Abstract
Analog-sensitive (AS) kinase technology is a powerful approach for studying phospho-signaling pathways in diverse organisms and physiological processes. The key feature of this technique is that a kinase-of-interest can be mutated to sensitize it to inhibitor analogs that do not target wild-type (WT) kinases. In theory, this enables specific inhibition of any kinase in cells and in mouse models of human disease. Typically these inhibitors are identified from a small library of molecules based on the pyrazolopyrimidine (PP) scaffold. However, we recently identified a subset of native human kinases, including the Ephrin A kinase family, that are sensitive to commonly used PP inhibitors. In an effort to develop a bioorthogonal AS-kinase inhibitor and to extend this technique to PP-sensitive kinases we sought an alternative inhibitor scaffold. Here we report the structure-based design of synthetically tractable, potent, and extremely selective AS-kinase inhibitors based on the natural product staurosporine. We demonstrate that these molecules, termed staralogs, potently target AS kinases in cells and we employ X-ray crystallography to elucidate their mechanism of efficacy. Finally, we demonstrate that staralogs target AS mutants of PP-sensitive kinases at concentrations where there is little to no inhibition of native human kinases. Thus, staralogs represent a new class of AS-kinase inhibitors and a core component of the chemical genetic tool kit for probing kinase-signaling pathways.
Introduction
Protein kinases are key signaling enzymes involved in nearly every physiological process in eukaryotes. A major challenge in biomedical research is to elucidate kinase-mediated signaling pathways and to understand how human diseases arise from their misregulation. Small molecule inhibitors are invaluable tools for studying kinase-signaling events as they allow rapid inactivation of kinase activity in cells and thereby enable the study of essential kinases1 and the dissection of events that occur on a time scale of seconds to minutes2. However, it remains difficult to identify specific inhibitors of individual kinases due to the high degree of conservation of the ATP-binding pocket throughout the human kinome (~520 kinases)3. Our lab has developed a systematic method for generating specific inhibitors of individual kinases by using genetic engineering to sensitize a single kinase to inhibition by a highly selective ATP-competitive molecule4,5. Specifically, the inhibitor is designed to form a steric clash with a conserved residue in the ATP-binding pocket, termed the gatekeeper, which prevents inhibitor binding. In contrast, the gatekeeper residue of the AS kinase is mutated from the larger native amino acid (Thr, Leu, Phe, etc) to glycine or alanine thereby sensitizing it to inhibition by the bulky analog. This chemical-genetic approach combines the specificity of genetics with the temporal resolution and reversibility of pharmacology.
A central design feature of this technology is the use of a bioorthogonal small molecule that does not bind to WT kinases but potently inhibits the activity of the AS mutant. We have demonstrated that pyrazolopyrimidine analogs fulfill these requirements for many kinases and have successfully applied this chemical genetic technique to more than 80 kinases from diverse species and kinase families. Recently, however, we and others discovered that the commonly used AS kinase inhibitors 1NA-PP1, 1NM-PP1, and 3MB-PP1 target a subset of WT kinases with weak to moderate potency6,7. This does not preclude application of the analog-sensitive approach to most kinases because potential off-target effects of these molecules are accounted for when WT cells are treated with the inhibitor. However, if the kinase-of-interest is, itself, sensitive to the PP, the window of selectivity for the AS allele may narrow and it may not be possible to completely abrogate AS kinase activity without also reducing that of the WT allele. Additionally, if a PP-sensitive kinase is involved in the same pathway as the AS kinase, unintentional inhibition might compromise or complicate a chemical genetic analysis. Thus we sought to identify a new inhibitor that retains potency and generality towards engineered AS kinases while remaining inactive toward all other human kinases.
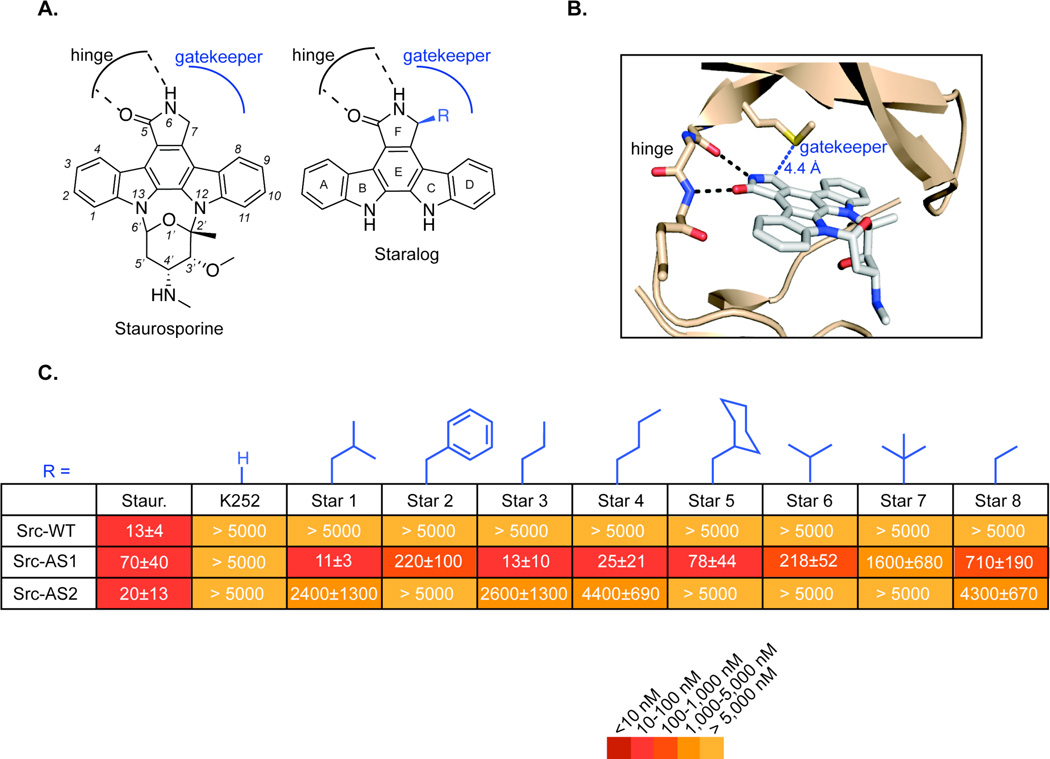
The ideal starting scaffold from which to design an AS-kinase inhibitor is a molecule that binds a maximum number of kinases but can be modified to introduce an orthogonality element that disrupts binding unless the kinase bears a complementary mutation. The indolocarbazole natural product staurosporine is an attractive option as it potently targets ~80% of protein kinases8. Staurosporine also adopts a well-defined and conserved binding mode in the ATP pocket of divergent kinases as evidenced by numerous X-ray co-crystal structures9. Furthermore, previous studies of the related molecule, K252a, have identified the C7 atom of the indolocarbazole as an ideal position at which functional groups can be introduced to prevent binding to WT kinases through steric clash with the gatekeeper residue (Figure 1A, 1B)5,10. Staurosporine and K252a (Figure S1) each contain a different carbohydrate moiety bridging the N12 and N13 atoms that contributes to the affinity of the molecules for the ATP-binding pocket of protein kinases. The different interactions of each sugar may account for the far greater number of kinases targeted by staurosporine8 and the poor generality of C7-modified K252a analogs towards diverse AS kinases (unpublished data). The carbohydrate functional groups of both staurosporine11 and K252a12 are challenging to synthesize and therefore not ideal for the construction of a panel of AS-kinase inhibitors or for the large-scale synthesis required for in vivo studies.
Figure 1.
Design of staralog inhibitors and structure-activity relationships of the C7-position. (A) Structure of staurosporine (left) and engineered staralog inhibitors (right) with kinase hinge-binding interaction and gatekeeper pocket illustrated in black and blue, respectively. (B) X-ray co-crystal structure of staurosporine bound to Protein Kinase Cθ14 with hinge interaction highlighted in black and the distance between C7-position of staurosporine and the Met gatekeeper side chain highlighted in blue. (C) IC50 values for staralog inhibitors against Src-WT, Src-AS1, and Src-AS2 kinases. IC50 values were determined from a seven-point dose-response. Each experiment was done in triplicate and repeated for a minimum total of three times for compounds that showed measureable inhibition. Compounds that showed no inhibition were measured twice. Reported values represent the mean and error represents the standard error of the mean. WT = native gatekeeper (Thr for Src), AS1 = Gly gatekeeper, AS2 = Ala gatekeeper
To develop a synthetically tractable surrogate for staurosporine, we asked if the non-glycosylated indolocarbazole could serve as the basis of a new scaffold for AS kinase inhibitors. Here we report the synthesis of carbohydrate-free staurosporine analogs, termed staralogs, and their structure-activity relationships toward a subset of AS kinases. We also show that simple polar groups at the N12 position form a novel interaction with a conserved asparagine residue and recover the potency that is lost by removal of the sugar group. Finally, we demonstrate that staralogs are uniquely selective inhibitors and are capable of targeting kinases that have thus far eluded the application of AS-kinase technology.
Results and Discussion
C7-substituted indolocarbazole derivatives complement the gatekeeper mutation in Src kinase
Removal of the staurosporine sugar ring (K252c) led to complete loss of activity towards the tyrosine kinase Src-WT, as well as Src-AS1 (glycine gatekeeper) and Src-AS2 (alanine gatekeeper), confirming previous reports that affinity interactions provided by the sugar are critical to inhibitor binding (Figure 1C)13. Despite this severe loss in potency for the parent aglycone, K252c, we reasoned that introduction of new substituents at C7 might complement the engineered pocket and enhance potency without the sugar moiety. Thus we prepared a panel of staralogs using a synthetic strategy whereby the (S)-configuration of the C7-substituent is introduced from commercially available L-amino acids (Scheme S1)10. We found that several alkyl or aryl groups indeed rescued activity toward Src-AS1 but not Src-AS2 or Src-WT. The most potent staralogs (Star 1, Star 3, Star 4) bear iso-butyl, n-butyl, and n-propyl groups at the C7 position. Larger groups, such as benzyl (Star 2) and cyclohexylmethyl (Star 5), were much less potent, presumably due to steric clash of these larger groups with the surface of the expanded gatekeeper pocket. Conversely, staralogs with C7 substituents that do not project as deeply into the expanded ATP binding pocket were also less potent (Figure 1C, Star 6 – 9). We interpret these structure-activity relationships as evidence that a complementary hydrophobic interaction exists between the C7 group of the staralog and the gatekeeper pocket. This interaction provides a significant and necessary contribution to inhibitor binding in the absence of sugar affinity elements.
Simple polar groups rescue activity of staralog inhibitors toward AS kinases bearing an alanine gatekeeper residue
Some kinases suffer decreased stability or activity upon mutation of the gatekeeper residue to glycine but are more tolerant of an alanine substitution15,16. Thus, an ideal AS kinase inhibitor must be able to target kinases with either a glycine (AS1) or alanine (AS2) gatekeeper. Although several molecules in our initial panel are potent Src-AS1 inhibitors, none were able to target Src-AS2. We hypothesized that the larger methyl side chain of the alanine gatekeeper decreases the size of the gatekeeper pocket thereby reducing the potential affinity of this hydrophobic interaction. We sought to identify other positions on the staralog scaffold that could be altered to increase inhibitor affinity and allow for targeting of AS kinases bearing an alanine gatekeeper. Since removal of the sugar group from staurosporine led to a significant decrease in potency towards Src-WT (Figure 1C), we asked if restoring one or more of the sugar affinity elements might be key to enhancing staralog potency towards kinases with a larger alanine gatekeeper.
We sought to retain the synthetic tractability of our simplified staralog derivatives while regaining the affinity interactions provided by the carbohydrate portion of staurosporine. We synthesized a new panel of staralogs that retain the C7-iso-butyl element of Star 1 but also contain simple alkylamine groups at either the N12 or N13 positions (Figure 2). These groups were designed to mimic the affinity elements of the staurosporine sugar as revealed by various co-crystal structures and could be installed in one or two synthetic steps (Scheme S2).
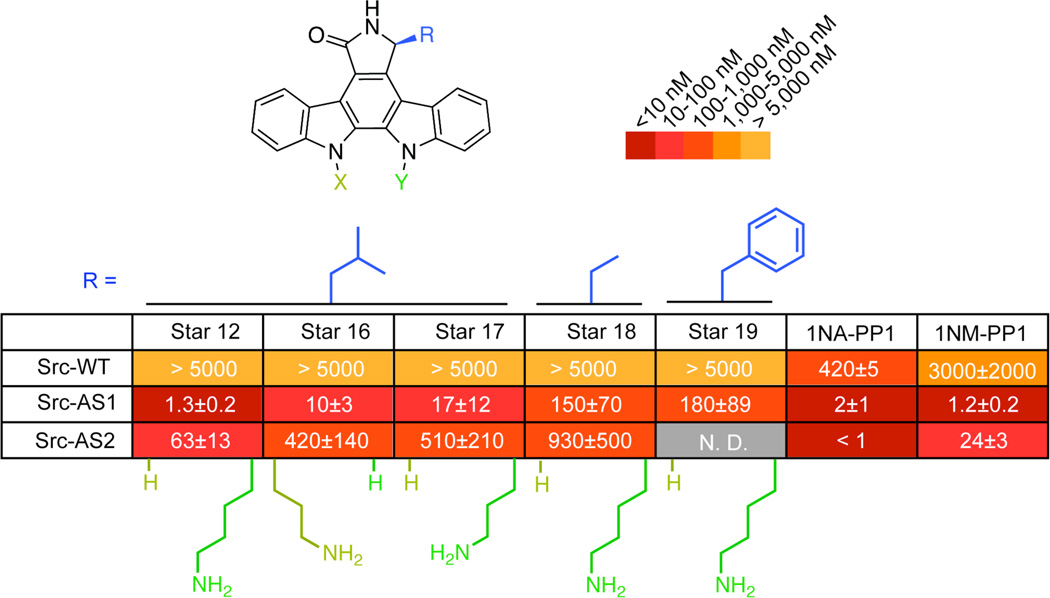
Figure 2.
Panel of staralogs bearing N12 or N13 substituents. IC50 values (nM) against Src-WT, Src-AS1, and Src-AS2. IC50 values were determined from a seven-point dose-response. Each experiment was done in triplicate and repeated for a minimum total of three times for compounds that showed measureable inhibition. Compounds that showed no inhibition were measured twice. Reported values represent the mean and error represents the standard error of the mean. WT = native gatekeeper (Thr for Src), AS1 = Gly gatekeeper, AS2 = Ala gatekeeper
We synthesized derivatives bearing alkyl amines to mimic potential affinity interactions provided by the C4’-methylamine, C3’-methylether, and O1’-cyclic ether of the sugar group. These derivatives retained selectivity for Src-AS1 over the wild-type kinase and some showed modest improvements in potency towards Src-AS2 (Figure 2). The N12-butylamine derivative (Star 12), however, yielded a marked improvement (~ ten-fold) in potency toward both Src-AS1 and Src-AS2, while remaining inactive (IC50 > 5µM) toward Src-WT. Importantly, Star 12 exhibited potency and selectivity that was similar or better than 1NA-PP1 and 1NM-PP1 against Src-AS1 and Src-AS2. Reducing the alkyl linker of Star 12 by one carbon (Star 17) decreased the potency of the inhibitor toward Src-AS2 by more than ten-fold. Such a precise structure-activity relationship suggests the N12-butylamine of Star 12 provides a specific affinity interaction with Src kinase and does not simply alter a general property, such as solubility, of staralogs.
We anticipate that individual AS kinases will exhibit varying sensitivity to the size and shape of the C7-orthogonality element. Thus, we asked if the affinity enhancement of the N12-butylamine group was unique to staralogs bearing the C7-iso-butyl modification or if it would enhance the potency of staralogs with other C7-substitutions. To test this we synthesized two molecules, each bearing the N12-butylamine in combination with a C7-group that is either smaller (ethyl, Star 18) or larger (benzyl, Star 19) than the iso-butyl group of Star 12 (Figure 2). The potency towards Src-AS1 was enhanced five-fold for Star 18 over Star 8 but did not significantly enhance Star 19 over Star 2. The precision of the structure-activity relationship at the N12-position and the enhancement of potency by N12-butylamine in diverse staralogs suggest that this group is a versatile affinity element for staralog inhibitors.
Structural basis of Star 12 binding
To understand the structural mechanism by which the N12-butylamine enhances inhibitor binding we solved a 2.7 Å X-ray co-crystal structure of Star 12 bound to the kinase domain of Src-AS1 (Figure 3, Table S1). In agreement with previously published structures of staurosporine bound to protein kinases, the lactam ring (Ring F) of the indolocarbazole forms hydrogen bond interactions with the backbone amides of E339, Y340, and M341 in the hinge region of the ATP binding pocket (Figure S2A). As predicted, the C7-iso-butyl group projects directly toward the gatekeeper pocket and occupies the cavity created by mutation of the threonine gatekeeper to glycine (Figure 3, S2A). An overlay of the Star 12/Src-AS1 structure with that of staurosporine bound to Src-WT7,14 reveals that Star 12 rings A, B and F occupy similar positions as those of staurosporine. Unexpectedly, the bulky C7-iso-butyl group of Star 12 forces rings E, C and D to project below the plane of staurosporine resulting in a binding mode that appears tilted to minimize steric clash with the gatekeeper pocket (Figure S2B).
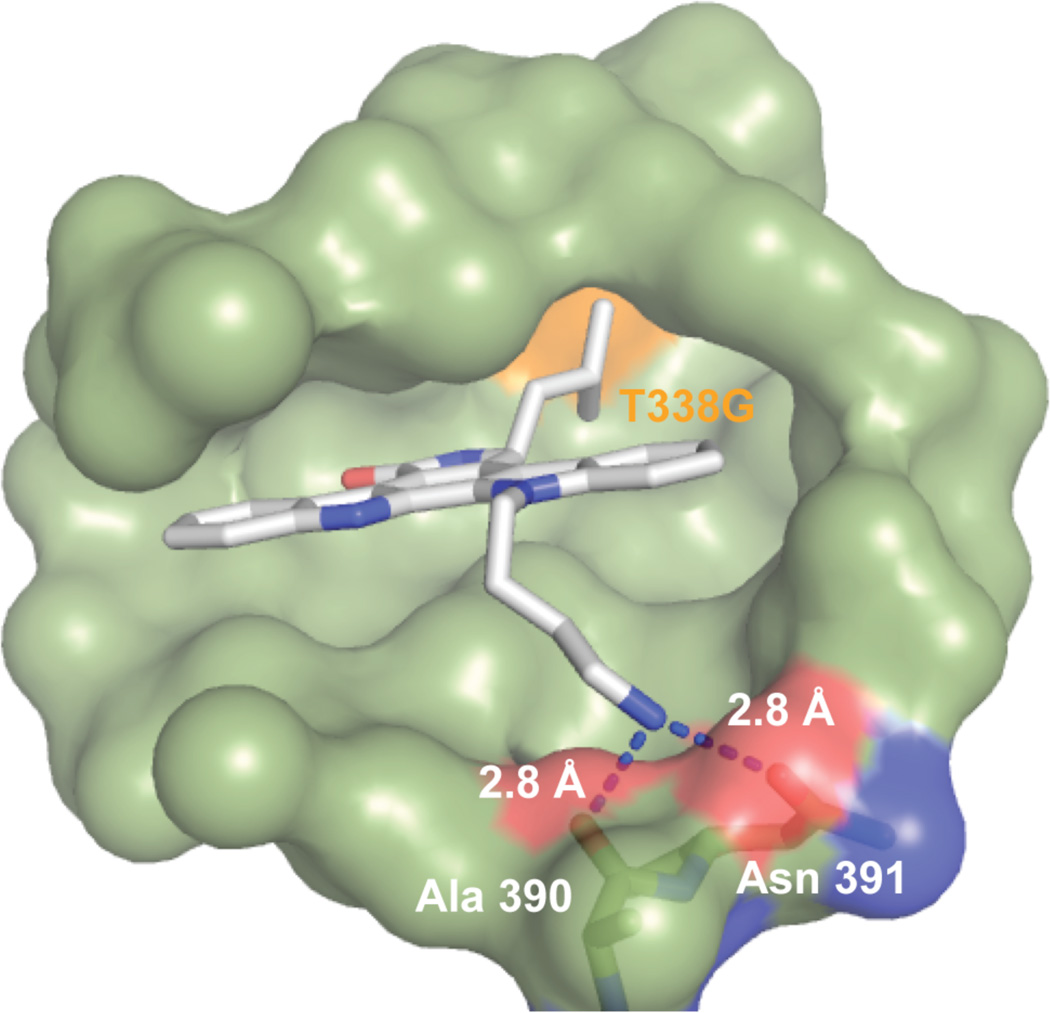
Figure 3.
X-ray co-crystal structure of Star 12 bound to the kinase domain of Src-AS1. The iso-butyl group of Star 12 projects towards the expanded gatekeeper pocket (orange). The N12-butylamine group forms hydrogen bonds with side chain of Asn 391 and the backbone carbonyl of Ala 390.
Despite the predicted flexibility of a fourmethylene linker, clear electron density reveals the N12-butylamine to be rigid and bent, allowing the formation of hydrogen bonds with the side chain of Asn391 and the backbone amide carbonyl of Ala390 (Figure 3). This observation is significant as it may explain why the butylamine group has such a profound influence on inhibitor affinity and why this effect is lost when the alkyl linker is shortened. We had initially installed polar groups at the N12 and N13 positions in an effort to mimic the affinity interactions of the staurosporine sugar. However, the A390/N391 hydrogen bonds observed are not found in any other crystal structure of staurosporine bound to a protein kinase. It is possible that this unique interaction is made possible by the tilted orientation of Star 12 relative to staurosporine. Importantly, Asn391 is completely conserved throughout protein kinases (Figure S3) suggesting this key affinity interaction may be a general feature applicable to staralog binding to any AS kinase.
Staralogs are bioorthogonal AS-kinase inhibitors
The primary goal of developing staralogs is to expand the scope of AS-kinase technology by creating more selective inhibitors that do not target WT-kinases inherently sensitive to PP analogs. To evaluate our progress towards this objective we measured in vitro IC50 values for Star 12 against a panel of PP-sensitive WT-kinases (Table 1). One or more of the three commonly used AS -kinase inhibitors, 1NA-PP1, 1NM-PP1 and 3MB-PP1, inhibit each of the PP-sensitive kinases at concentrations between one and several hundred nanomolar7,9. Strikingly, Star 12 exhibits enhanced selectivity and does not potently target any of theses PP-sensitive WT kinases. The observation that Star 12 is a potent inhibitor of AS kinases but is innocuous towards PP-sensitive WT kinases suggests it may be capable of selectively targeting AS alleles of PP-sensitive kinases, thereby expanding this technology to include a previously intractable subset of enzymes.
Table 1.
IC50 values for Star 12 towards PP-sensitive kinases
| CK1ε | CKδ | Pkd1 | Pkd2 | Ack | EphA1 | Ptk6 | Ret | |
|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | >5,000 | >5,000 | 3,300±1700 | >5,000 | 1,300±340 | >5,000 | >5,000 | >5,000 |
IC50 values were determined from a seven-point dose-response. Each experiment was done in triplicate, repeated once, and the mean values are reported. Error represents the standard error of the mean.
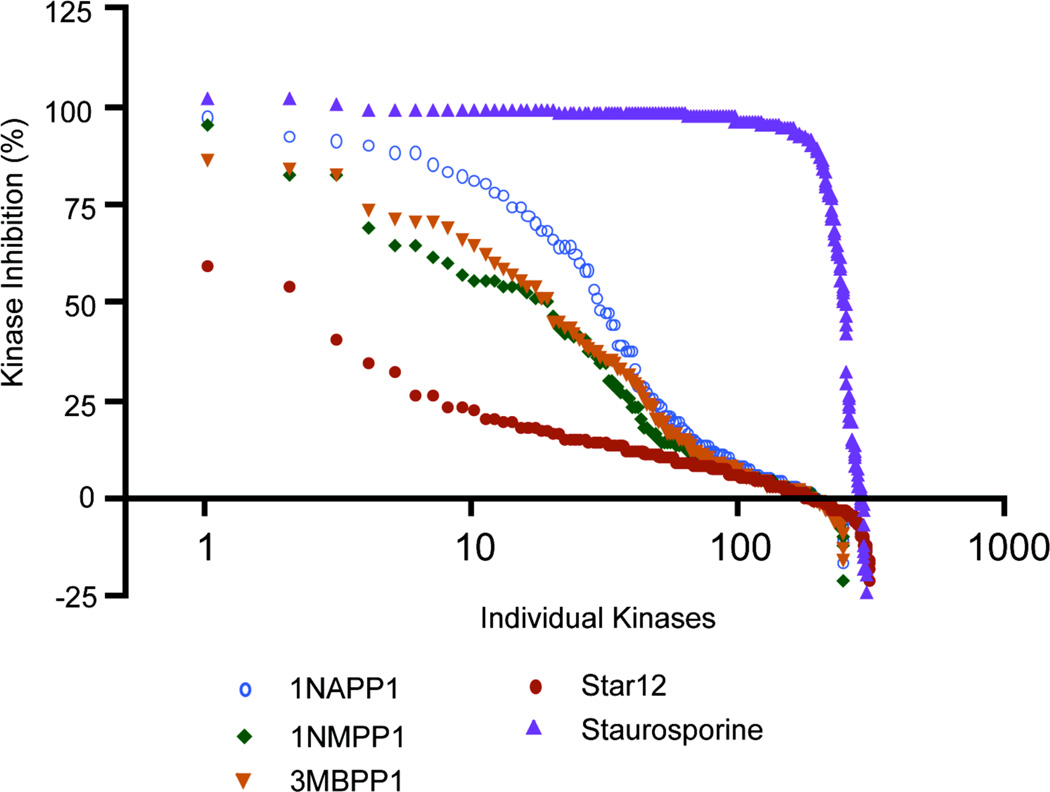
Although Star 12 does not target PP-sensitive WT kinases, we examined the possibility that a new subset of the kinome might be inherently sensitive to C7-modified staralogs. For example, staurosporine was originally reported to be a potent inhibitor of Protein Kinase C7,9, and thus we asked if staralogs might retain activity against some WT kinases with intrinsic sensitivity to a staurosporine-derived molecule. We analyzed 308 human kinases by measuring the fraction of activity inhibited by 1µM Star 12 (Figure 4, Table S2). Strikingly, a concentration of 1 µM Star 12 did not inhibit a single kinase by more than 80% and thus displayed remarkable and unprecedented selectivity for an AS kinase inhibitor. By comparison, 1NA-PP1, 1NM-PP1, and 3MB-PP1 inhibit 4.5%, 1.2% and 1.2% of kinases, respectively7. Using the same metric, staurosporine and K252a target 72% and 49%, respectively, of all protein kinases at a concentration of 500 nM8.
Figure 4.
Inhibition profiles of Star 12 (red), staurosporine (purple), 1NA-PP1 (brown), 1NM-PP1 (orange), and 3MB-PP1 (green) towards ~300 kinases. Kinases were ordered from most-to-least inhibited for each drug and plotted with the X-axis formatted in log-scale.
Staralogs expand the scope of AS kinase technology
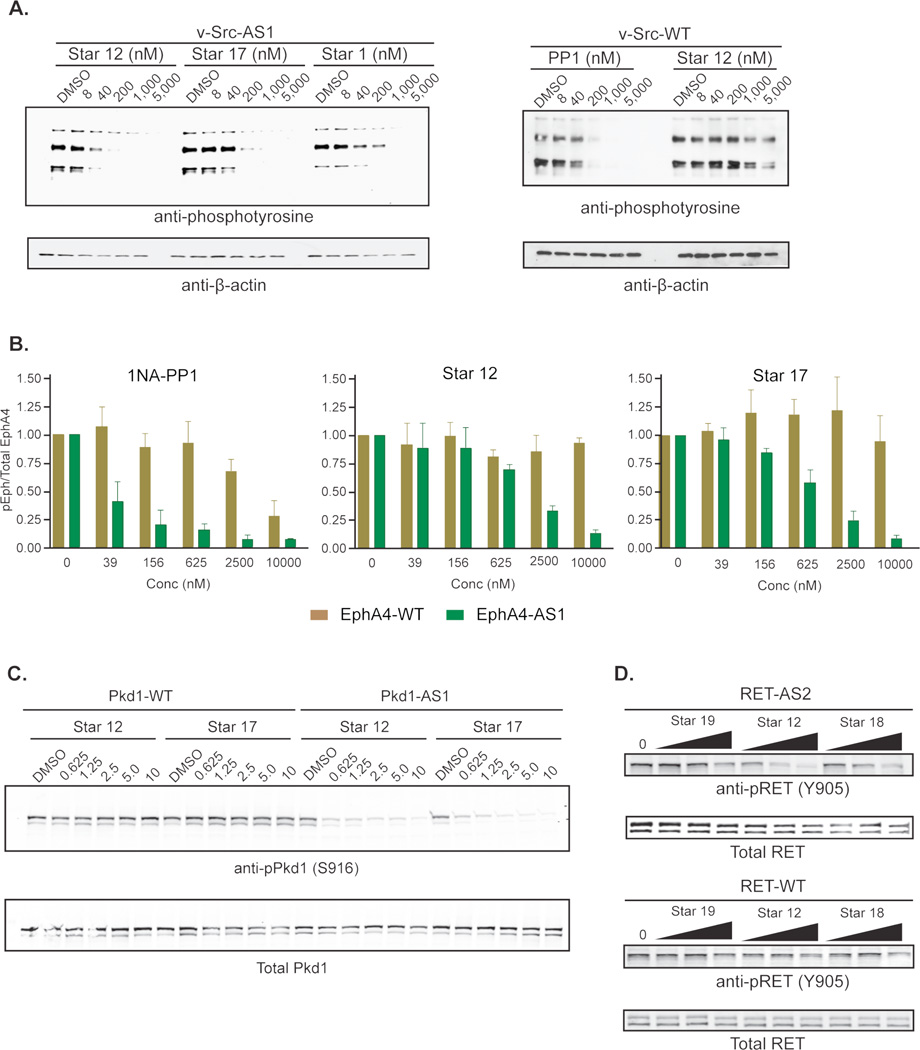
An important application of AS -kinase technology is to decipher the roles of individual kinases in cellular signaling pathways relevant to human diseases. Thus, it is critical a new inhibitor scaffold be membrane-permeable and capable of targeting AS kinases in intact mammalian cells. To determine if staralogs meet this requirement we measured the ability of Star 1, Star 17, and Star 12 to inhibit v-Src-WT or v-Src-AS1 in stably transfected mouse 3T3 cells (Figure 5A). All three staralogs robustly inhibit v-Src-AS1 mediated phosphorylation in cells at concentrations below 1µM. Star 12 was the most potent inhibitor, while Star 17 exhibited intermediate potency, and Star 1 was the weakest. We also directly compared Star 12 and 1NM-PP1 and found they have similar potency against v-Src-AS1 (Figure S4). These observations indicate that the structural changes that enhance inhibitor affinity in vitro do not have counterproductive effects on the cell-permeability of the molecules. Unlike the promiscuous tyrosine kinase inhibitor PP1, Star 12 had little effect on v-Src-WT-mediated phosphorylation in, confirming the selectivity of Star 12 extends to cellular conditions.
Figure 5.
Staralogs are potent and selective inhibitors of AS kinases in cells. (A.) 3T3 mouse cells over-expressing v-Src-AS1 or v-Src-WT were treated with varying concentrations and analyzed for inhibition of tyrosine phosphorylation. (B.) HEK-293T cells were transfected with EphA4-WT or EphA4-AS1, treated with inhibitors and analyzed for EphA4 auto-phosphorylation. Error bars are the SEM of three separate experiments. (C.) HEK-293T cells were transfected with Pkd1-WT or Pkd1-AS1, treated with indicated inhibitor (625nM - 10µM), and analyzed for Pkd auto-phosphorylation at S916. (D.) 3T3 mouse cells expressing Kifb-Ret-AS2 or Kif5b-Ret-WT were treated with the indicated inhibitors (10µM, 3.3µM, and 1.1µM) and analyzed for Ret auto-phosphorylation at Y905.
EphA4 is a receptor tyrosine kinase involved in neuronal development and axon guidance in mammals17,18 and has recently been identified as a potential therapeutic target for the treatment of amyotrophic lateral sclerosis (ALS)17,19. EphA4 belongs to the ephrin (Eph) receptor family of 16 tyrosine kinases, a branch of the kinome that is inherently sensitive to the PP inhibitor scaffold. Some family members, such as EphB1, EphB2, and EphB3, retain a differential of sensitivity for the AS allele over the WT allele that is sufficient to allow the application of our chemical genetic technique7,19. Meanwhile, others, such as EphA4, have eluded AS technology due to their inherent sensitivity to PP inhibitors. To determine if staralogs are capable of extending AS-kinase technology to EphA4, we measured the ability of several staralogs to target EphA4-AS1 in HEK-293T cells (Figure 5B). While Star 12 exhibited activity towards EphA4-AS1, we found that Star 17 was in fact the most potent staralog derivative, inhibiting greater than 90% of AS kinase activity without affecting the WT kinase at a concentration of 10µM (Figure 5B). 1NA-PP1, in contrast, inhibited EphA4-WT activity at concentrations (≥2.5µM) at which inhibition of the AS allele was complete (>90%). Thus, the enhanced selectivity of staralogs may allow for the extension of AS-kinase technology to EphA4 and potentially the entire Eph family of kinases. In particular, staralog inhibitors may be used to target EphA4-AS1 in ALS mouse models to clarify the potential of EphA4 as a therapeutic drug target.
To explore the generality of staralog inhibitors we asked if we could identify potent inhibitors of two other PP-sensitive kinases, Pkd1 and Ret. Although AS versions of these kinases have been successfully targeted with 1NA-PP1, the WT kinases are also weakly inhibited at higher drug concentrations7,20. Thus, staralogs might serve as complementary or superior alternative inhibitors when studying these kinases and their orthologs. Although Star 18, Star 12, and Star 19 did not show activity towards Pkd1-AS2, Star 12 was able to target over-expressed Pkd1-AS1 in HEK-293T cells (Figure S5). Since the C7-isobutyl group is optimal for Pkd-AS1, we next showed that both Star 12 and Star 17 are potent Pkd-AS1 inhibitors and do not affect Pkd1-WT activity (Figure 5C). We also found that Star 12 but not Star 19 is capable of inhibiting Ret-AS2 autophosphorylation in 3T3 mouse cells (Figure S6). Since Ret-AS2 contains a larger alanine gatekeeper, we next examined whether a smaller C7-ethyl group of Star 18 might target this kinase more potently. However, as with our earlier studies with Src-AS2, Star 18 was less active than Star 12 (Figure 5D). Furthermore, Star 18 showed weak activity against Ret-WT. Thus, from a small panel of staralogs we identified potent inhibitors of Src-AS1, Src-AS2, EphA4-AS1, Pkd1-AS1 and Ret-AS2. These molecules represent a versatile inhibitor scaffold that may enable the application of AS-kinase technology to previously intractable PP-sensitive kinases such as EphA4.
Conclusion
Orthogonal small molecule inhibitors of engineered AS kinases are powerful tools for elucidating the cellular roles of individual kinases and for deciphering paradoxical effects of clinical therapeutics20. The potential impact of the AS approach grows commensurately with advances4,21 in gene-editing technology as they allow for replacement of WT-kinase genes with AS alleles in cells and in vivo. The ability to target AS kinases with a selective inhibitor provides the unique advantages of a chemical tool (reversibility, temporal resolution, etc.) while retaining the specificity of a genetic manipulation. An essential feature of AS kinase technology is that the small molecule inhibitor must potently target the engineered kinase at a concentration at which the WT enzyme is unaffected. Although PP inhibitors satisfy this requirement for the majority of kinases, a small subset has been identified that are inherently sensitive to PP inhibitors. In this report we have introduced a panel of (S)-C7-substituted indolocarbazoles, termed staralogs, which overcome the limited selectivity of PP inhibitors and expand the potential scope of AS kinase technology to PP-sensitive kinases such as EphA4. Furthermore, the generality of Star 12 allows for the study of diverse AS kinases without the complications of off-target inhibition of PP-sensitive kinases. For example, Star 12 should be used to target an AS kinase if the experiment requires that CK1ε retain activity since this kinase is potently targeted by PP analogs. Thus staralogs are useful tools for studying AS mutants of PP-sensitive kinases as well as for targeting AS-kinases involved in pathways that may be affected by unintentional inhibition of PP-sensitive native kinases. The inhibitors described here now form a core piece of the repertoire of the chemical tool kit necessary for the chemical genetic investigation of protein kinases.
Supplementary Material
ACKNOWLEDGMENT
We thank Professor Chao Zhang and members of the Shokat laboratory for thoughtful comments and suggestions. M.S.L. is supported by a National Science Foundation Graduate Research Fellowship. MLS is a fellow of the International Association for the Study of Lung Cancer (IASLC) and receives a Young Investigator Award of the Prostate Cancer Foundation (PCF). We kindly thank Dr. Takashi Kohno for providing the KIF5B-RET construct and Professor Michael Greenberg and Dr. Michael Soskis for providing the EphA4-WT and EphA4-AS1 constructs.
ABBREVIATIONS
- AS
Analog-Sensitive
- WT
Wild Type
- ATP
Adenosine Triphosphate
- PP
pyrazolo[3,4-d]pyrimidine
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental procedures, inhibitor profiling data, X-ray crystallography data collection and refinement statistics, and compound characterization. This material is available freely of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 2.Lampson MA, Kapoor TM. Nat. Chem. Biol. 2006;2:19–27. doi: 10.1038/nchembio757. [DOI] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Bishop AC, Kung C, Shah K, Witucki LJ. Am. Chem Soc. 1999;121:627–631. [Google Scholar]
- 5.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 6.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, Mclauchlan H, Klevernic I, Arthur JSC, Alessi DR, Cohen P. Biochem. J. 2007;408:297. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Lopez MS, Dar AC, Ladow E, Finkbeiner S, Yun C-H, Eck MJ, Shokat K. ACS Chem. Biol. 2013;8:1931–1938. doi: 10.1021/cb400376p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Nat. Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamaoki T, Nomoto H, Takahashi I, Kato Y. Biochem. Biophys. Res. Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 10.Wood JL, Petsch DT, Stoltz BM, Hawkins EM. Synthesis. 1999;1:1529–1533. [Google Scholar]
- 11.Link JT, Raghavan S, Danishefsky SJ. J. Am. Chem. Soc. 1995;111:552–553. [Google Scholar]
- 12.Wood JL, Stoltz BM, Dietrich HJ. J. Am. Chem. Soc. 1995;117:10413–10414. [Google Scholar]
- 13.Tanramluk D, Schreyer A, Pitt WR, Blundell TL. Chem Biol Drug Des. 2009;74:16–24. doi: 10.1111/j.1747-0285.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z-B, Chaudhary D, Olland S, Wolfrom S, Czerwinski R, Malakian K, Lin L, Stahl ML, Joseph-McCarthy D, Benander C, Fitz L, Greco R, Somers WS, Mosyak L. J. Biol. Chem. 2004;279:50401–50409. doi: 10.1074/jbc.M409216200. [DOI] [PubMed] [Google Scholar]
- 15.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Nat. Struct. Mol. Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garske AL, Peters U, Cortesi AT, Perez JL, Shokat KM. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15046–15052. doi: 10.1073/pnas.1111239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, Peeters E, Philips T, Goris A, Dubois B, Andersen PM, Al-Chalabi A, Thijs V, Turnley AM, van Vught PW, Veldink JH, Hardiman O, Van Den Bosch L, Gonzalez-Perez P, Van Damme P, Brown RH, van den Berg LH, Robberecht W. Nat. Med. 2012;18:1418–1422. doi: 10.1038/nm.2901. [DOI] [PubMed] [Google Scholar]
- 18.Egea J, Nissen UV, Dufour A, Sahin M, Greer P, Kullander K, Mrsic-Flogel TD, Greenberg ME, Kiehn O, Vanderhaeghen P, Klein R. Neuron. 2005;47:515–528. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 19.Soskis MJ, Ho H-YH, Bloodgood BL, Robichaux MA, Malik AN, Ataman B, Rubin AA, Zieg J, Zhang C, Shokat KM, Sharma N, Cowan CW, Greenberg ME. Nat. Neurosci. 2012;15:1645–1654. doi: 10.1038/nn.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Nat. Chem. Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joung JK, Sander JD. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.