Biomarkers play a crucial and growing role in diagnosing and managing cardiovascular disease. Currently, there are several well established biomarkers employing Elisa detection of serum peptides such as Troponin I (TnI), Troponin T (TnT), Natriuretic Peptide B (BNP), C-Reactive Protein (CRP), Creatine Kinase (CK-MB), Myeloperoxidase (MPO), Lipoprotein(a), and Myoglobin (Mgb). While RNA signature-based biomarkers have become standard of care in cancer, notably breast cancer1, this approach has yet to fully emerge in cardiovascular medicine, although studies in patient tissues2-5_ENREF_2, preclinical models, and recent clinical studies6, 7 support their utility.
To date clinical transcriptomic cardiac studies have employed gene/mRNA arrays2-5, 8-10, exonic arrays11, 12_ENREF_6, microRNA arrays13-17_ENREF_8_ENREF_6, and it is expected that data on sequencing of long non-coding RNAs (lncRNAs) will emerge and grow rapidly in the coming few years. Whereas, RNAs in these studies were extracted either directly from heart tissue or peripheral blood, few studies have compared simultaneously global transcript profiles from heart tissue with peripheral blood, to determine whether there is a sufficient correlation between heart and blood transcriptomics to support the use of RNA blood biomarkers for diseases of the myocardium.
In the current issue of JACC Heart Failure, Gerling et al18 address this important issue by comparing the global mRNA expression profiles from heart tissue to peripheral blood mononuclear cells (PBMCs) in an aldosterone rat model of heart failure. Their findings in gene expression and molecular pathway analysis supported a correlation between the blood and heart transcriptomics. The mRNA data was also supported by similar correlation in the increase of cytosolic calcium and zinc cations and the elevation of 8-isoprotane in cardiac myocytes and PBMCs. These findings add an important data point to the discussion of whether RNA blood biomarkers can serve as an appropriate surrogate for cardiovascular disease.
Several studies have shown that expression profiles obtained from myocardium provide highly accurate biomarkers of disease etiology and prognosis. Almost a decade ago, Kittleson and colleagues performed microarray analysis on tissue obtained from explanted hearts and revealed that ischemic cardiomyopathy (ICM) could be distinguished from non-ischemic cardiomyopathy (NICM) and that that the hearts of patients with NICM who do not undergo LVAD implantation resemble non-failing (NF) hearts more than those of the sicker NICM patients who require an LVAD before cardiac transplantation2, 3_ENREF_2. Heidecker and co-workers identified a unique myocardial gene signature that distinguished patients with myocarditis with 100% sensitivity and specificity among a broad range of secondary cardiomyopathies including stress-induced cardiomyopathy, sarcoidosis, peripartum cardiomyopathy, arrhythmogenic right ventricular dysplasia, giant-cell myocarditis, and systemic lupus erythematosus5. Other investigators have shown the value of transcriptomic biomarkers for a variety of other cardiovascular disorders, including atherosclerotic coronary artery disease10 and asymptomatic left ventricular dysfunction (ALVD)9. The question remains, however, can blood based transcriptomic biomarkers accurately substitute for those obtained directly by the affected tissue. Due to its amorphous nature, blood is rarely referred to as tissue. In reality, blood is tissue that is in direct physical contact with all organs (except the brain). Unsurprisingly, in a thoughtful study by Liew et al19 who queried the absolute transcript levels of global mRNAs from 248 human blood samples on 248 microarray chips, and compared the results with publicly available microarray data from different human tissues, blood was shown to express tissue-specific transcripts. For example, the β-MHC transcript which is heart specific, was found to be expressed in the blood (Figure 1). Similarly, Adachi et al20 reported based on global miRNA profiling of various human tissue and miRNA qPCR of cardiac patient sera, that miR-499 is heart specific and is up-regulated in the plasma of Myocardial Infarct (MI) patients, respectively.
Figure 1. Heart specific transcripts expressed in blood.
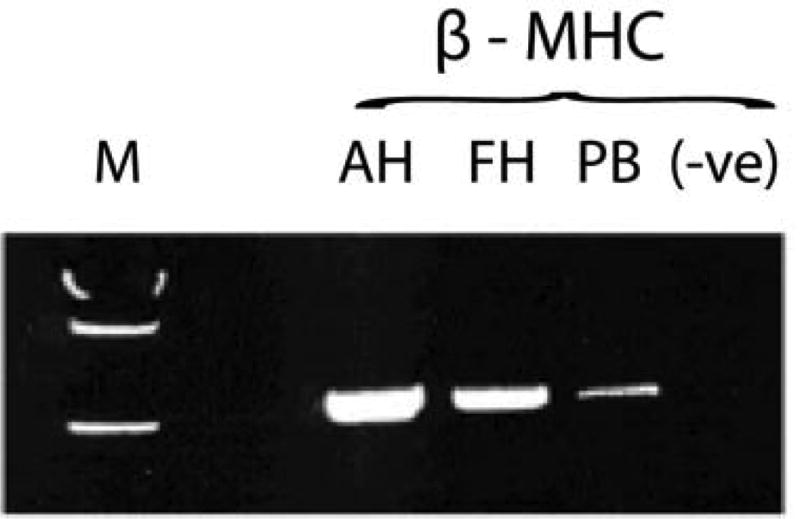
β-MHC transcripts were detected in human peripheral blood (PB). Positive controls used were human adult and fetal heart tissue (AH and FH respectively). No template/blank (-ve) was used as a negative control. M =molecular weight marker. Adapted from Liew et al (2006).
In order for RNA transcripts to become clinically useful blood biomarkers in the future (Figure 2), there are several important studies that need to be performed:
Figure 2. A look into the future.
RNA blood transcripts will be translated into scores that can predict the kind and degree of cardiovascular disease.
1. Comparison between heart and blood transcripts in cardiac patients
Such studies will be ground breaking in screening for and identifying the transcripts that are potential biomarkers. It is possible that the transcripts to be identified could be previously unrelated to cardiac disease. In a study comparing the transcriptomics of brains and blood in Parkinson's disease patients, we identified an RNA splicing molecule among others to be dysregulated in both the brain and blood21. Taken into consideration the blood brain barrier, we anticipate that the comparison in heart disease to be much more direct and informative. In addition, these studies should not be limited to gene/mRNA expression, but rather include global miRNA expression, exonic expression/alternative splicing, and sequencing of lncRNAs.
2. The transcriptomic data from cardiac patients need to be correlated with clinical/functional measures
As in the informative study by Smih et al9 where blood transcriptomic microarray data from ALVD patients was correlated with echocardiography data to predict clinical outcome, it will be important to use clinical data as the basis for hierarchical clustering of the transcriptomic data.
Otherwise, there will continue to be a gap between transcriptomic data and its clinical translation. Additionally, transcriptomic data can be correlated with the currently acceptable standard clinical assays used for cardiac diseases. For, example, Cheng et al22 found that plasma miR-1 transcripts were correlated with plasma CK-BM levels in patients with MI.
3. Mathematical models need to be created to fit the blood transcriptomic data and the clinical data
Mathematical models need to be created integrating clinical data and the newly identified blood transcripts (which hopefully at this level would be screened down to hundreds rather than tens of thousands of genes or isoforms, and tens rather than hundreds of microRNAs and lncRNAs). Based on the combinatorial values of the absolute transcript levels, different models would be built to fit the expression data onto the clinical data. The final desired outcome is a simple readout/score of blood transcript data that, based on the built models, can predict the kind and level of cardiac disease as in Figure 2. These mathematical models will require validation in clinical populations, in a manner similar to the transcriptomic biomarkers developed using myocardial tissue2-5, 17.
In conclusion, while further studies are warranted to make blood RNA transcripts as clinical biomarkers for cardiac diseases, we agree with Liew et al19 that “blood cells can act as sentinels of disease”, and therefore we could capitalize on this property of blood for the diagnosis/prognosis of cardiac diseases. The current study by Gerling et al18_ENREF_15_ENREF_15_ENREF_15 provides additional supportive data for this concept. While direct sampling of myocardium might offer advantages for biomarker application, using peripheral blood has obvious benefit in terms of broader application and generalizability of transcriptomic biomarkers as they emerge in cardiovascular medicine.
Acknowledgments
Dr. Hare is funded by the National Institutes of Health (NIH) grants (RO1 HL094849, RO1 HL084275, RO1 HL107110, RO1 HL110737 and UM1HL113460), and Dr. Shehadeh is funded by the NIH (K01 AG040468), State of Florida (3KN05), and Florida Heart Research Institute.
Footnotes
Disclosure – Dr. Hare discloses equity in Heart Genomics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- 1.Abba MC, Lacunza E, Butti M, Aldaz CM. Breast cancer biomarker discovery in the functional genomic age: A systematic review of 42 gene expression signatures. Biomark Insights. 2010;5:103–118. doi: 10.4137/BMI.S5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kittleson MM, Ye SQ, Irizarry RA, Minhas KM, Edness G, Conte JV, Parmigiani G, Miller LW, Chen Y, Hall JL, Garcia JG, Hare JM. Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation. 2004;110:3444–3451. doi: 10.1161/01.CIR.0000148178.19465.11. [DOI] [PubMed] [Google Scholar]
- 3.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, Conte JV, Tomaselli G, Garcia JG, Hare JM. Gene expression analysis of ischemic and nonischemic cardiomyopathy: Shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 4.Heidecker B, Kasper EK, Wittstein IS, Champion HC, Breton E, Russell SD, Kittleson MM, Baughman KL, Hare JM. Transcriptomic biomarkers for individual risk assessment in new-onset heart failure. Circulation. 2008;118:238–246. doi: 10.1161/CIRCULATIONAHA.107.756544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidecker B, Kittleson MM, Kasper EK, Wittstein IS, Champion HC, Russell SD, Hruban RH, Rodriguez ER, Baughman KL, Hare JM. Transcriptomic biomarkers for the accurate diagnosis of myocarditis. Circulation. 2011;123:1174–1184. doi: 10.1161/CIRCULATIONAHA.110.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas GS, Voros S, McPherson JA, Lansky AJ, Winn ME, Bateman TM, Elashoff MR, Lieu HD, Johnson AM, Daniels SE, Ladapo JA, Phelps CE, Douglas PS, Rosenberg S. A blood-based gene expression test for obstructive coronary artery disease tested in symptomatic nondiabetic patients referred for myocardial perfusion imaging the compass study. Circ Cardiovasc Genet. 2013;6:154–162. doi: 10.1161/CIRCGENETICS.112.964015. [DOI] [PubMed] [Google Scholar]
- 7.Cadeiras M, von Bayern M, Sinha A, Shahzad K, Latif F, Lim WK, Grenett H, Tabak E, Klingler T, Califano A, Deng MC. Drawing networks of rejection - a systems biological approach to the identification of candidate genes in heart transplantation. J Cell Mol Med. 2011;15:949–956. doi: 10.1111/j.1582-4934.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappuzzello C, Napolitano M, Arcelli D, Melillo G, Melchionna R, Di Vito L, Carlini D, Silvestri L, Brugaletta S, Liuzzo G, Crea F, Capogrossi MC. Gene expression profiles in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2009;38:233–240. doi: 10.1152/physiolgenomics.90364.2008. [DOI] [PubMed] [Google Scholar]
- 9.Smih F, Desmoulin F, Berry M, Turkieh A, Harmancey R, Iacovoni J, Trouillet C, Delmas C, Pathak A, Lairez O, Koukoui F, Massabuau P, Ferrieres J, Galinier M, Rouet P. Blood signature of pre-heart failure: A microarrays study. PLoS ONE. 2011;6:e20414. doi: 10.1371/journal.pone.0020414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinnaeve PR, Donahue MP, Grass P, Seo D, Vonderscher J, Chibout SD, Kraus WE, Sketch M, Jr, Nelson C, Ginsburg GS, Goldschmidt-Clermont PJ, Granger CB. Gene expression patterns in peripheral blood correlate with the extent of coronary artery disease. PLoS ONE. 2009;4:e7037. doi: 10.1371/journal.pone.0007037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong SW, Hu YW, Ho JW, Ikeda S, Polster S, John R, Hall JL, Bisping E, Pieske B, dos Remedios CG, Pu WT. Heart failure-associated changes in rna splicing of sarcomere genes. Circ Cardiovasc Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricci M, Xu Y, Hammond HL, Willoughby DA, Nathanson L, Rodriguez MM, Vatta M, Lipshultz SE, Lincoln J. Myocardial alternative rna splicing and gene expression profiling in early stage hypoplastic left heart syndrome. PLoS ONE. 2012;7:e29784. doi: 10.1371/journal.pone.0029784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC. Circulating micrornas are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. Micrornas in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 15.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, Melillo G, Rigolini R, Costa E, Crea F, Capogrossi MC, Napolitano M, Martelli F. Microrna signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010;42:420–426. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 16.Meder B, Keller A, Vogel B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J, Leidinger P, Meese E, Katus HA, Rottbauer W. Microrna signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol. 2011;106:13–23. doi: 10.1007/s00395-010-0123-2. [DOI] [PubMed] [Google Scholar]
- 17.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial micrornas and messenger rna in human cardiomyopathy and reversal of the microrna signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerling Iea. Gene expression profiles of peripheral blood mononuclear cells reveal transcriptional signatures as novel biomarkers for cardiac remodeling. JACC Heart Failure. 2013 doi: 10.1016/j.jchf.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, Nonogi H, Iwai N. Plasma microrna 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183–1185. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]
- 21.Shehadeh LA, Yu K, Wang L, Guevara A, Singer C, Vance J, Papapetropoulos S. Srrm2, a potential blood biomarker revealing high alternative splicing in parkinson's disease. PLoS ONE. 2010;5:e9104. doi: 10.1371/journal.pone.0009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, Dong X, Qin S, Zhang C. A translational study of circulating cell-free microrna-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]


