Abstract
A method to determine the content and composition of total fatty acids present in microalgae is described. Fatty acids are a major constituent of microalgal biomass. These fatty acids can be present in different acyl-lipid classes. Especially the fatty acids present in triacylglycerol (TAG) are of commercial interest, because they can be used for production of transportation fuels, bulk chemicals, nutraceuticals (ω-3 fatty acids), and food commodities. To develop commercial applications, reliable analytical methods for quantification of fatty acid content and composition are needed. Microalgae are single cells surrounded by a rigid cell wall. A fatty acid analysis method should provide sufficient cell disruption to liberate all acyl lipids and the extraction procedure used should be able to extract all acyl lipid classes.
With the method presented here all fatty acids present in microalgae can be accurately and reproducibly identified and quantified using small amounts of sample (5 mg) independent of their chain length, degree of unsaturation, or the lipid class they are part of.
This method does not provide information about the relative abundance of different lipid classes, but can be extended to separate lipid classes from each other.
The method is based on a sequence of mechanical cell disruption, solvent based lipid extraction, transesterification of fatty acids to fatty acid methyl esters (FAMEs), and quantification and identification of FAMEs using gas chromatography (GC-FID). A TAG internal standard (tripentadecanoin) is added prior to the analytical procedure to correct for losses during extraction and incomplete transesterification.
Keywords: Environmental Sciences, Issue 80, chemical analysis techniques, Microalgae, fatty acid, triacylglycerol, lipid, gas chromatography, cell disruption
Introduction
Fatty acids are one of the major constituents of microalgal biomass and typically make up between 5-50% of the cell dry weight1-3. They are mainly present in the form of glycerolipids. These glycerolipids in turn mainly consist of phospholipids, glycolipids, and triacylglycerol (TAG). Especially the fatty acids present in TAG are of commercial interest, because they can be used as a resource for production of transportation fuels, bulk chemicals, nutraceuticals (ω-3 fatty acids), and food commodities3-6. Microalgae can grow in sea water based cultivation media, can have a much higher areal productivity than terrestrial plants, and can be cultivated in photobioreactors at locations that are unsuitable for agriculture, possibly even offshore. For these reasons, microalgae are often considered a promising alternative to terrestrial plants for the production of biodiesel and other bulk products3-6. Potentially no agricultural land or fresh water (when cultivated in closed photobioreactors or when marine microalgae are used) is needed for their production. Therefore, biofuels derived from microalgae are considered 3rd-generation biofuels.
The total cellular content of fatty acids (% of dry weight), the lipid class composition, as well as the fatty acid length and degree of saturation are highly variable between microalgae species. Furthermore, these properties vary with cultivation conditions such as nutrient availability, temperature, pH, and light intensity1,2. For example, when exposed to nitrogen starvation, microalgae can accumulate large quantities of TAG. Under optimal growth conditions TAG typically constitutes less than 2% of dry weight, but when exposed to nitrogen starvation TAG content can increase to up to 40% of the microalgal dry weight1.
Microalgae mainly produce fatty acids with chain lengths of 16 and 18 carbon atoms, but some species can make fatty acids of up to 24 carbon atoms in length. Both saturated as well as highly unsaturated fatty acids are produced by microalgae. The latter include fatty acids with nutritional benefits (ω-3 fatty acids) like C20:5 (eicosapentaenoic acid; EPA) and C22:6 (docosahexaenoic acid; DHA) for which no vegetable alternatives exist1,2,4,7. The (distribution of) fatty acid chain length and degree of saturation also determines the properties and quality of algae-derived biofuels and edible oils4,8.
To develop commercial applications of microalgal derived fatty acids, reliable analytical methods for quantification of fatty acid content and composition are needed.
As also pointed out by Ryckebosch et al.9, analysis of fatty acids in microalgae distinguishes itself from other substrates (e.g. vegetable oil, food products, animal tissues etc.) because 1) microalgae are single cells surrounded by rigid cell walls, complicating lipid extraction; 2) microalgae contain a wide variety of lipid classes and the lipid class distribution is highly variable7. These different lipid classes have a wide variety in chemical structure and properties such as polarity. Also, lipid classes other than acyl lipids are produced; 3) microalgae contain a wide variety of fatty acids, ranging from 12-24 carbon atoms in length and containing both saturated as well as highly unsaturated fatty acids. Therefore, methods developed to analyze fatty acids in substrates other than microalgae, might not be suitable to analyze fatty acids in microalgae.
As reviewed by Ryckebosch et al.9, the main difference between commonly used lipid extraction procedures is in the solvent-systems that are used. Because of the large variety of lipid classes present in microalgae, each varying in polarity, the extracted lipid quantity will vary with solvents used10-12. This leads to the inconsistencies in the lipid content and composition presented in literature9,10. Depending on the solvent system used, methods based on solvent extraction without cell disruption through, for example, bead beating or sonication, might not extract all lipids because of the rigid structure of some microalgae species9,13. In the case of incomplete lipid extraction, the extraction efficiency of the different lipid classes can vary14. This can also have an effect on the measured fatty acid composition, because the fatty acid composition is variable among lipid classes7.
Our method is based on a sequence of mechanical cell disruption, solvent based lipid extraction, transesterification of fatty acids to fatty acid methyl esters (FAMEs), and quantification and identification of FAMEs using gas chromatography in combination with a flame ionization detector (GC-FID). An internal standard in the form of a triacylglycerol (tripentadecanoin) is added prior to the analytical procedure. Possible losses during extraction and incomplete transesterification can then be corrected for. The method can be used to determine the content as well as the composition of the fatty acids present in microalgal biomass. All fatty acids present in the different acyl-lipid classes, including storage (TAG) as well as membrane lipids (glycolipids, phospholipids), are detected, identified and quantified accurately and reproducibly by this method using only small amounts of sample (5 mg). This method does not provide information about the relative abundance of different lipid classes. However, the method can be extended to separate lipid classes from each other1. The concentration and fatty acid composition of the different lipid classes can then be determined individually.
In literature several other methods are described to analyze lipids in microalgae. Some methods focus on total lipophilic components15, whereas other methods focus on total fatty acids9,16. These alternatives include gravimetric determination of total extracted lipids, direct trans-esterification of fatty acids combined with quantification using chromatography, and staining cells with lipophilic fluorescent dyes.
A commonly used alternative to quantification of fatty acids using chromatography is quantification of lipids using a gravimetric determination17,18. Advantages of a gravimetric determination are the lack of requirements for advanced and expensive equipment like a gas chromatograph; ease to set up the procedure, because of the availability of standardized analytical equipment (e.g. Soxhlet); and a gravimetric determination is less time-consuming than chromatography based methods. The major advantage of using chromatography based methods on the other hand is that in such a method only the fatty acids are measured. In a gravimetric determination the non-fatty acid containing lipids, like pigments or steroids, are also included in the determination. These non-fatty acid containing lipids can make up a large proportion (>50%) of total lipids. If one is only interested in the fatty acid content (for example for biodiesel applications), it will be overestimated when a gravimetric determination is used. In addition, in a gravimetric determination the accuracy of the analytical balance used to weigh the extracted lipids determines the sample size that needs to be used. This quantity is typically much more than the amount needed when chromatography is used. Finally, another advantage of using chromatography over gravimetric determination is that chromatography provides information about the fatty acid composition.
Another alternative to our presented method is direct transesterification16,19,20. In this method lipid extraction and transesterification of fatty acids to FAMEs are combined in one step. This method is quicker than a separate extraction and transesterification step, but combining these steps limits the solvents that can be used for extraction. This might negatively influence extraction efficiency. Another advantage of a separate lipid extraction and transesterification step is that it allows for an additional lipid class separation between these steps1. This is not possible when direct transesterification is used.
Other commonly used methods to determine the lipid content in microalgae include staining the biomass with lipophilic fluorescent stains such as Nile red or BODIPY and measuring the fluorescence signal21,22. An advantage of these methods is that they are less laborious than alternative methods. A disadvantage of these methods is that the fluorescent response is, for various reasons, variable between species, cultivation conditions, lipid classes, and analytical procedures. As an example, several of these variations are caused by differences in uptake of the dye by the microalgae. Calibration using another quantitative method is therefore needed, preferably performed for all the different cultivation conditions and growth stages. Finally, this method does not provide information about the fatty acid composition and is less accurate and reproducible than chromatography based methods.
The presented method is based on the method described by Lamers et al.23 and Santos et al.24 and has also been applied by various other authors1,25-27. Also other methods are available that are based on the same principles and could provide similar results9,28.
Protocol
1. Sample Preparation
There are two alternate protocols for sample preparation included as steps 1.1 and 1.2. Both methods are equally suitable and give similar results, but if a limited amount of algae culture volume is available, method 1.1 is recommended.
NOTE: For either protocol, prepare two additional bead beater tubes according to the entire protocol but without adding algae to them to be used as a blank. In this way, peaks in the GC chromatogram resulting from extraction of components from materials used can be identified and quantified.
1.1. Sample Preparation Protocol Option 1: Recommended When a Limited Amount of Algae Culture is Available
Determine the algae dry weight concentration (g/L) in the culture broth, for example as described by Breuer et al.1.
Transfer a volume of culture broth that contains 5-10 mg algae dry weight to a glass centrifuge tube. Calculate the exact amount of biomass transferred using the biomass concentration determined in step 1.1.1.
Centrifuge 5 min at 1,200 x g.
Discard part of the supernatant, leave approximately 0.25 ml in the tube.
Re-suspend the algae in the remaining supernatant by gentle pipetting the pellet up and down and transfer the complete cell pellet to a bead beater tube using a 200 μl pipette.
Rinse the centrifuge tube and glass pipette with ±0.15 ml milliQ water and transfer the liquid to the same bead beater tube.
Centrifuge bead beater tubes for one minute at maximum rpm to make sure no air bubbles remain in the bottom of the tubes. It is possible to store closed bead beater tubes at -80 °C.
Lyophilize the bead beater tubes containing the sample. It is possible to store the closed bead beater tubes at -80 °C.
1.2. Sample Preparation Protocol Option 2
Centrifuge an undetermined volume of algae culture broth and discard supernatant. It is not necessary to determine the biomass concentration or to measure the volume used.
Measure or calculate the osmolarity of the culture medium using the concentration of the main salts present in the culture medium.
Wash the cell pellet by resuspending the cell pellet in the same volume, as used in step 1.2.1, of an ammonium formate solution, which approximates the osmolarity of the culture medium. An equiosmolar ammonium formate solution prevents cell lyses during washing of the cells.
NOTE: Washing the cells is necessary to remove salts present in the culture medium. This is especially important for fatty acid analysis in marine microalgae because of the high salt concentrations in their culture medium. If salts would still be present, this would cause overestimation of the amount of cell dry weight which will be determined later in this protocol. Ammonium formate is used to wash cells because this solution will completely evaporate during lyophilization and not leave any residue.
Centrifuge algae suspension and discard supernatant.
Lyophilize the cell pellet.
Weigh 5-10 mg lyophilized microalgae powder into a bead beater tube. Record the exact weight.
2. Cell Disruption and Lipid Extraction
NOTE: This section describes an extensive cell disruption procedure. Possibly, some of the cell disruption steps are redundant and less extensive cell disruption might yield the same results for some microalgae species or for biomass derived from certain cultivation conditions. This would need validation for each specific situation however. Therefore the proposed extensive disruption protocol is recommended as a universal method suitable for all microalgal material.
Weigh an exact amount of tripentadecanoin (triacylglycerol containing three C15:0 fatty acids) and add it to an exactly known amount of 4:5 (v/v) chloroform:methanol. In this way the concentration of tripentadecanoin is exactly known. Aim for a concentration of 50 mg/L. Tripentadecanoin serves as the internal standard for fatty acid quantification.
Add 1 ml chloroform:methanol 4:5 (v/v) containing tripentadecanoin to each beating tube (specified in the reagents and equipment table). Check that there are no beads remaining inside the cap of the beadbeater tube, this will result in leaking of the tubes. Use a positive displacement pipette for accurate addition of solvents.
Bead beat the bead beater tubes 8x at 2,500 rpm for 60 sec, each time with a 120 sec internal between each beating.
Transfer the solution from the bead beater tubes to a clean heat-resistant 15 ml glass centrifuge tube with Teflon insert screw-caps. Be sure to transfer all the beads from the bead beater tube as well.
To wash the bead beater tube, add 1 ml chloroform:methanol 4:5 (v/v) containing tripentadecanoin into the bead beater tube, close cap and mix, and transfer this solution to the same glass tube from step 2.4. Wash the tube three times (in total 4 ml of chloroform:methanol 4:5 (v/v) containing tripentadecanoin is used per sample - 1 ml added in step 2.2 and 3 ml for 3 washing steps).
Vortex the glass tubes for 5 sec and sonicate in a sonication bath for 10 min.
Add 2.5 ml of MilliQ water, containing 50 mM 2-amino-2-hydroxymethyl-propane-1,3-diol (Tris) and 1 M NaCl, which is set to pH 7 using a HCl solution. This will cause a phase separation between chloroform and methanol:water. 1 M NaCl is used to enhance the equilibrium of lipids towards the chloroform phase.
Vortex for 5 sec and then sonicate for 10 min.
Centrifuge for 5 min at 1,200 x g.
Transfer the entire chloroform phase (bottom phase) to a clean glass tube using a glass Pasteur pipette. Make sure not to transfer any interphase or top phase.
Re-extract the sample by adding 1 ml chloroform to old tube (containing the methanol:water solution).
Vortex for 5 sec and then sonicate for 10 min.
Centrifuge for 5 min at 1,200 x g.
Collect the chloroform phase (bottom phase) using a glass Pasteur pipette and pool with the first chloroform fraction from step 2.10.
If difficulties are experienced in collecting the entire chloroform fraction in the previous step, repeat steps 2.11-2.14. Otherwise proceed with step 2.16.
Evaporate the chloroform from the tube in a nitrogen gas stream. After this step samples can be stored at -20 °C under a nitrogen gas atmosphere.
3. Transesterification to Fatty Acid Methyl Esters
Add 3 ml methanol containing 5% (v/v) sulfuric acid to the tube containing the dried extracted lipids (tube originally containing the chloroform fraction) and close the tube tightly.
Vortex for 5 sec.
Incubate samples for 3 hr at 70 °C in a block heater or water bath. Periodically (every ±30 min) ensure that the samples are not boiling (caused by an improperly closed lid) and vortex tubes. During this reaction fatty acids are methylated to their fatty acid methyl esters (FAMEs).
Cool samples to room temperature and add 3 ml MilliQ water and 3 ml n-hexane.
Vortex for 5 sec and mix for 15 min with a test tube rotator.
Centrifuge for 5 min at 1,200 x g.
Collect 2 ml of the hexane (top) phase and put in fresh glass tube using a glass Pasteur pipette.
Add 2 ml MilliQ water to the collected hexane phase to wash it.
Vortex for 5 sec and centrifuge for 5 min at 1,200 x g. After this step it is possible to store the samples at -20 °C under nitrogen gas atmosphere (phase separation not necessary).
4. Quantification of FAMEs Using Gas Chromatography
Fill GC vials with the hexane phase (upper phase) using a glass Pasteur pipette.
Place the caps on the GC vials. Make sure the caps are completely sealed, and cannot be turned, to prevent evaporation from the vial.
Refill the GC wash solvents (hexane), empty the waste vials, and put sample vials in auto-sampler. Before running the actual samples on the GC, first run 2 blanks containing only hexane on the GC.
Run the samples on a GC-FID with a Nukol column (30 m x 0.53 mm x 1.0 μm). Use inlet temperature of 250 °C and use helium as carrier gas, pressure of 81.7 kPa and split ratio of 0.1:1, total flow rate: 20 ml/min. FID detector temperature of 270 °C with H2 flow rate of 40 ml/min, air flow rate of 400 ml/min and He flow rate of 9.3 ml/min. Set injection volume to 1 μl/sample. Pre and post wash injector with n-hexane. Initial oven temperature is set to 90 °C for 0.5 min and then raised with 20 °C/min until an oven temperature of 200 °C is reached. Oven temperature is then maintained at 200 °C for 39 min. Total run time is 45 min.
Representative Results
A typical chromatogram that is obtained via this process is shown in Figure 1. FAMEs are separated by size and degree of saturation by the GC column and protocol used. Shorter chain length fatty acids and more saturated fatty acids (fewer double bonds) have shorter retention times. The used GC column and protocol do not intend to separate fatty acid isomers (same chain length and degree of saturation, but different positions of double bonds), but this could be achieved by using a different GC column and protocol.
The fatty acid concentration and composition can be calculated using the area of the GC peaks, internal standard concentration (mg/L) (step 2.1), and the amount of biomass added to the sample (mg) (step 1.1.2 or 1.2.6, depending on which sample preparation protocol was chosen).
The content of each individual fatty acid in the biomass (mg fatty acid/g dry weight) can be determined by
![]() With Area of individual FAME as determined by integration of the GC chromatogram (Figure 1); Area of C15:0 FAME as determined by integration of the GC chromatogram (Figure 1);
With Area of individual FAME as determined by integration of the GC chromatogram (Figure 1); Area of C15:0 FAME as determined by integration of the GC chromatogram (Figure 1);
![]() With [IS] the concentration of tripentadecanoin in the chloroform:methanol solution as determined in step 2.1; MWC15:0 FA the molecular weight of the C15:0 fatty acid (242.4 g/mol); MWC15:0 TAG the molecular weight of tripentadecanoin (765.24 g/mol);
With [IS] the concentration of tripentadecanoin in the chloroform:methanol solution as determined in step 2.1; MWC15:0 FA the molecular weight of the C15:0 fatty acid (242.4 g/mol); MWC15:0 TAG the molecular weight of tripentadecanoin (765.24 g/mol);
![]() As determined by running calibration solutions on the GC, constituting out of mixtures of C15:0 FAME and FAMEs of all individual fatty acids expected in the sample, with [C15:0 FAME in calibration solution] the concentration of C15:0 FAME in the calibration solution (mg/L); [individual FAME in calibration solution] the concentration of the individual FAME in the calibration solution (mg/L); and the areas as determined by integration of the GC chromatogram of the calibration solution.
As determined by running calibration solutions on the GC, constituting out of mixtures of C15:0 FAME and FAMEs of all individual fatty acids expected in the sample, with [C15:0 FAME in calibration solution] the concentration of C15:0 FAME in the calibration solution (mg/L); [individual FAME in calibration solution] the concentration of the individual FAME in the calibration solution (mg/L); and the areas as determined by integration of the GC chromatogram of the calibration solution.
The total fatty acid content can be calculated as the sum of all individual fatty acids. The relative abundance of each fatty acid can be calculated by dividing the concentration of each individual fatty acid by the total fatty acid content.
The fatty acid composition and content of Scenedesmus obliquus UTEX393 (Chlorophyceae) under both nitrogen replete and deplete conditions is shown in Figure 2. Fatty acid composition and content are highly affected by nitrogen starvation. In S. obliquus, C16:0 (palmitic acid) and C18:1 (oleic acid) are the two most abundant fatty acids.
The fatty acid composition and content of Phaeodactylum tricornutum UTEX640 (diatom) under both nitrogen replete and deplete conditions are shown in Figure 3. Similar to S. obliquus, the fatty acid content and composition are highly affected by nitrogen starvation. P. tricornutum also produces substantial amounts of highly unsaturated fatty acids such as C20:5 (eicosapentaenoic acid, EPA) as well as very long chain fatty acids (lignoceric acid, C24:0) that can be detected by this method.
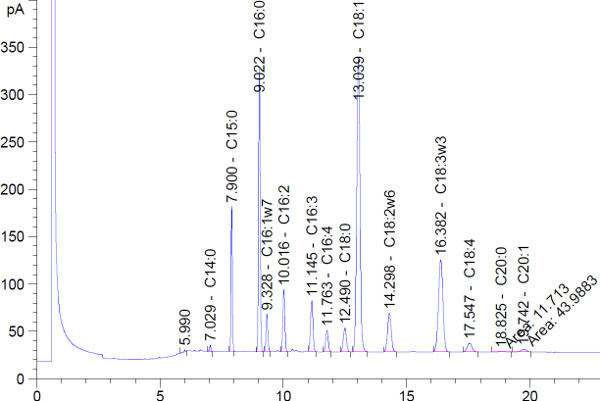 Figure 1. Representative GC-FID chromatogram. A sample derived from Scenedesmus obliquus exposed to nitrogen starvation was used. Only the first 20 min of the chromatogram are shown. After 20 min no peaks were detected in this sample.
Figure 1. Representative GC-FID chromatogram. A sample derived from Scenedesmus obliquus exposed to nitrogen starvation was used. Only the first 20 min of the chromatogram are shown. After 20 min no peaks were detected in this sample.
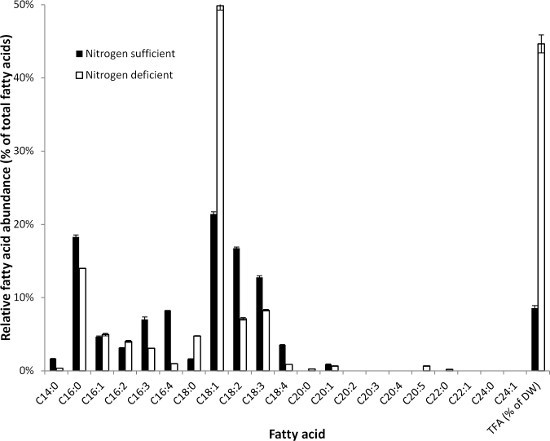 Figure 2. Fatty acid composition (relative abundance of fatty acids) of Scenedesmus obliquus (Chlorophyceae) under both nitrogen sufficient (black bars) and nitrogen deprived cultivation conditions (white bars). The total fatty acid contents were 8.6±0.5% and 44.7±1.7% (% of dry weight) under nitrogen sufficient and nitrogen deprived cultivation conditions, respectively. Error bars indicate the highest and lowest value of two analytical replicates.
Figure 2. Fatty acid composition (relative abundance of fatty acids) of Scenedesmus obliquus (Chlorophyceae) under both nitrogen sufficient (black bars) and nitrogen deprived cultivation conditions (white bars). The total fatty acid contents were 8.6±0.5% and 44.7±1.7% (% of dry weight) under nitrogen sufficient and nitrogen deprived cultivation conditions, respectively. Error bars indicate the highest and lowest value of two analytical replicates.
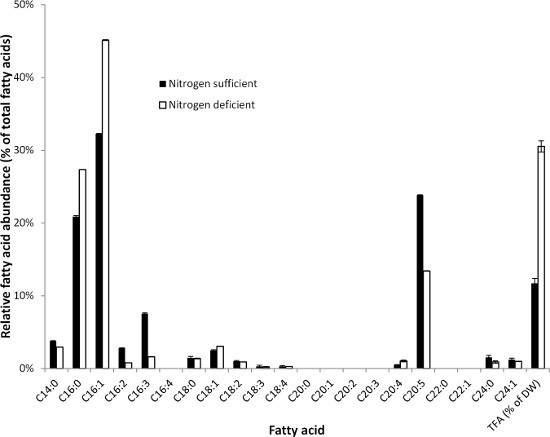 Figure 3. Fatty acid composition (relative abundance of fatty acids) of Phaeodactylum tricornutum (diatom) under both nitrogen sufficient (black bars) and nitrogen deprived cultivation conditions (white bars). The total fatty acid contents were 11.7±0.9% and 30.5±1.1% (% of dry weight) under nitrogen sufficient and nitrogen deprived cultivation conditions, respectively. Error bars indicate the highest and lowest value of two analytical replicates.
Figure 3. Fatty acid composition (relative abundance of fatty acids) of Phaeodactylum tricornutum (diatom) under both nitrogen sufficient (black bars) and nitrogen deprived cultivation conditions (white bars). The total fatty acid contents were 11.7±0.9% and 30.5±1.1% (% of dry weight) under nitrogen sufficient and nitrogen deprived cultivation conditions, respectively. Error bars indicate the highest and lowest value of two analytical replicates.
Discussion
The described method can be used to determine the content as well as the composition of total fatty acids present in microalgal biomass. Fatty acids derived from all lipid classes, including storage (TAG) as well as membrane lipids (phospholipids and glycolipids), are detected. All fatty acid chain lengths and degrees of saturation that are present in the microalgae can be detected and distinguished. The method is based on mechanical cell disruption, solvent based lipid extraction, transesterification of fatty acids to FAMEs, and quantification of FAMEs using gas chromatography in combination with a flame ionization detector (GC-FID). This method requires small amounts of sample (5 mg), is accurate and reproducible and can be used among a wide variety of microalgae species. This method is intended to be used for analytical purposes and is specifically tailored for using small sample sizes. The method is not intended to be scaled-up and used for extracting large quantities of microalgal fatty acids. A limitation of this method is that it is laborious and it can take a few days from starting the procedure to obtaining results. Especially in the case of process monitoring for industrial microalgal fatty acid production this might be considered a limitation. If faster results are required, other options such as staining with lipophilic fluorescent dyes might be more suitable. The following recommendations are made with regard to performing the analytical procedure:
Extraction and transesterification procedure are standardized by using a triacylglycerol (tripentadecanoin) as an internal standard. The use of an internal standard is commonly used for quantification of FAMEs in samples using gas chromatography, but most often a FAME internal standard is used. It is recommended to use a TAG internal standard rather than a FAME internal standard and to add it before cell disruption and solvent extraction. Possible losses during lipid extraction or incomplete transesterification can then be corrected for, since it is expected that similar losses would occur to the internal standard. An internal standard should be used that does not co-elute with fatty acids produced by microalgae. Because microalgae only produce fatty acids with even carbon numbers, a TAG containing a fatty acid with an odd number of carbon atoms is a good candidate. Because the GC column and protocol not only separate on chain length but also on degree of unsaturation, a saturated odd numbered fatty acid could hypothetically co-elute with an unsaturated even numbered fatty acid. It should be validated that the used internal standard does not co-elute with fatty acids naturally present in the sample. In our experience with various microalgae, tripentadecanoin (TAG containing three C15:0 fatty acids) does not co-elute with fatty acids naturally present in microalgae.
Saturated and highly unsaturated fatty acids give different GC-FID responses (peak areas) at identical concentrations. Therefore standards of FAMEs should be used for calibration and determination of relative response factors of individual fatty acids as described in the representative results section. Standards of FAMEs can also be used to determine retention times of individual fatty acids. Alternatively, GC-MS can be used for fatty acid identification.
Both the retention time and relative response factor of individual fatty acids will change with GC equipment age. Frequent calibration of retention time and relative response factor is therefore recommended.
Mechanical cell disruption is an important step in this method. It is realized by a combination of lyophilization, bead beating and sonication. Without cell disruption it is possible that not all lipids are extracted9,13. Possibly, some of the cell disruption steps are redundant and less extensive cell disruption (for example shorter bead beating times) might yield the same results for some microalgae species or for biomass derived from certain cultivation conditions. This would need validation for each specific situation however, therefore the proposed extensive disruption protocol is recommended.
During the analytical procedure, the solvents used might extract components from the plastic tubes and pipette tips used. These extracted components might interfere with detection of the FAMEs using GC-FID. Especially, long bead-beat times and high temperature during bead-beating will result in contaminating components. Our experience is that components extracted from the bead-beat tubes will result in a few additional peaks in the GC-FID chromatogram, some of which overlay with algae derived FAMEs (mainly the saturated fatty acids). When the method is executed as proposed, this is never more than 1-2% of the total peak area. Cooling of the bead-beater tubes during bead-beating and using larger amounts of biomass per sample will reduce the relative peak area of these plastic derived peaks. It is recommended to use glass pipettes and glass tubes as much as possible throughout the protocol. Furthermore, the use of blanks to check and correct for the components extracted from plastics, but also to check and correct for contaminations in for example the used solvents, is recommended.
During the bead-beating process, both the bead-beater and the bead-beater tubes will heat up. When bead-beating for a much longer time or much higher rpm than proposed in this method, and when no cooling is applied in between, this can result in boiling of the solvents and eventually rupture of the tubes. This will result in a loss of sample. In our experience, this situation occurs when bead-beating for more than 6 min at 6,000 rpm.
Microalgae can produce highly unsaturated fatty acids, which can be prone to oxidation. Ryckebosch et al.9 observed no change in fatty acid composition caused by oxidation of unsaturated fatty acids during various extraction procedures. It was proposed that antioxidants naturally occurring in microalgae protect these unsaturated fatty acids from oxidation. If it is suspected that oxidation might occur, a possibility would be to add a synthetic antioxidant such as butylhydroxytoluene (BHT) as long as the antioxidant does not co-elute with FAMEs, and as long as the amount of antioxidant used does not interfere with the chromatography9. To prevent oxidation during long term storage, it is recommended to store samples at -80 °C under a nitrogen gas atmosphere.
Lipases present in microalgae can potentially hydrolyze lipids during the analytical procedure and therefore affect the results. Ryckebosch et al.9 found that lyophilization inactivates lipases. Moreover, lipases do not degrade fatty acids, but just detach them from the glycerol group of glycerolipids. Hence, lipase activity should not interfere with the presented fatty acid protocol. Enzymes involved in fatty acid degradation act on Coenzyme A activated fatty acids and the occurrence of such degradation during the extraction thus does not seem likely. Nevertheless, in case it is suspected that enzymatic breakdown of fatty acids from glycerolipids might affect results, a possibility would be to add a lipase inhibitor, such as isopropanol, as long as the inhibitor does not interfere with the analytical procedure itself9.
This method could be extended to quantify and determine the fatty acid composition of neutral lipids (TAG) and polar lipids (phospholipids, glycolipids) by using a solid phase extraction (SPE) separation step between step 2 (lipid extraction) and step 3 (transesterification) as described by for example Breuer et al.1. Alternatively, a Thin Layer Chromatography (TLC) step can be used to separate various polar lipid classes, for example as described by Wang and Benning28.
This method could be extended to be used in, for example, plants, animal tissues and other micro-organisms. Because of the emphasis on cell disruption, this method is mainly suitable for analysis of fatty acids in materials that are very difficult to extract.
The extraction part of the method (steps 1 and 2) could be used for extraction of pigments and other lipophilic components23,25. The cell disruption procedure could be used for extraction of other components such as starch or carbohydrates by using other solvent systems (including water).
Disclosures
Authors have nothing to disclose.
Acknowledgments
A part of this work was financially supported by the Institute for the Promotion of Innovation by Science and Technology—Strategic Basic Research (IWT-SBO) project Sunlight and Biosolar cells. Erik Bolder and BackKim Nguyen are acknowledged for their contribution to the optimization of the bead beating procedure.
References
- Breuer G, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresource Technology. 2012;124:217–226. doi: 10.1016/j.biortech.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, et al. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. The Plant Journal. 2008;54(4):621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnology Advances. 2007;25(3):294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Draaisma RB, Wijffels RH, et al. Food commodities from microalgae. Curr. Opin. Biotechnol. 2012;24(2):169–177. doi: 10.1016/j.copbio.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329(5993):796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ, Eppink MHM. Microalgae for the production of bulk chemicals and biofuels. Biofuels, Bioprod. Bioref. 2010;4(3):287–295. [Google Scholar]
- Guschina IA, Harwood JL. Lipids and lipid metabolism in eukaryotic algae. Progress in Lipid Research. 2006;45(2):160–186. doi: 10.1016/j.plipres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Thomas-Hall SR, et al. Second Generation Biofuels: High-Efficiency Microalgae for Biodiesel Production. BioEnergy Research. 2008;1(1):20–43. [Google Scholar]
- Ryckebosch E, Muylaert K, Foubert I. Optimization of an Analytical Procedure for Extraction of Lipids from Microalgae. Journal of the American Oil Chemists' Society. 2011;89(2):189–198. [Google Scholar]
- Laurens LML, Dempster TA, et al. Algal Biomass Constituent Analysis: Method Uncertainties and Investigation of the Underlying Measuring Chemistries. Analytical Chemistry. 2012;84(4):1879–1887. doi: 10.1021/ac202668c. [DOI] [PubMed] [Google Scholar]
- Iverson SJ, Lang SLC, Cooper MH. Comparison of the bligh and dyer and folch methods for total lipid determination in a broad range of marine tissue. Lipids. 2001;36(11):1283–1287. doi: 10.1007/s11745-001-0843-0. [DOI] [PubMed] [Google Scholar]
- Grima EM, Medina AR, et al. Comparison Between Extraction of Lipids and Fatty Acids from microalgal biomass. JAOCS. 1994;71(9):955–959. [Google Scholar]
- Lee JY, Yoo C, Jun SY, Ahn CY, Oh HM. Comparison of several methods for effective lipid extraction from microalgae. Bioresour Technol. 2010;101(1 Supplement):75–77. doi: 10.1016/j.biortech.2009.03.058. [DOI] [PubMed] [Google Scholar]
- Guckert JB, Cooksey KE, Jackson LL. lipid solvent systems are not equivalent for analysis of lipid classes in the micro eukaryotic green alga. Journal of Microbiological Methods. 1988;8:139–149. [Google Scholar]
- Pruvost J, Van Vooren G, Cogne G, Legrand J. Investigation of biomass and lipids production with Neochloris oleoabundans in photobioreactor. Bioresource Technology. 2009;100(23):5988–5995. doi: 10.1016/j.biortech.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Griffiths MJ, Hille RP, Harrison STL. Selection of Direct Transesterification as the Preferred Method for Assay of Fatty Acid Content of Microalgae. 2010;45(11):1053–1060. doi: 10.1007/s11745-010-3468-2. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. J Biol. Chem. 1956;226:497–509. [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Lepage G, Roy CC. Improved recovery of fatty acid through direct transesterification without prior extraction or purification. Journal of Lipid research. 1984;25:1391–1396. [PubMed] [Google Scholar]
- Welch RW. A micro-method for the estimation of oil content and composition in seed crops. J. Sci. Food Agric. 1002;28(7):635–638. [Google Scholar]
- Chen W, Zhang C, Song L, Sommerfeld M, Hu Q. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. Journal of Microbiological Methods. 2009;77(1):41–47. doi: 10.1016/j.mimet.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Cooper MS, Hardin WR, Petersen TW, Cattolico RA. Visualizing "green oil" in live algal cells. Journal of Bioscience and Bioengineering. 2010;109(2):198–201. doi: 10.1016/j.jbiosc.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Lamers PP, van de Laak CC, et al. Carotenoid and fatty acid metabolism in light-stressed Dunaliella salina. Biotechnology and Bioengineering. 2010;106(4):638–648. doi: 10.1002/bit.22725. [DOI] [PubMed] [Google Scholar]
- Santos AM, Janssen M, Lamers PP, Evers WA, Wijffels RH. Growth of oil accumulating microalga Neochloris oleoabundans under alkaline-saline conditions. Bioresour Technol. 2012;104:593–599. doi: 10.1016/j.biortech.2011.10.084. [DOI] [PubMed] [Google Scholar]
- Mulders KJM, Weesepoel Y, et al. Growth and pigment accumulation in nutrient-depleted Isochrysis aff. galbana T-ISO. J. Appl. Phycol. 2012.
- Kliphuis AM, Klok AJ, et al. Metabolic modeling of Chlamydomonas reinhardtii: energy requirements for photoautotrophic growth and maintenance. J. Appl. Phycol. 2012;24(2):253–266. doi: 10.1007/s10811-011-9674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers PP, Janssen M, De Vos RCH, Bino RJ, Wijffels RH. Carotenoid and fatty acid metabolism in nitrogen-starved Dunaliella salina, a unicellular green microalga. Journal of Biotechnology. 2012;162(1):21–27. doi: 10.1016/j.jbiotec.2012.04.018. [DOI] [PubMed] [Google Scholar]
- Wang Z, Benning C. Arabidopsis thaliana Polar Glycerolipid Profiling by Thin Layer Chromatography (TLC) Coupled with Gas-Liquid Chromatography (GLC) J. Vis. Exp. 2011. p. e2518. [DOI] [PMC free article] [PubMed]


