling in the adult. Stem Cells. 2012;30:732–740. doi: 10.1002/stem.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soskis MJ, Ho H-YH, Bloodgood BL, Robichaux MA, Malik AN, Ataman B, Rubin AA, Zieg J, Zhang C, Shokat KM, Sharma N, Cowan CW, Greenberg ME. A chemical genetic approach reveals distinct EphB signaling mechanisms during brain development. Nat. Neurosci. 2012;15:1645–1654. doi: 10.1038/nn.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarcsay A, Keserű GM. Contributions of molecular properties to drug promiscuity. J. Med. Chem. 2013;56:1789–1795. doi: 10.1021/jm301514n. [DOI] [PubMed] [Google Scholar]
- 28.McGovern SL, Helfand BT, Feng B, Shoichet BK. A Specific Mechanism of Nonspecific Inhibition. J. Med. Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 29.Witucki LA, Huang X, Shah K, Liu Y, Kyin S, Eck MJ, Shokat KM. Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme. Chem. Biol. 2002;9:25–33. doi: 10.1016/s1074-5521(02)00091-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.