Abstract
Natural killer (NK) cells, large granular lymphocytes comprising a key cellular subset of innate immunity, were originally named for their capacity to elicit potent cytotoxicity against tumor cells independent of prior sensitization or gene rearrangement. This process is facilitated through the expression of activating and inhibitory receptors that provide for NK cell “education” and a subsequent ability to survey, recognize and lyse infected or transformed cells, especially those lacking or possessing mutated major histocompatibility complex (MHC) class I expression. Since these original observations were made, how NK cells recognize candidate target cells continues to be the topic of ongoing investigation. It is now appreciated that NK cells express a diverse repertoire of activating and inhibitory receptors of which killer immunoglobulin-like receptors (KIR) appear to play a critical role in mediating self-tolerance as well as facilitating cytotoxicity against infected or transformed cells. Additionally, in the presence of an activating signal, the absence or mismatch of MHC class I molecules on such targets (which serve as inhibitory KIR ligands) promotes NK cell-mediated lysis. An increasing understanding of the complexities of KIR biology has provided recent opportunities to leverage the NK cell versus tumor effect as a novel avenue of therapeutic immunotherapy for cancer. The present review seeks to summarize the current understanding of KIR expression and function and highlight ongoing efforts to translate these discoveries into novel NK cell-mediated immunotherapies for cancer.
Keywords: Natural Killer cells, KIR, tumor immunology
Introduction
Natural killer (NK) cells are traditionally defined as CD56+CD3− large granular lymphocytes that can exhibit cytotoxicity against malignant and infected cells and secrete cytokines to shape and direct an immune response (1). NK cells were first described as such because of their ability to kill a target cell “naturally,” that is, without the requirement of prior sensitization, co-stimulatory signal, gene rearrangement, or presentation of an antigen via major histocompatibility complex (MHC) proteins (2). In fact, many tumor cells down-regulate expression of MHC class I molecules to facilitate escape from cytolytic T cell attack (3); however, this process increases their susceptibility to NK cell-mediated lysis, a concept referred to as “missing self” recognition (4). Building on these initial discoveries, it is now understood that NK cells display a complex array of activating and inhibitory receptors that interact with constitutively expressed as well as inducible ligands on candidate target cells (5). The interactions between members of the killer immunoglobulin-like receptor (KIR) family expressed by NK cells with MHC class I molecules on target cells play a key role in modulating NK cell immune surveillance and NK cell-mediated cytotoxicity (6).
KIR nomenclature is based on structure and function
KIR names are based on structure in regards to the number of extracellular immunoglobulin-like domains (D) and the length (Long or Short) of the intracytoplasmic tail (or if the KIR is a Pseudogene). A digit is added to indicate the number of the gene encoding a KIR with the ascribed structure, and in one case (KIR2DL5A and KIR2DL5B), a final letter (A or B) indicates close sequence similarity, perhaps due to a recent gene duplication event (7). A total of 17 KIR genes (including two pseudogenes) have been discovered which reside in the leukocyte receptor complex, a ~150 kb segment on chromosome 19q13.14 (8).
From this nomenclature system, information on cognate ligands and function may be deduced for individual KIRs. For example, KIRs with two extracellular domains interact with human leukocyte antigen (HLA)-C allotypes whereas KIR with three extracelluar domains interacts with HLA-A and –B allotypes (6). All seven KIRs with a short intracytoplasmic tail have activating function, and generally most of the eight KIRs with a long intracytoplasmic tail have inhibitory function. KIR2DL4 is the notable exception to this pattern as its ligation leads to an activation signal promoting cytokine production (9).
Inhibitory KIRs contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs) which are phosphorylated by Src family kinases upon KIR interaction with their cognate class I MHC molecules (10). This leads to recruitment of SHP-1 and SHP-2 phosphatases and subsequent dominant suppression of activation signals (10). On the other hand, activating KIRs do not possess ITIM sequences, and rather, associate with DAP12 or FcεRI-γ to signal through immunoreceptor tyrosine-based activating motifs (ITAMs) via Syk and ZAP-70 kinases upon ligation with their cognate class I MHC molecules (11). It appears that every NK cell capable of cytotoxicity must express at least one inhibitory receptor recognizing a class I MHC molecule, and generally, activating KIRs bind with less avidity than inhibitory KIRs (12,13). In the presence of an activating signal, the absence or mismatch of MHC class I molecules on target cells (which serve as ligands for inhibitory KIRs) promotes NK cell-mediated lysis.
KIRs are expressed in a diverse repertoire
KIR expression results from a complicated, multifactorial process. First, KIR genes are inherited in diverse allelic combinations. Haplotype A, represented in about 50% of humans, encodes mostly inhibitory receptors: KIR2DL1-3 and KIR3DL1-3. Haplotype A also includes the activating receptors KIR2DS4 and KIR2DL4; however, KIR2DS4 lacks function in a majority of instances and many KIR2DL4 allelic gene products are not displayed on the cell surface (14). Haplotype B includes KIR2DL genes encoding for inhibitory receptors that recognize both HLA-C1 and HLA–C2 ligands, but typically includes activating receptors encoded by KIR2DS and KIR3DS genes (14). The inherited copy number of a particular KIR gene may also influence its ultimate expression on NK cells.
Second, a number of polymorphisms have been reported in each KIR gene that affect ligand affinity and NK cell surface expression that in turn may affect NK cell function. Although inherited independently on chromosome 6, the presence of cognate MHC class I KIR ligands is associated with an increase in the frequency of NK cells expressing matching KIR and a decrease in the incidence of NK cells expressing KIR that do not recognize these ligands (15).
Third, only a fraction of all KIRs are expressed on each individual NK cell which is, in part, the result of gene silencing by DNA methylation (16). As a consequence of all these factors, a complex repertoire of circulating NK cells is capable of recognizing even minute alterations in MHC class I expression (17).
KIRs recognize specific class I MHC molecules
Each individual KIR has specific HLA class I allotypes as ligands. Polymorphisms at positions 77 and 80 of the HLA-C heavy chain α1-domain yield distinct allotypes recognized by inhibitory KIR2DL1 and KIR2DL2/3. KIR2DL1 binds HLA-Cw2, -Cw4, -Cw5, and -Cw6 (referred to as “C2”) and KIR2DL2/3 binds HLA-Cw1, -Cw3, -Cw7, and -Cw8 (referred to as “C1”; ref 17). Therefore, as a group, KIR2DL1/2/3 can inhibit NK cell lysis of targets expressing any HLA-C allotype. Inhibitory KIR3DL1 binds HLA-Bw4 (present in 40–50% of known HLA-B allotypes) and inhibitory KIR3DL2 binds HLA-A3 and HLA-A11 (18). Interestingly, recognition of HLA-A3 and HLA-A11 appears to be peptide-specific (19), and from an evolutionary standpoint, HLA-C appears to have originated more recently than HLA-A and HLA-B to serve in a more specialized role for KIR binding (20).
As mentioned above, KIR and MHC class I KIR ligands are inherited independently, and this process may impact selective evolutionary pressures as well as NK cell-mediated susceptibility towards infection and disease. For example, the incidence of reproductive disorders such as pre-eclampsia and spontaneous abortion occur at a higher rate in a mother who is homozygous for group A KIR haplotypes with a fetus bearing HLA-C2 (21). The evolution of the group B KIR haplotype appears to have occurred uniquely in humans as compared to other simian primates to facilitate reproductive success. In a similar manner, inheritance of specific KIR/HLA combinations (termed “compound genotypes”) has been associated with susceptibility or resistance to viral infection, autoimmunity, and cancer (13). KIR/HLA combinations that result in a lower activation threshold have been associated with a pro-inflammatory state leading to susceptibility to autoimmune disease, but less inhibition may also be protective against viral infections (13).
KIRs mediate NK cell education
The potential exists for the generation of "autoreactive" NK cells (which are not capable of self-recognition) for two reasons. First, KIR and KIR ligands are inherited independently and second, the expression of activating and inhibitory receptors on individual NK cells appears to be a random process. Indeed, circulating NK cells that lack inhibitory KIR for self-recognition are observed, and one might predict these NK cells could mediate autoimmunity (22). Instead of being clonally deleted, such cells are rendered hyporesponsive through a "licensing" process during development (6,23). Although a complete understanding of NK cell education is lacking, NK cell responsiveness is acquired in a finely regulated manner through KIR - KIR ligand interactions during development. For example, NK cells expressing two self-recognizing inhibitory KIRs will receive stronger inhibitory signals in surveying a self-target; however, such cells will also elicit more potent cytotoxicity when encountering a "missing self" target as compared with NK cells expressing one or no self-recognizing inhibitory KIR (24). This process is dynamic as NK cells from an MHC class I-deficient environment can be licensed to kill upon transfer to an MHC class I-sufficient environment (25).
KIR-ligand relationships impart cancer susceptibility
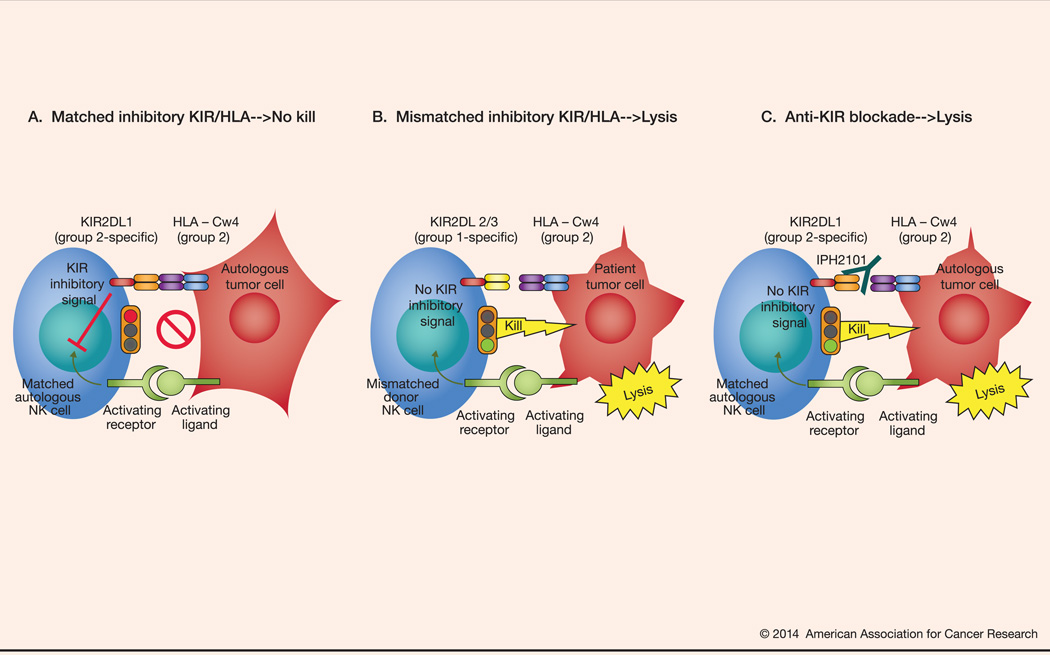
A number of studies have shown that KIR/HLA relationships may be associated with the incidence and course of both solid tumors and hematologic malignancies (13). In these observations, KIR/HLA relationships that are particularly inhibitory in nature are overrepresented in patients with melanoma, breast cancer, acute and chronic leukemias and Hodgkin lymphoma; this can be explained in part as matched autologous inhibitory KIR/HLA interactions prevent lysis of cancerous target cells (Figure 1A). In a complementary manner, the expression of activating KIRs in some circumstances is associated with better outcomes in various malignancies (26,27).
Figure 1.
Illustration of the concept of haploidentical allogeneic stem cell transplantation. NK cell cytotoxicity requires the presence of an activating signal and the absence or mismatch of MHC class I molecules on the target cell. (A). Autologous NK cell vs tumor cell, baseline with no lysis: On the left, a patient-derived, autologous NK cell with matched inhibitory KIR to the MHC class I self-molecule on the autologous tumor cell on the right; the NK cell receives an activating signal through the activating receptor-ligand interaction with a tumor cell; however, the NK cell is incapable of initiating a cytotoxic response due to the inhibitory signal resulting from an inhibitory KIR interaction with its cognate MHC class I self-molecule. (B) Mismatched inhibitory KIR/HLA→tumor cell lysis: On the left, a donor-derived NK cell with mismatched inhibitory KIR to the MHC class I ligand on the patient tumor cell on the right. The. mismatched NK cell lyses the target tumor cell in response to the activating signal because no inhibitory signal is delivered as a result of KIR/HLA ligand mismatch. (C). Anti-KIR blockade→tumor cell lysis: On the left, a patient-derived, autologous NK cell with matched inhibitory KIR to the MHC class I ligand on the autologous tumor cell. However, the presence of IPH2101, a monoclonal blocking antibody directed against the inhibitory KIR interrupts its binding to the cognate MHC class I ligand on the target cell resulting in a loss of the inhibitory signal thereby allowing for NK cell recognition and lysis of the tumor cell.
KIR-ligand modulation for cancer therapy
The therapeutic manipulation of KIR-ligand relationships was first demonstrated in the setting of T-cell depleted, haploidentical hematopoietic stem cell transplantation for hematologic malignancies. Patients with acute leukemia first received an extensive preparative, conditioning regimen followed by an infusion of a high dose of CD34+ hematopoietic stem cells without graft versus host disease prophylaxis (28,29). Outcomes were studied as a function of NK cell alloreactivity defined as a donor having: MHC class I ligand expression that was not found in the recipient (i.e., ligand-ligand mismatch), KIR genes for the missing self-recognition MHC class I ligands on recipient target cells, i.e., KIR that did not recognize the recipient’s MHC class I molecules, and the presence of alloreactive NK cell clones against patient leukemia cells (Figure 1B). Patients in complete remission who received an NK cell alloreactive graft experienced a durable event-free survival rate of 67% (compared to 18% in patients receiving NK cell non-alloreactive grafts, p = 0.02). Even in patients with chemotherapy-refractory leukemia, the event-free survival rate was 34% with NK cell alloreactive grafts compared to 6% for the NK cell non-alloreactive grafts (p = 0.04). In a similar manner, KIR-ligand mismatch may also improve outcomes in multiple myeloma (MM) (30). Such results have been interpreted to support the “missing self” (or “ligand-ligand mismatch”) model.
More recently, a second model based on receptor-ligand mismatch (or “missing ligand” model) has been proposed to account for NK cell alloreactivity specifically for the recipient lacking a MHC class I cognate ligand to a KIR expressed on donor NK cells. While not necessarily a mutually exclusive explanation to “missing self”, one unique implication of the “missing ligand” model is that HLA mismatch may not be necessary for NK cell alloreactivity. This notion is based on observations in both the allogeneic and autologous transplant settings where the early post-transplant microenvironment which is conducive to a loss of self-tolerance and where normally hyporesponsive NK cells expressing inhibitory KIRs to non-self ligands unexpectedly exhibit functional competence (31–33).
Extending on these results, the direct infusion of alloreactive, haploidentical KIR-ligand mismatched NK cells has been studied in a number of malignancies. In one trial, a 50% complete response rate was observed in heavily pre-treated patients with advanced MM who received infusions of haploidentical, T-cell depleted KIR-ligand mismatched NK cells shown to display potent in vitro cytotoxicity against patient tumor cells (34). Encouraging results with such an approach have also been observed in adult and pediatric leukemia (35,36).
In a broader context, while the inhibitory KIR-ligand relationship (in the absence of other inhibitory receptor-ligand interaction, such as NKG2A/HLA-E) seems to be foundational in establishing NK cell alloreactive potential, it is important to note that the balance of activating and inhibitory signals received ultimately appears to direct cytotoxicity. NK cells also express a diverse repertoire of activating receptors that are capable of recognizing cognate ligands expressed on stressed, infected, or transformed cells. Thus, the mere absence of inhibition is insufficient to elicit NK cell-mediated cytotoxicity. Rather, the absence of inhibition in the presence of an activating signal “tips the balance” towards killing (Figure 1B). Indeed, a number of studies have elucidated a role for activating KIR to modulate alloreactivity. For instance, KIR2DS1 appears to contribute to improved outcomes in an HLA-C dependent manner in patients undergoing allogeneic transplantation for leukemia (37). The presence of KIR2DS1 may, in fact, override inhibitory signaling via NKG2A/HLA-E interaction perhaps defining a particularly alloreactive NK cell subset (38). In addition, donors with group B KIR haplotypes which are associated with a relatively greater presence of activating KIR have been shown to improve survival for patients with leukemia receiving allografts (39).
Despite the promising results observed in several trials of haploidentical, T-cell depleted allogeneic transplantation, this approach remains largely investigational in part due to its complexity as well as the potential for infectious morbidity and mortality related to the profound immunosuppression. However, extending on the concept of KIR-ligand mismatch, therapeutic monoclonal antibodies directed against common inhibitory KIRs are in development which may provide an opportunity to extrapolate to the autologous setting the results obtained from the haploidentical, allogeneic transplant studies (Figure 1C). The first agent in this class is IPH2101 (formerly, 1-7F9), a human IgG4 monoclonal antibody (mAb) directed against KIR2DL-1, -2, and -3 (39). In binding to these KIRs, IPH2101 was shown to prevent inhibitory signaling and enhance NK cell cytokine production and cytotoxicity against tumor cell targets bearing cognate HLA-C ligands. Blocking inhibitory KIR with IPH2101 also facilitated in vitro antibody-dependent cellular cytotoxicity (ADCC), and enhanced patient-derived NK cell cytotoxicity preferentially against autologous leukemia and MM tumor cell targets but not against normal autologous cells (39,40). Results from three in vivo systems also showed the addition of IPH2101 improved NK cell-mediated antitumor effects (39,40). Single-agent, dose-escalation phase I clinical trials of IPH2101 in acute myeloid leukemia (AML) and MM have been published (41,42). Both studies demonstrated the approach to be safe and well-tolerated, achieving the biologic endpoint of full KIR occupancy over the entire 28-day dosing interval without an identified dose-limiting toxicity. Results are currently pending from several trials of IPH2101 in additional settings (e.g., smoldering MM, as maintenance in MM and AML, and in combination with lenalidomide for MM). Lirilumab, a second generation anti-KIR mAb (BMS-986015, formerly IPH2102), is being evaluated currently as a single agent in AML, and in combination with ipilimumab (anti-CTLA-4 mAb) or with nivolumab (BMS-936558, anti-PD-1 mAb) in patients with advanced solid tumors.
Summary and future directions
Historically, most attempts at immune-mediated therapy for cancer may be regarded as strategies to stimulate or augment immune function (e.g., cytokines, tumor-directed mAbs, vaccination strategies, etc.) In a complementary manner however, more recently, so called “checkpoint-blocking” approaches to cancer immunotherapy (e.g., CTLA-4 and PD-1) that target T cell-associated inhibitory receptor/ligand axes and related signaling have shown efficacy and generated increasing interest in, and appreciation for, the concept of balance in immunity (43). These studies, in combination with the data reviewed above from experience modulating inhibitory KIR-MHC class I ligand relationships, suggest that effective immunotherapy for cancer may have as much or more to do with releasing the immune system from inhibition as with stimulating or promoting immune activation directly. To wit, combinations of an immunostimulatory agent with an anti-inhibitory therapy are already in clinical testing (44).
As our understanding of the complexities of KIR biology expands, a number of new observations provide additional opportunities to translate KIR ligand biology into novel approaches to cancer therapy. First, a specific polymorphism in KIR2DL1 has been identified where an arginine at position 245 (KIR2DL1-R245) confers stronger inhibitory function than the KIR2DL1 allele bearing cysteine at the same position (KIR2DL1-C245) (45). More recently, this polymorphism was associated with important clinical relevance, as patients undergoing allogeneic transplantation for pediatric leukemia who received grafts from KIR2DL1-R245 donors experienced superior survival outcomes compared to those receiving grafts from KIR2DL1-C245 donors (p = 0.0004) (46). The best survival (p = 0.00003) and lowest risk of leukemia relapse (p = 0.0005) were observed in patients who received a KIR2DL1-R245 graft coupled with HLA-C receptor-ligand mismatch (46). These findings suggest that the intensity of inhibitory signaling involved in NK cell education correlates directly with the magnitude of the NK cell cytotoxic response to a target determined to be non-self. Second, it appears that specific KIR-MHC class I ligand relationships may contribute to particular aspects of NK cell education. A number of studies have shown that the KIR3DL1/HLA-Bw4 interaction licenses NK cell function in regards to CD16-mediated ADCC (47). Third, KIRs may have additional functional roles beyond determining self and non-self through ligand interaction. Interestingly, KIR3DL2 appears to bind microbial CpG oligodeoxynucleotides that in turn internalize to promote encounter with TLR9 in endosomes leading to cytokine production and enhanced cytotoxicity (48). This process occurs independent of the initiation of inhibitory signaling typically observed as a consequent to HLA-A3 or HLA-A11 ligand binding and may facilitate the early immune response to infectious pathogens. Such a finding raises the question as to whether or not KIRs may be capable of detecting other stimuli, such as in the tumor microenvironment.
In summary, increasing knowledge in KIR-related basic biology as well as a growing experience in clinical translation of KIR ligand modulation have set the stage for significant advances in NK cell-mediated cancer therapy. The near future may hold opportunity to personalize highly effective, anti-cancer therapy based on knowledge of tumor target ligand expression, important microenvironmental influences, and refined principles of NK cell alloreactivity mediated by KIRs.
Acknowledgments
Supported by: National Cancer Institute CA095426 (MAC, DMB); CA163205, CA68458 (MAC) American Cancer Society RSG-13-249-01 (DMB)
Disclosures: DMB has received research funding from Innate Pharma and has served on a scientific advisory board to Bristol Myers Squibb. MAC serves on the Supervisory Board of Innate Pharma and has served on a scientific advisory board to Bristol Myers Squibb.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors, II: characterization of effector cells. Int J Cancer. 1975;16:230–239. doi: 10.1002/ijc.2910160205. [DOI] [PubMed] [Google Scholar]
- 3.Restifo NP, Kawakami Y, Marincola F, Shamamian P, Taggarse A, Esquivel F, et al. Molecular mechanisms used by tumors to escape immune recognition: immunogenetherapy and the cell biology of major histocompatibility complex class I. J Immunother Emphasis Tumor Immunol. 1993;14:182–190. doi: 10.1097/00002371-199310000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karre K, Ljunggren HC, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 5.Moretta L, Locatelli F, Pende D, Sivori S, Falco M, Bottino C, et al. Human NK receptors: from the molecules to the therapy of high risk leukemias. FEBS Lett. 2011;585:1563–1567. doi: 10.1016/j.febslet.2011.04.061. [DOI] [PubMed] [Google Scholar]
- 6.Thielens A, Viver E, Romagne F. NK cell MHC class I specific receptors (KIR): from biology to clinical intervention. Curr Opin Immunol. 2012;24:239–245. doi: 10.1016/j.coi.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Lozano N, Gardiner CM, Parham P, Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;53:314–319. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- 8.Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP. Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.14. Immunogenetics. 2000;51:659–669. doi: 10.1007/s002510000183. [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2 regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFNgamma production. J Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 10.Yusa S, Campbell KS. Src homology region 2-containing protein tyrosine phosphates-2 (SHP-2) can play a direct role in the inhibitory function of killer cell Ig-like receptors in human NK cells. J Immunol. 2003;170:4539–4547. doi: 10.4049/jimmunol.170.9.4539. [DOI] [PubMed] [Google Scholar]
- 11.McVicar DW, Taylor LS, Gosselin P, Willette-Brown J, Mikhael AI, Geahlen RL, et al. DAP12-mediated signal transduction in natural killer cells: a dominant role for the Syk protein-tyrosine kinase. J Biol Chem. 1998;273:32934–32942. doi: 10.1074/jbc.273.49.32934. [DOI] [PubMed] [Google Scholar]
- 12.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenback K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 13.Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer Ig-like receptors (KIR) Cancer Biol Ther. 2009;8:13–22. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chain HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 18.Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132:315–325. doi: 10.1111/j.1365-2567.2010.03398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLa-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 20.Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185:4238–4251. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of material KIR and fetal HLA-C genes influence the risk of pre-eclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, French AR, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 436:709–713. doi: 10.1038/nature03847. 205. [DOI] [PubMed] [Google Scholar]
- 24.Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol. 2009;182:4572–4580. doi: 10.4049/jimmunol.0803900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joncker NT, Shifrin N, Delebecque F, Raulet DH. Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J Exp Med. 2010;207:2065–2072. doi: 10.1084/jem.20100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besson C, Roetynck S, Williams F, Orsi L, Amiel C, Lependeven C, et al. Association of killer immunoglobulin-like receptor genes with Hodgkin’s lymphoma in a familial study. PLoS One. 2007;2:e406. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18:2002–2007. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 28.Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–440. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 30.Kroger N, Shaw B, Iacobelli S, Zabelina T, Peggs K, Shimoni A, et al. Comparison between antithymocyte globulin and alemtuzumab and the possible impact of KIR-ligand mismatch after dose-reduced conditioning and unrelated stem cell transplantation in patients with multiple myeloma. Brit J Haematol. 2005;129:631–643. doi: 10.1111/j.1365-2141.2005.05513.x. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Venstrom JM, Liu XR, Pring J, Hasan RS, O’Reilly RJ, Hsu KC. Breaking tolerance to self, circulating natural killer cells expressing inhibitory KIR for non-self HLA exhibit effector function after T cell-depleted allogeneic hematopoietic cell transplantation. Blood. 2009;113:3875–3884. doi: 10.1182/blood-2008-09-177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung W, Handgretinger R, Iyengar R, Turner V, Holladay MS, Hale GA. Inhibitory KIR-HLA mismatch in autologous haematopoietic stem cell transplantation for solid tumour and lymphoma. Brit J Cancer. 2007;97:539–552. doi: 10.1038/sj.bjc.6603913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ksu KC, Keever-Taylor CA, Wilton A, Pinto C, Heller G, Arkun K, et al. Improved outcome in HLA-identical sibling hematopoietic stem cell transplantation for acute myeloid leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, Tricot G, Szmania S, Rosen N, Garg TK, Malaviarachchi PA, et al. Infusion of haplo-identical killer immunoglobulin-like receptor ligand mismatched NK cells for relapsed myeloma in the setting of autologous stem cell transplantation. Brit J Haematol. 2008;143:641–653. doi: 10.1111/j.1365-2141.2008.07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105:3051–3057. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 36.Rubnitz JE, Inaba H, Ribeiro RC, Pounds S, Rooney B, Bell T, et al. NKAML: a pilot study to determine the safety and feasibility of haploidentical natural killer cell transplantation in childhood acute myeloid leukemia. J Clin Oncol. 2010;20:955–959. doi: 10.1200/JCO.2009.24.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venstrom JM, Pittari G, Gooley TA, Chewning JH, Haagenson M, Gallagher MM, et al. HLA-C dependent prevention of leukemia relapse by donor activating KIR2DS1. New Eng J Med. 2012;367:805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pende D, Marcenaro S, Falco M, Martini S, Bernardo ME, Montagna D, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009;113:3119–3129. doi: 10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 38.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113:726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romagne F, Andre P, Spee P, Zahn S, Anfossi N, Gauthier L, et al. Preclinical characterization of 1–7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediating killing of tumor cells. Blood. 2009;114:2667–2677. doi: 10.1182/blood-2009-02-206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson DM, Bakan CE, Zhang S, Collins SM, Liang J, Srivastava S, et al. IPH2101, a novel anti-inhibitory KIR antibody, and lenalidomide combine to enhance the natural killer cell versus multiple myeloma effect. Blood. 2011;118:6387–6391. doi: 10.1182/blood-2011-06-360255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson DM, Hofmeister CC, Padmanabham S, Suvannasankha A, Jagannath S, Abonour R, et al. A phase 1 trial of the anti-KIR antibody IPH2101 in patients with relapsed/refractory multiple myeloma. Blood. 2012;120:4324–4333. doi: 10.1182/blood-2012-06-438028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vey N, Bourhis JH, Boissel N, Bordessoule D, Prebet T, Charbonnier A, et al. A phase 1 trial of the anti-inhibitory KIR mAb IPH2101 for AML in complete remission. Blood. 2012;120:4317–4323. doi: 10.1182/blood-2012-06-437558. [DOI] [PubMed] [Google Scholar]
- 43.Callahan MK, Wolchok JD. At the bedside: CTLA-4 and PD-1 blocking antibodies in cancer immunotherapy. J Leukoc Biol. 2013;94:41–53. doi: 10.1189/jlb.1212631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benson DM, Cohen A, Munshi NC, Jagannath S, Spitzer G, Hofmeister CC, et al. A phase I trial of the anti-inhibitory KIR antibody, IPH2101, and lenalidomide in multiple myeloma: interim results. Blood. 2012;120:4058a. [Google Scholar]
- 45.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Gupta ND, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bari R, Rujkijyanont P, Sullivan E, Kang G, Turner V, Gan K, Leung W. Effect of donor KIR2DL1 allelic polymorphism on the outcome of pediatric allogeneic hematopoietic stem cell transplantation. J Clin Oncol. 2013;31:3782–3790. doi: 10.1200/JCO.2012.47.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsons MS, Zipperlen K, Gallant M, Grant M. Killer cell immunoglobulin-like receptor 3DL1 licenses CD16-mediated effector functions of natural killer cells. J Leukoc Biol. 2010;88:905–912. doi: 10.1189/jlb.1009687. [DOI] [PubMed] [Google Scholar]
- 48.Sivori S, Falco M, Carlomango S, Romeo E, Soldani C, Bensussan A, et al. A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood. 2010;116:1637–1647. doi: 10.1182/blood-2009-12-256586. [DOI] [PubMed] [Google Scholar]