Abstract
Mobile elements compose more than half of the human genome, but until recently their large-scale detection was time-consuming and challenging. With the development of new high-throughput sequencing technologies, the complete spectrum of mobile element variation in humans can now be identified and analyzed. Thousands of new mobile element insertions have been discovered, yielding new insights into mobile element biology, evolution, and genomic variation. We review several high-throughput methods, with an emphasis on techniques that specifically target mobile element insertions in humans, and we highlight recent applications of these methods in evolutionary studies and in the analysis of somatic alterations in human cancers.
Keywords: retrotransposon, mobile DNA element, high-throughput sequencing, polymorphism, somatic insertion, cancer
Mobile DNA elements in the human genome
Mobile DNA elements, or transposable DNA elements, are discrete DNA fragments that can be moved or copied to other regions of a genome. They are present in virtually all organisms and compose up to two-thirds of the human genome [1, 2]. Several types of mobile elements, including Alu, L1, and SVA, have been actively transposing during human evolution history and remain active today (Box 1). These elements have had profound influence on genomic architecture, and the active elements continue to change the human genomic landscape, sometimes causing human diseases in the process (See recent reviews [3–5] for more details). In addition to their genomic impact, some Mobile Element Insertions (MEIs) have occurred so recently that they are present in some individuals but not others. These polymorphic MEIs (pMEIs) are ideal markers for studying human evolutionary history and population relationships because of their known ancestral state, lack of homoplasy, and other properties [6, 7]. In the past two decades, pMEIs have been used in a large number of population genetic and phylogenetic studies in humans as well as in other species [8–11].
Box1: Mobile DNA elements in the human genome.
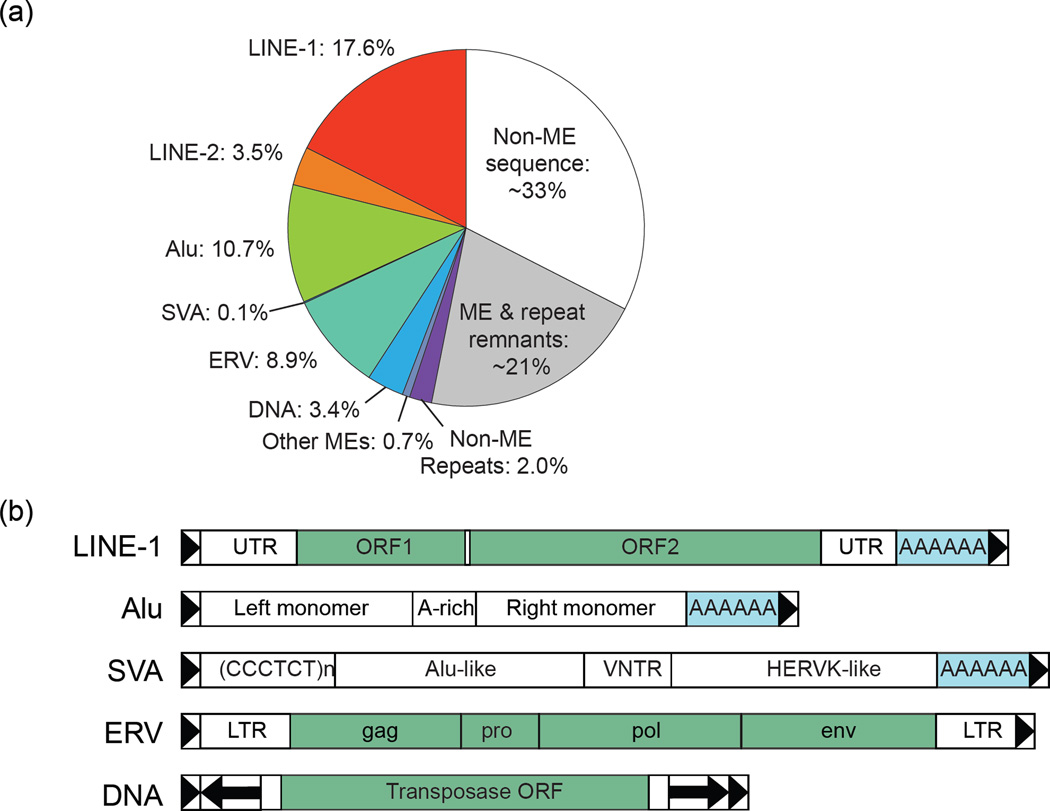
Almost half of the human genome (47.8%) is composed of readily recognizable mobile element insertions and their fragments (Figure Ia). The most prevalent are the type I retrotransposons, including non-LTR retrotransposon LINE (Long INterspersed Elements) families (e.g., LINE-1 and LINE-2); the SINE (Short INterspersed Elements) Alu family; the LTR retrotransposon ERV (endogenous retroviruses); and SVA elements, a small but still active family. The type II "cut and paste" DNA transposons of the Tc1-Mariner, hAT, and piggyBac families are also present in the human genome. An additional ~20% of the genome is made up of short remnants of ancient mobile elements and other repeats [2].
The structures of major mobile elements in the human genome are illustrated in Figure Ib. The LINE-1 (L1) element is the most prevalent autonomous non-LTR retrotransposon (about half a million copies). Full-length autonomous L1 elements are ~6 kb in length and contain two open reading frames (ORFs) that encode the proteins necessary to catalyze L1 retrotransposition. The 5' UTR contains an RNA Pol II promoter. Upon insertion, L1 elements generate a variable-length poly-A tail at the 3' end and short target site duplications (TSDs) at the insertion site. The vast majority of L1s are nonautonomous due to accumulated mutations and 5' truncations that often occur upon retrotransposition. Approximately 100 “hot” L1 elements are active today [32, 62]. Present at more than one million copies, Alu is the most prevalent nonautonomous non-LTR retrotransposon. Canonical Alu elements are ~300 bp in length, composed of two halves (right and left monomers joined by an A-rich spacer) derived from a 7SL RNA. Alu elements do not encode any proteins but do carry an internal RNA Pol III promoter in the left monomer. Alu elements replicate by hijacking the L1 retrotransposition machinery and thus generate similar TSDs and a 3' poly-A tail upon insertion [63]. Most Alu insertions are ancient and inactive, but some subfamilies (notably AluYa and AluYb) are still active. SVA (SINE-VNTR-Alu) elements (~5000 copies) are nonautonomous and rely on L1 proteins for their retrotransposition [64]. The canonical SVA is ~2 kb in length and is composed of a hexameric repeat region, a stretch of Alu-derived sequence, a variable number of tandem repeats (VNTR) of 35–50 bp, a segment derived from the LTR of a human endogenous retrovirus (HERV), and a polyadenylation signal followed by a poly-A tail. Autonomous ERV copies are ~8 kb long and encode the proteins gag, protease (pro), pol, and env, which are required for reverse transcription of their RNAs and integration into the genome [65, 66]. DNA elements typically encode a transposase protein that recognizes the element by its inverted terminal repeats (longer boxed arrows in the diagram), then excises and reinserts it elsewhere in the genome. DNA elements in the human genome are ancient remnants, nonautonomous, and inactive.
Since the release of the human genome reference sequence, many efforts have been made to identify pMEIs in the human genome. This, however, has been a labor intensive task. For example, the initial release of the human Retrotransposon Insertion Polymorphisms database (dbRIPs, http://dbrip.brocku.ca) contained ~2100 pMEIs [12], representing all pMEIs identified in more than 50 studies over the past several decades. Hundreds of additional pMEI loci were identified when additional individual genome sequences were published [13–16]. All these efforts have been dwarfed by recent advances in high-throughput technologies, which have dramatically changed the landscape of whole-genome, population-level MEI studies. Several high-throughput methods have been developed specifically for human pMEI detection, identifying thousands of novel pMEIs that are not present in the human reference genome. In this review, we first describe these methods and then summarize some recent major findings using whole-genome MEI data. We conclude by outlining new questions that can be addressed in the near future using these technologies.
Detecting mobile elements at the whole-genome level
There are two primary types of methods for profiling pMEIs at the whole-genome level: (i) targeted methods in which DNA fragments related to a MEI are experimentally enriched before sequencing/genotyping and (ii) post-sequencing bioinformatic methods in which pMEIs are identified using whole-genome sequencing data. Although the focus of this review is on the detection of MEIs in humans, similar high-throughput MEI identification methods have recently been applied to non-human species (Box 2).
Box 2: High-throughput pMEI identification in other organisms.
The basic targeted MEI sequencing approach is completely flexible: one need only replace the primers specific to one mobile element family in one species to assay insertions of another family in another species. Indeed, the Transposon Display (TD) method on which the sequencing approaches are based was developed to detect dTph1 polymorphisms in Petunia hybrida [17] and has since been used on many MEI families and in many species (most recently in mosquitos, maize, and rice [67–70]). Moreover, two non-human MEI families – mPing elements in rice [71] and Minos elements in wheat [72] – have been investigated by HTS-based, targeted MEI sequencing methods similar to those used in humans. In human studies, analyses of the sequence data have been aided by the high-quality human reference genome, but the reference genome is not required. Just as MEIs were identified through TD by the positions of their corresponding products on a polyacrylamide gel, they are now more uniquely identified by the genomic sequences at their insertion sites. By either means, the presence or absence of pMEIs in an individual can be determined. This allows pMEIs to be used as genetic markers and for the study of population dynamics even in the absence of a reference genome, although analyses of the genomic contexts of MEIs would be hampered. The successful study of Minos elements in wheat [72] is a case in point, as the wheat genome remains largely unassembled due to its large size, hexaploid composition, and high repeat content [73].
The opportunity to apply targeted MEI sequencing to non-human MEIs is ripe. High-throughput library preparation methods have become more general, robust, and flexible, enabling rapid development of targeting strategies that address the peculiarities of different MEI types. The use of mechanical DNA fragmentation, as in ME-Scan [25], eliminates concerns about the distributions of random hexamers and restriction enzyme sites. The availability, capacity and diversity of HTS platforms have increased as sequencing costs continue to drop. MEI families with lower per-genome copy numbers can be assayed on medium-throughput platforms, or assayed in many more individuals for similar cost by multiplexing samples. Longer read lengths will allow better identification and mapping of MEIs as well as insights into the internal element sequences themselves. The lack of information about family-specific sequence characteristics of non-human MEIs may introduce uncertainty into the MEI-specific primer design process. However, the extremely high capacity of current sequencing platforms will allow researchers to either characterize the MEI families with low-pass genome sequencing or to simply overcome the uncertainty by obtaining very high coverage using permissive or mixed primer designs. The same strategy will allow fast and reliable identification of MEIs in a family across closely related species for use as phylogenetic or species-specific markers [74].
Targeted MEI sequencing methods
One of the most commonly used MEI selection methods is termed Transposon Display (TD). This gel-based technique was developed initially to visualize pMEIs in Petunia species [17] and has since been adapted for studying Alus [18, 19], L1s [20–22], and HERVs [23] in the human genome. Several recent techniques have adapted the TD method for high-throughput sequencing (HTS) (Table 1) [24–26], and the basic principle of the method is illustrated in Figure 1. The key step for MEI enrichment is a PCR step using one primer complementary to the MEI sequence and another primer complementing the linker sequence (Figure 1d). Sometimes referred as a “hemi-specific PCR”, only DNA fragments that contain the targeted MEIs will be amplified. In the “Transposon-Seq” method [24], two rounds of nested hemi-specific PCR are used to increase the specificity and to allow the sequence read to cover the junction between the MEI and the flanking genomic region (MEI-genome junction). In Mobile Element Scanning (ME-Scan) [25], the MEI-specific primer for the first-round hemi-specific PCR is biotinylated, which allows the amplification products to be easily isolated. In a slight variation of the original protocol, a single-primer extension step rather than PCR is used to amplify L1 insertions [26]. Next, anchored degenerate primers are used in separate PCR reactions to further reduce genomic complexity. Unlike methods based on TD, the RC-Seq (Retrotransposon capture sequencing) method enriches MEI-containing DNA fragments by microarray-hybridization [27]. Using probes that are complementary to full-length retrotransposons (Alu, L1, and SVA), genomic DNA fragments containing MEIs are captured by probes and subsequently sequenced using HTS.
Table 1.
Targeted methods for high-throughput MEI identification
| Name | Fragmentation | Enrichment Method |
Data Generation Method |
Index for pooling |
Advantages | Disadvantages | Ref |
|---|---|---|---|---|---|---|---|
| Transposon-Seq | Enzymatic Digestion | Hemi-specific PCR | HTS | 2nd round PCR | High throughput, high MEI family specificity, individuals can be pooled after indexing to reduce cost | Only MEI close to restriction digestion sites can be analyzed, different type of MEIs require separate PCR experiments with different MEI-specific primers. | [24] |
| ME-Scan | Mechanical Shearing | Hemi-specific PCR with biotin enrichment | HTS | Ligation | High throughput, high MEI family specificity, individuals can be pooled after indexing to reduce cost | Different type of MEIs require separate PCR experiments with different MEI-specific primers. | [25] |
| L1 Display | - | primer-extension and hemi-specific PCR with radom hexamers | HTS | 2nd round PCR | High throughput, high L1-specificity, individuals can be pooled after indexing to reduce cost | Certain hexamer only amplify a subset of genome. | [26] |
| RC-Seq | Mechanical Shearing | Hybridization | HTS | Ligation | High throughput, individuals can be pooled after indexing to reduce cost | Different type of MEIs require separate PCR experiments with different MEI-specific primers. | [27] |
| TIP-Chip | Enzymatic Digestion | Hemi-specific PCR | Tiling-array | N/A | High specificity, different MEI families can be examined on the same chip, a subset of the genome can be examined | Relatively low throughput, only one individual can be analyzed per chip, only MEI close to restriction digestion sites can be analyzed | [30] |
| AIP | Enzymatic Digestion | primer-extension with biotin enrichment and hemi-specific PCR | Tiling-array | N/A | High specificity, a subset of the genome can be examined | Rrelatively low throughput, only one individual can be analyzed per chip, only MEI close to restriction digestion sites can be analyzed | [31] |
| fosmid-based paired-end sequencing | Fosmid library | allele-specific oligonucleotide hybridization, Southern blot | Fosmid sequencing/ATLAS | N/A | Specifically identify full-length L1 insertions | Relatively low throughput. Can only apply to full-length L1s or MEIs larger than 6kb in size. | [32] |
Figure 1. Identification of MEIs using targeted high-throughput sequencing.
(a) Genomic DNA (gray) containing a MEI (green). Targeted site duplications (TSDs) are shown as boxed arrows. (b) DNA is fragmented by mechanical shearing or enzymatic digestion, generating multiple fragments that span the junction of the MEI and the flanking unique genomic sequence. (c) If necessary, adapter oligonucleotides (blue) are ligated onto them. (d) A primer that anneals to the mobile element family of interest and an adapter primer are used to carry out hemi-specific MEI PCR. Here the targeting primer is biotinylated (star) to facilitate the specific capture and enrichment of fragments carrying the targeted MEIs. This is the method used in ME-Scan [25]. Other methods use hybridization to enrich insertion-carrying fragments, whereas some use carefully designed PCR strategies that amplify only the fragments of interest. (e) High-throughput sequencing is used to generate reads from unique genomic sequences immediately adjacent to the insertions in the enriched library or spanning their junctions; both types of reads are depicted.
Overall, HTS-based methods share several advantages. Hundreds of millions of sequencing reads can be generated in one experiment, which allows for indexing and pooling of multiple individuals to reduce cost. The hemi-specific PCR-based methods are usually highly specific, and pMEIs from a specific MEI family or subfamily can be detected with high sensitivity. A disadvantage of HTS methods is that MEI-specific primers must be designed for each MEI family. It may also be difficult to specifically target MEI families that differ from related families or remnant copies of older families by only a few diagnostic nucleotide sites. Separate amplification experiments are needed for different MEI families, and in general pMEI identification can only be performed at the whole-genome level (with the exception of RC-Seq).
In addition to HTS-based approaches, microarray-based methods related to TIP-chip (Transposon Insertion Profiling by microarray) [28, 29] have been developed and applied in human MEI detection [30, 31]. DNA fragments containing MEIs are first enriched by hemi-specific PCR and then fluorescently labeled and hybridized to a genome-tiling array to identify genomic regions adjacent to MEIs [30, 31]. Compared with HTS-based methods, microarray-based methods have relatively low throughput, and only one individual can be examined on one microarray. Nevertheless, if a certain subset of the genome (e.g., a single chromosome) is the focus of a project, a customized array can be used to provide coverage for the specific genomic regions more easily than with HTS methods.
Another method for identifying full-length L1 insertions [32] is an adaptation of the fosmid-based paired-end sequencing method that was designed to detect fine-scale (kb level) structural variation (SV) [33]. (A fosmid is a cloning vector that usually carries ~40 kb of genomic DNA insertion.) Candidate loci are identified by comparing the size of the fosmid inserts to the reference genome. Inserts that are ~6 kb larger than the reference potentially harbor a non-reference full-length L1 insertion. L1 5’UTR-specific oligonucleotide hybridization and Southern blotting are then used to identify fosmid inserts containing full-length L1 insertions. Although this method has high sensitivity for detecting full-length L1s, the size limitation of the fosmid vectors prevents it from detecting pMEIs that are smaller than 6kb in length, including Alu, SVA, and ~99.9% of L1s.
Post-sequencing Bioinformatics Methods
With the advent of affordable whole-genome sequencing, large-scale detection of pMEIs can be accomplished computationally using whole-genome data. Many algorithms designed to identify SVs from whole-genome data can identify pMEIs without any knowledge of MEI characteristics. Read-pair and split-read mapping (Box 3) can both be used in this context. However, standard SV detection methods are not ideal for MEI detection because they do not explicitly take information about MEIs into account. To address this issue, methods have been developed to specifically identify MEIs from whole-genome data. For example, VariationHunter [34] discovers MEIs using a maximum-parsimony algorithm. After initial SV detection, it uses the consensus sequence of MEI families to determine if a MEI is the mostly likely cause of a putative SV locus.
Box3: Read-pair (RP) and split-read (SR) methods for structural variation (SV) detection.
The RP algorithm uses paired-end reads that are sequenced from the two ends of a DNA fragment. The size of the DNA fragment is usually larger than the length of the two sequencing reads; therefore, the middle of the DNA fragment is usually not sequenced. After the reads are mapped to a reference genome, the reads that have a non-standard mapping pattern (e.g., too far apart, not in the correct direction, etc.) are used to infer SV in the genome. In the SR algorithm, the goal is to identify sequence reads that contain the exact breakpoints of SVs. The reads are generally split into two sections and mapped separately to a reference genome. In contrast to the RP algorithm, the SR algorithm does not require paired-end reads. However, long sequencing reads are needed for accurately mapping a section of the read to the reference. Therefore, the appropriate use of these algorithms partly depends on the format of the HTS data. For current HTS systems, reads from the Roche’s 454 GS FLX+ and Pacific Biosciences’ PacBio RS are more suitable for the SR algorithm, whereas reads from Illumina’s HiSeq 2000 and Life technology’s SOLiD 5500 are more suitable for the RP algorithm.
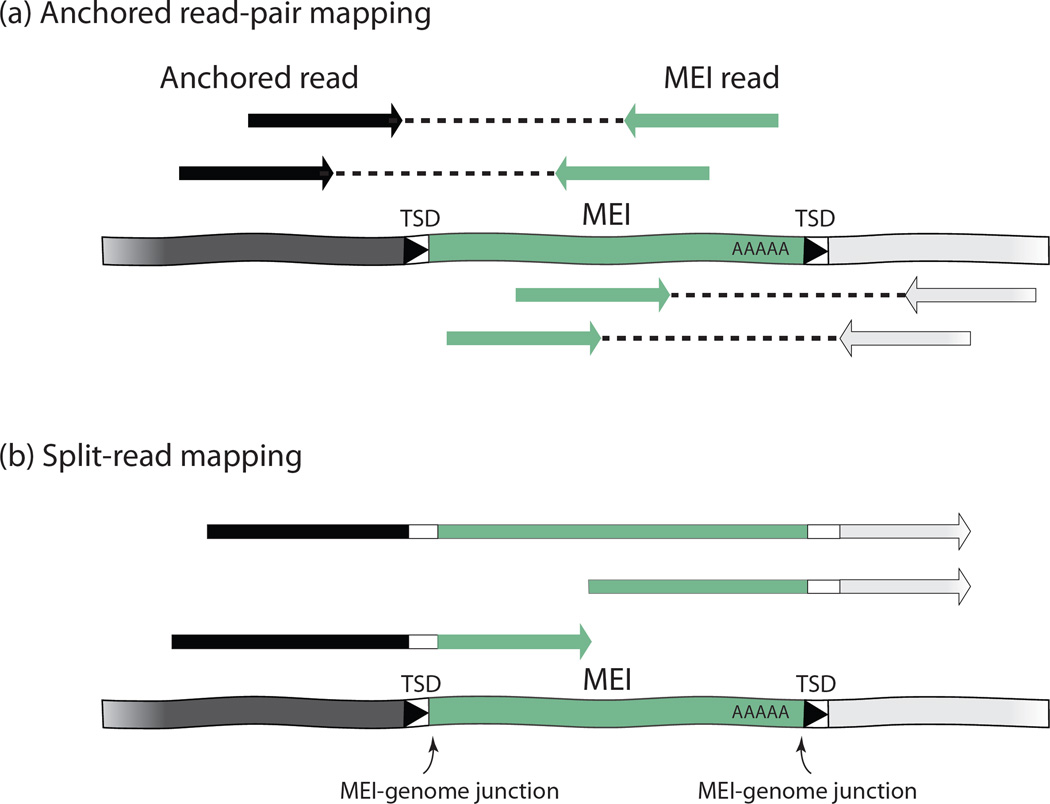
One type of commonly used algorithm, termed “anchored read-pair mapping”, infers MEIs using paired-end reads (Figure 2a). First, the paired-end reads are mapped to the reference genome. Read pairs in which one read maps to a unique genomic position (i.e., “anchored”) and the other maps to an MEI consensus sequence indicate the presence of an MEI. Because the exact MEI-genome junction is usually not retrieved in this process, multiple read pairs surrounding one region are required to support identification of an MEI. Implementations of this algorithm include RetroSeq [35] and PAIR (polymorphic Alu insertion recognition) [36]. Another algorithm, termed “split-read mapping,” is designed to determine the exact MEI position using reads that overlap the MEI-genome junction (Figure 2b). If one part of a read can be mapped to a unique position of the genome and the rest of the read is mapped to the MEI sequence, the position of an MEI can be determined. This type of algorithm generally requires longer read lengths than the anchored read-pair methods because it relies on unambiguous mapping of a portion of a read. With long sequence reads, both ends of the MEI-genome junction, as well as the whole MEI sequence, can be recovered from a single read. This greatly increases the accuracy of MEI inference. In the 1000 Genomes project, for example, the split-read method identified more than 4000 non-reference MEIs in 24 individuals using relatively long reads from the Roche 454 sequencer [37].
Figure 2. Bioinformatic MEI identification methods.
A) Anchored read-pair mapping. Targeted MEIs are shown as green boxes. The flanking genomic sequence is shown as black bar (5’) or grey bar (3’), respectively. The paired-end reads are shown in arrows with patterns matching the corresponding genomic regions. The arrows indicate the direction of the read. The middle of the DNA fragment that is not sequenced is shown as dotted lines. B) Split-read mapping. Reads are shown in arrows with patterns matching the corresponding genomic regions. The top read illustrates a long sequence read that covers the whole MEI and both TSDs. This type of read provides the highest confidence in MEI inference.
With paired-end whole-genome sequencing, reads that can provide information for either anchored read-pair mapping or split-read mapping are usually present in a single dataset. Therefore, some MEI discovery methods attempt to integrate these two algorithms, along with other characteristics of MEIs. One example is the Tea (TE analyzer) pipeline [38], in which anchored read-pair mapping is used to infer the MEI candidate and split-reading mapping is used to identify the exact MEI-genome junction. In addition, the presence of an MEI is inferred from signatures of a typical retrotransposition event, such as target site duplications (TSDs) and poly(A) stretches. Several recent analyses, including the 1000 Genomes Project, have employed similar combinations of algorithms in their MEI discovery pipelines [37, 39].
Whole-genome versus targeted sequencing
Although whole-genome analyses have provided an overview of all types of pMEIs, targeted sequencing has a powerful advantage over whole-genome sequencing when the goal is to identify rare pMEIs across many DNA samples. To detect heterozygous MEIs with high sensitivity using whole-genome sequencing, each human individual must be sequenced at high coverage, which requires about 90×109 bp of sequencing at 30× coverage (30×3×109 bp). By contrast, accurate MEI detection using targeted sequencing requires far less sequencing per individual. For example, the Yb8/9 Alu subfamily, which has roughly 6,500 copies per individual, accounts for approximately 30% of active mobile elements in the human genome. Assuming paired 100 bp reads and aiming for 120× coverage per MEI copy to allow for variation of coverage across loci, targeted sequencing can detect these elements with high sensitivity using only about 0.2% of the reads required for whole-genome sequencing (120×6,500×2×100 = 0.16×109 bp). To fully exploit this advantage requires indexing and pooling samples in groups of ~200. This is no longer a limiting factor with current technology, as efficient pooling of more than 500 samples has been demonstrated recently [40]. Furthermore, because most library preparation steps are carried out after pooling, the per-sample cost remains low. The throughput advantage of targeted methods – roughly two orders of magnitude – may be used to quickly process hundreds or thousands of samples, or to assay fewer samples with greatly increased sensitivity, a critical factor for detecting low-prevalence insertions in somatic tissues or tumors.
Human population history studies using genome-wide, population-level MEI data
High-throughput MEI-profiling methods have opened the door to a new dimension in the study of mobile element biology and human population genetics. Although the sequencing of a single genome provided insight into the activity of mobile elements in our evolutionary past (e.g., [1, 16]), it could not illuminate the patterns of genetic variation that mobile elements create across individuals and populations. Limited numbers of polymorphic Alu and L1 insertion loci, ascertained first in European samples, helped to shed light on the pattern of MEI-generated genetic diversity across populations [8, 41]. Compared to these earlier efforts, high-throughput MEI-profiling approaches can identify thousands of pMEIs of all frequency classes across hundreds of individuals without ascertainment bias, with a much greater potential to infer evolutionary history [24–26, 39, 42]. These studies have provided an unprecedented view of the breadth of variation generated by pMEIs.
An analysis of the data generated by the 1000 Genomes Project from 185 individuals identified 7,380 polymorphic MEIs, including 5,370 insertions that are not present in the human reference genome [37]. About a third of these (2,649) had not been observed in previous studies. Approximately 85% are due to Alu insertions, 12% are L1 elements, and 3% are SVAs. These proportions mirror the relative retrotransposition rates estimated for the three element classes (0.039, 0.0056, and 0.002 insertions per genome per generation, respectively). MEI frequency spectra for African, Asian, and European samples show more pMEIs in African samples, especially at the lower-frequency end. Principal components analyses grouped individuals according to their continents of origin, indicating stratification of MEI allele frequencies across populations. The 1000 Genomes analysis relied mainly on pooled low-coverage sequencing data (1–3× per individual) from many individuals for MEI identification. This approach is biased towards common insertions because an insertion allele present in multiple individuals will effectively receive high coverage across the pooled data set. Despite this detection bias, the majority of MEIs detected in this study are rare (<10% allele frequency).
In another HTS-based analysis, 4,342 non-reference Alu insertions were identified in eight individuals [42]. The majority (79%) of these insertions had not been previously observed, suggesting that they are mostly very rare and could only be detected with the higher sensitivity allowed by high sequencing coverage. Similar to [37], more novel insertions were observed in African individuals than non-Africans. A complementary study [39] extended the computational search for L1 insertions in the 1000 Genomes Project data set to 310 individuals. Of the 1,016 L1 insertions resulting from this and other studies, 104 were present only in African samples. Such a large number of private alleles (more than detected in any other population) is consistent with the "Out of Africa" hypothesis, which posits that modern humans originated in Africa and then migrated to populate the rest of the world. The migrating populations experienced a population size bottleneck and therefore lost genetic diversity, leading to the observed pattern of higher genetic diversity in African vs. non-African populations [43].
Whole-genome sequence data allow simultaneous analysis of all mobile element types and are unaffected by modest element truncations and mutations that can interfere with targeted approaches. However, cost imposes sharp limits on this approach, especially if high sensitivity for rare insertions - and thus high coverage sequencing - is required. The lower cost, efficiency, and sensitivity of targeted MEI sequencing approaches has allowed researchers to rapidly scan samples of their choosing and to identify thousands of additional pMEIs.
As an example of this approach, ME-Scan was used in a proof-of-principle analysis of two active Alu element subfamilies (AluYb8 and AluYb9) in four Asian individuals. The scan identified 5,053 Alu insertions, 487 of which had not been previously observed [25]. In Table 2, data from a subsequent application of ME-Scan (3,228 AluYb8/9 insertions identified in 102 individuals) shows that AluYb8/9 insertions in Europeans are largely a subset of those observed in African samples. Although rare insertions are more likely to be population-specific than common ones, rare African insertions are much more likely to be observed only in African populations. A study targeting active L1 subfamilies in 25 individuals from Africa, Europe, and Japan identified 797 L1 insertion loci [26]. The between-population frequency differences of these pMEIs enabled a parsimony-based analysis to group the samples correctly according to their geographic origin.
Table 2.
Allele sharing and allele frequency for 3,228 Alu insertion polymorphisms
| Allele frequency (a.f.) | 0 < a.f. < 0.05 | 0.05 < a.f. < 0.10 | a.f. > 0.10 |
|---|---|---|---|
| Probability of observing an Alu outside Africa, given ascertainment in Africa | 0.09 | 0.25 | 0.80 |
| Probability of observing an Alu in Africa, given ascertainment outside Africa | 0.41 | 0.76 | 0.97 |
Samples: African: 53 Bantu and Pygmy; non-African: 49 HapMap TSI and Indian Brahmin.
A consistent overall pattern emerges from these studies: In each one, hundreds to thousands of novel pMEIs are detected. Most novel pMEIs are rare, as expected for derived alleles under a neutral model of drift in a population. Many MEIs are stratified by population, and some appear to be population-specific, particularly those in African populations. African populations have more pMEI loci than non-African populations, and pMEIs tend to have higher frequencies in Africans, resulting in higher heterozygosity in African populations. The higher genetic diversity among Africans is expected under the "Out of Africa" hypothesis as described above.
Among the thousands of novel MEIs detected, almost no insertions interrupt protein-coding exons. The 1000 Genomes study confirmed the presence of just two Alu elements inside coding exons out of 5,370 non-reference MEIs detected [37]. This implies a rate that is 46-fold lower than expected for randomly distributed insertions. In another study, the TIP-Chip method was used to search for novel L1 insertions on the X chromosome in 69 males with presumptively X-linked intellectual disability [30]. No insertions in coding exons were found, although two novel insertions were found in introns of genes related to intellectual disability. The absence of even very rare pMEIs from protein-coding exons implies that MEIs are highly disruptive of gene function and eliminated quickly by natural selection.
Another unique property of MEIs is that they have a very low probability of inserting at any particular position in the genome per generation (although certain insertion “hotspots” have been reported, e.g., [44, 45]). Thus, the presence of a pMEI marks the surrounding DNA sequence as a reservoir of ancient genetic diversity. Analysis of many such regions, flagged by polymorphic Alu insertions, demonstrated that the effective population size of human ancestors living ~2 million years ago was ~20,000 [46] (age modified according to improved SNP mutation rate estimates [47]). This is a remarkably small number for a species that left remains and artifacts across the entire Old World.
Somatic MEIs in cancers
Retrotransposons have long been known to play a role in cancer etiology (reviewed in [3]). With high-throughput methods, we can now compare tumor and normal tissue pairs from the same individual at the whole-genome level. This allows us to identify tumor-specific MEIs and assess their roles in cancer initiation and progression. Several types of tumors have been examined using high-throughput methods, and they have revealed important insights into retrotransposon activity in tumors [24, 27, 38, 48].
For example, one study examined Alu and L1 insertions in 20 non-small cell lung tumors and their matched normal adjacent tissues as well as 10 brain tumors with matched blood leukocytes [24]. Nine tumor-specific insertions were detected in lung tumors, but none were found in brain tumors. Interestingly, the tumors’ tolerance of somatic MEIs was correlated with their methylation status: all six tumors that contained somatic L1 insertions were hypomethylated compared to the tumor samples without L1 insertions.
In another study [38], 194 putative tumor-specific MEIs (mostly L1s) were identified using whole-genome sequences from 43 tumors and blood samples from the same individuals, including samples from multiple myeloma, glioblastoma, and colorectal, prostate, and ovarian cancer. Strikingly, all 194 insertions were found in epithelial tumors (colorectal, prostate, and ovarian cancers), whereas no insertions were found in blood or brain tumors. Almost all of the newly identified L1 insertions were heavily truncated, with an average length of 545bp, omitting their coding regions. Somatic L1 insertions were identified in 62 annotated genes, including genes with potential tumor suppressor function and genes frequently mutated in colorectal tumors. Although no somatic L1s were identified in the coding region, genes containing L1 insertions in the sense direction showed a significant reduction in expression level, suggesting that the new insertions have a functional impact. Consistent with [24], the insertions appear to be enriched in regions of the genome that are hypomethylated in tumors. Because MEI insertion rates may vary among tissues, a comparison of tumor tissue with blood cells could overestimate the number of tumor-specific MEIs. Ideally, such studies should compare tumor tissue with normal surrounding tissue to control for potential tissue-specific variation.
In a recent study of colorectal tumors [48], the L1-Seq [26] and RC-Seq methods [27] were used to identify tumor-specific L1 insertions among 16 pairs of tumor/normal matched colorectal samples. Among the L1 insertions that are present in the tumor but not in normal tissues, 73 were validated by locus-specific PCR. Similar to the other studies, these tumor-specific L1s were found in many genes that are frequently mutated in cancers and were heavily truncated (mean size 585 bp). This study also showed that the new L1 insertions likely occurred after cancer initiation (single-cell stage) and were heterozygous among cancer tissues.
These studies have revealed several important features common to tumor-specific somatic MEIs: most tumor-specific somatic L1 insertions are heavily truncated; hypomethylation is correlated with the increase in somatic MEI insertions; and different cancer types/samples vary greatly in their receptiveness to retrotransposon insertions. Somatic insertions might play a role in tumor progression by affecting the expression of genes that have tumor repressor function. However, tumor initiation and the global changes that allow somatic retrotransposition might have happened before these somatic MEIs appeared.
Somatic MEIs in normal tissues
Before high-throughput methods became available, somatic retrotransposition activity was studied primarily using engineered retrotransposon constructs in cell culture systems or transgenic mice/rats. This research suggested somatic retrotransposon activity in early embryogenesis [49–52], tumor cell lines [53, 54], and neural progenitor cells [55–57], but little is known about retrotransposon activity in normal somatic tissues. More recently, HTS data have been used to examine several tissue types for somatic MEIs, including brain [27, 48, 58], liver, and testis [48]. Thus far, somatic insertions have not been identified in most tissues, with the exception of the brain [27, 58]. For example, a recent report based on low-coverage sequencing suggests substantial somatic L1 retrotransposition activity in the hippocampus [27]. However, most of the somatic insertion candidates are singletons, and low-coverage candidates from HTS have high false-positive rates. Site-specific validation PCRs of ~30 somatic insertion candidates were carried out using a modified two-round PCR protocol on the enriched library, not the original DNA samples. The additional rounds of PCR, in the presence of large numbers of DNA templates, could introduce artifacts that inflate the validation rate.
Another recent study examined L1 insertions in neuronal cells from the cerebral cortex and caudate nucleus [58]. Individual neuronal cells were first isolated from postmortem brains of three neurologically normal individuals. A combination of single-cell multiple-displacement amplification and an L1 profiling procedure similar to L1-Seq [26] identified L1 insertions in 300 neuronal cells (50 cells from the cerebral cortex and 50 from the caudate nucleus for each individual). After excluding known L1 polymorphisms and insertions that are present in other tissues from the same individual, the remaining somatic insertion candidates were subjected to PCR validation. The validation results suggest that most somatic insertion candidates are artifacts, and only five insertions were validated in the 300 cells. After correcting for sensitivity, this result extrapolates to an L1 neuronal somatic insertion rate of one insertion per 25 cells, consistent with the rate of insertions per cell division in the germline. The low somatic insertion rate argues against an essential role for L1s in generating neuronal diversity in these brain regions.
Future directions for high-throughput mobile element biology
The development of high-throughput technologies, along with new bioinformatics tools, has transformed the study of mobile elements in the human genome. As sequencing read lengths continue to increase with improved technology, we can expect further improvement in the accuracy of MEI identification. One pressing question is the sensitivity of current methods to detect insertions that are present in only a small number of cells. Several methods show the potential to identify MEIs present in a single cell or only a small proportion of cells, but carefully designed experiments with different proportions of MEI cells/fragments will be needed to fully address this question.
Results from recent studies also highlight the necessity of validation for pMEI candidates, especially for those that are supported by only a few or even one sequence read. In our experience, previously unknown (novel) MEIs observed in only one individual of a sample (singletons) and supported by only by a few sequencing reads (≤10) have a validation rate of ~20% (by locus-specific PCR). A similarly poor replication rate was reported in an study of somatic pMEIs in single cells [58]. Novel MEIs supported by just one sequencing read in a very high-coverage experiment are very likely to be false positives, presumably reflecting rare chimeras generated during library preparation. For germline insertions that are present in all somatic tissues of an individual, validation and high-throughput replication studies will determine thresholds that clearly distinguish between true and false positives. Such validation will be more difficult for somatic insertions that are present in only portions of a tissue or even in a single cell.
With population-level, genome-wide pMEI profiles, a number of long-standing questions related to mobile element biology will soon be answered. One of these is the de novo insertion rate of mobile elements, which has been estimated indirectly using phylogenetic and population methods [16, 26, 59]. However, these approaches will not detect MEIs that were lost soon after integration, and such insertions may have important functional consequences. The de novo insertion rate can be directly obtained using trio data, counting new insertions that are present in the offspring but absent in both parents. Because the rate of retrotransposon insertion is likely to be low, a large number of trios must be analyzed to obtain a reliable rate. Methods described here allow simultaneous analysis of hundreds of individuals in a cost-effective fashion, enabling this question to be answered.
Another important issue is the role of somatic MEIs in cancer etiology and brain development. The cancer MEI studies reviewed here provide critical information about the characteristics of cancer-specific MEIs and suggest high retrotransposition activity in certain regions of the brain. Nevertheless, only a few types of cancers and several brain samples were examined, and the numbers of documented MEIs remain relatively small. Do cancer-specific MEIs play a pivotal role in cancer initiation/progression in certain type of cancers? If so, what is the mechanism behind it? What are the frequency and impact of brain-specific MEIs? Do these insertions happen in certain stage(s) of development or certain areas of the brain? These questions may be answered with surveys of large panels of different samples, in combination with in vitro analysis of the genes affected by somatic MEIs.
Lastly, whole-genome MEI profiling will provide the groundwork for a better understanding of the genomic control of mobile elements. It is known that host factors, methylation, and small RNAs (small interfering RNAs and piRNAs) all play important roles in governing retrotransposon activities in the genome (Reviewed in [5, 60, 61]). Recent studies have demonstrated great variability of pMEIs among human individuals as well as within populations [37, 39]. Therefore, it is likely that retrotransposon regulation activity also varies among human individuals/populations. With population-level MEI profiling, we can begin to explore the correlation between retrotransposon activity and host controlling mechanisms, eventually elucidating how mobile elements are regulated in the human genome.
Figure I.
(a) Types and prevalence of mobile elements in the human genome. The pie chart is based on the human genome reference sequence (hg19). ME: mobile element. (b) Structure of the most prevalent mobile elements in the human genome. All these elements create short TSDs upon insertion into the genome, represented by boxed arrows.
Acknowledgments
The authors thank the three anonymous reviewers for their constructive and valuable comments. This study was supported by a grant from the National Institutes of Health (GM059290). JX is supported by the National Human Genome Research Institute (R00HG005846).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.de Koning AP, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS genetics. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Current opinion in genetics & development. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature reviews. Genetics. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishihara H, Okada N. Retroposons: genetic footprints on the evolutionary paths of life. Methods Mol Biol. 2008;422:201–225. doi: 10.1007/978-1-59745-581-7_13. [DOI] [PubMed] [Google Scholar]
- 7.Ray DA, et al. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- 8.Watkins WS, et al. Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome research. 2003;13:1607–1618. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing J, et al. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol Suppl. 2007;45:2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- 10.Kriegs JO, et al. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimamura M, et al. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, et al. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Human mutation. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing J, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome research. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van den Broeck D, et al. Transposon Display identifies individual transposable elements in high copy number lines. The Plant journal : for cell and molecular biology. 1998;13:121–129. doi: 10.1046/j.1365-313X.1998.00004.x. [DOI] [PubMed] [Google Scholar]
- 18.Roy AM, et al. Recently integrated human Alu repeats: finding needles in the haystack. Genetica. 1999;107:149–161. [PubMed] [Google Scholar]
- 19.Mamedov IZ, et al. Whole-genome experimental identification of insertion/deletion polymorphisms of interspersed repeats by a new general approach. Nucleic acids research. 2005;33:e16. doi: 10.1093/nar/gni018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheen FM, et al. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome research. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovchinnikov I, et al. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome research. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badge RM, et al. ATLAS: a system to selectively identify human-specific L1 insertions. American journal of human genetics. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzdin A, et al. A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics. 2002;79:413–422. doi: 10.1006/geno.2002.6705. [DOI] [PubMed] [Google Scholar]
- 24.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witherspoon DJ, et al. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome research. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wheelan SJ, et al. Transposon insertion site profiling chip (TIP-chip) Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17632–17637. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabriel A, et al. Global mapping of transposon location. PLoS genetics. 2006;2:e212. doi: 10.1371/journal.pgen.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang CR, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardelli M, et al. Alu insertion profiling: Array-based methods to detect Alu insertions in the human genome. Genomics. 2012;99:340–346. doi: 10.1016/j.ygeno.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuzun E, et al. Fine-scale structural variation of the human genome. Nature genetics. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 34.Hormozdiari F, et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics. 2010;26:i350–i357. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong K, et al. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome biology. 2010;11:R128. doi: 10.1186/gb-2010-11-12-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sveinbjornsson JI, Halldorsson BV. PAIR: polymorphic Alu insertion recognition. BMC bioinformatics. 2012;13(Suppl 6):S7. doi: 10.1186/1471-2105-13-S6-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart C, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS genetics. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee E, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome research. 2011;21:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes A, et al. IS-seq: a novel high throughput survey of in vivo IS6110 transposition in multiple Mycobacterium tuberculosis genomes. BMC genomics. 2012;13:249. doi: 10.1186/1471-2164-13-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witherspoon DJ, et al. Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum Hered. 2006;62:30–46. doi: 10.1159/000095851. [DOI] [PubMed] [Google Scholar]
- 42.Hormozdiari F, et al. Alu repeat discovery and characterization within human genomes. Genome research. 2011;21:840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annual review of genomics and human genetics. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 44.Wimmer K, et al. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS genetics. 2011;7:e1002371. doi: 10.1371/journal.pgen.1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conley ME, et al. Two independent retrotransposon insertions at the same site within the coding region of BTK. Human mutation. 2005;25:324–325. doi: 10.1002/humu.9321. [DOI] [PubMed] [Google Scholar]
- 46.Huff CD, et al. Mobile elements reveal small population size in the ancient ancestors of Homo sapiens. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2147–2152. doi: 10.1073/pnas.0909000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solyom S, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome research. 2012 doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nature genetics. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 50.Prak ET, et al. Tracking an embryonic L1 retrotransposition event. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes & development. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Perez JL, et al. LINE-1 retrotransposition in human embryonic stem cells. Human molecular genetics. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Perez JL, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 55.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 58.Evrony GD, et al. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordaux R, et al. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nature reviews. Genetics. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castaneda J, et al. piRNAs, transposon silencing, and germline genome integrity. Mutation research. 2011;714:95–104. doi: 10.1016/j.mrfmmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dewannieux M, et al. LINE-mediated retrotransposition of marked Alu sequences. Nature genetics. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 64.Hancks DC, et al. Retrotransposition of marked SVA elements by human L1s in cultured cells. Human molecular genetics. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dewannieux M, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome research. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS pathogens. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang G, et al. ATon, abundant novel nonautonomous mobile genetic elements in yellow fever mosquito (Aedes aegypti) BMC genomics. 2012;13:283. doi: 10.1186/1471-2164-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith AM, et al. TCUP: A Novel hAT Transposon Active in Maize Tissue Culture. Frontiers in plant science. 2012;3:6. doi: 10.3389/fpls.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zerjal T, et al. Maize genetic diversity and association mapping using transposable element insertion polymorphisms. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik. 2012;124:1521–1537. doi: 10.1007/s00122-012-1807-9. [DOI] [PubMed] [Google Scholar]
- 70.Dong HT, et al. A Gaijin-like miniature inverted repeat transposable element is mobilized in rice during cell differentiation. BMC genomics. 2012;13:135. doi: 10.1186/1471-2164-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 72.Yaakov B, Kashkush K. Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant molecular biology. 2012;80:419–427. doi: 10.1007/s11103-012-9957-3. [DOI] [PubMed] [Google Scholar]
- 73.Winfield MO, et al. Targeted re-sequencing of the allohexaploid wheat exome. Plant biotechnology journal. 2012;10:733–742. doi: 10.1111/j.1467-7652.2012.00713.x. [DOI] [PubMed] [Google Scholar]
- 74.Grzebelus D, et al. Dynamics of Vulmar/VulMITE group of transposable elements in Chenopodiaceae subfamily Betoideae. Genetica. 2011;139:1209–1216. doi: 10.1007/s10709-011-9622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]