Abstract
P450s are heme thiolate enzymes that catalyze the regio- and stereoselective functionalization of unactivated C-H bonds using molecular dioxygen and two electrons delivered by the reductase. We have developed hybrid P450 BM3 heme domains containing a covalently attached Ru(II) photosensitizer in order to circumvent the dependency on the reductase and perform P450 reactions upon visible light irradiation. A highly active hybrid enzyme with improved stability and a modified Ru(II) photosensitizer is able to catalyze the light-driven hydroxylation of lauric acid with total turnover numbers of 935 and initial reaction rate of 125 mol product / mol enzyme / min.
The field of direct C-H functionalization has seen great advances in recent decades with diverse emerging approaches,1 including the use of photoredox catalysts,2 to activate rather inert bonds toward the construction of complex molecular scaffolds. Nevertheless, selective functionalization of unactivated C-H bonds using molecular dioxygen as the sole oxidant still remains one of the hallmarks of catalysis.3 While few synthetic examples of both homogeneous and heterogeneous catalysts are emerging, enzymatic processes continue to be of considerable importance. In particular, the Cytochrome P450 superfamily of heme-thiolate enzymes, which catalyze a myriad of chemical reactions often with high regio and stereo selectivity, has received great attention.4 Activation of molecular dioxygen at the heme center generates a high-valent Fe(IV)-oxo porphyrin radical species, namely Compound I, capable of performing the desired C-H bond functionalization.5 Among the large and diverse family of P450 enzymes, the fatty acid hydroxylase CYP102A1 or P450 BM3 from Bacillus megaterium shows the highest catalytic hydroxylation rate ever measured for P450 enzymes.6 Its high catalytic activity is attributed to the efficient coupling between the delivery of electrons from the reductase and the activation of oxygen at the heme center. The singularity of P450 BM3 is that the NADPH-dependent reductase and the heme domain are fused together in a single peptide chain. The fused reductase in conjunction with the high catalytic activity have rendered P450 BM3 an effective model to study P450 monooxygenase and an important design platform for biocatalysis.6 However, in order for P450 BM3 and P450s in general to become valuable biocatalysts, certain challenges such as the requirement of the expensive NAD(P)H cofactor and the dependency on redox partners must be addressed.7
Current strategies to overcome these limitations include the use of NAD(P)H recycling systems,8 surrogate oxygen atom donors9 as well as chemical,10 electrochemical,11 enzymatic12 and light-activated systems13 to deliver the reducing equivalents to the heme domain. The most successful examples have achieved high P450 catalytic activity using co-factor regeneration systems, fusion P450 systems, and electrochemistry. The vast majority of these approaches are, however, still dependent on native or non-physiological redox partners for high activity. This dependency is limiting their broader application due to the specificity of each reductase.12 On the other hand, very few alternative approaches that use only the P450 heme domain,10 including those using terminal oxidants9b,11e–f result in efficient biocatalysts despite successful heme reduction achieved in certain cases.11a
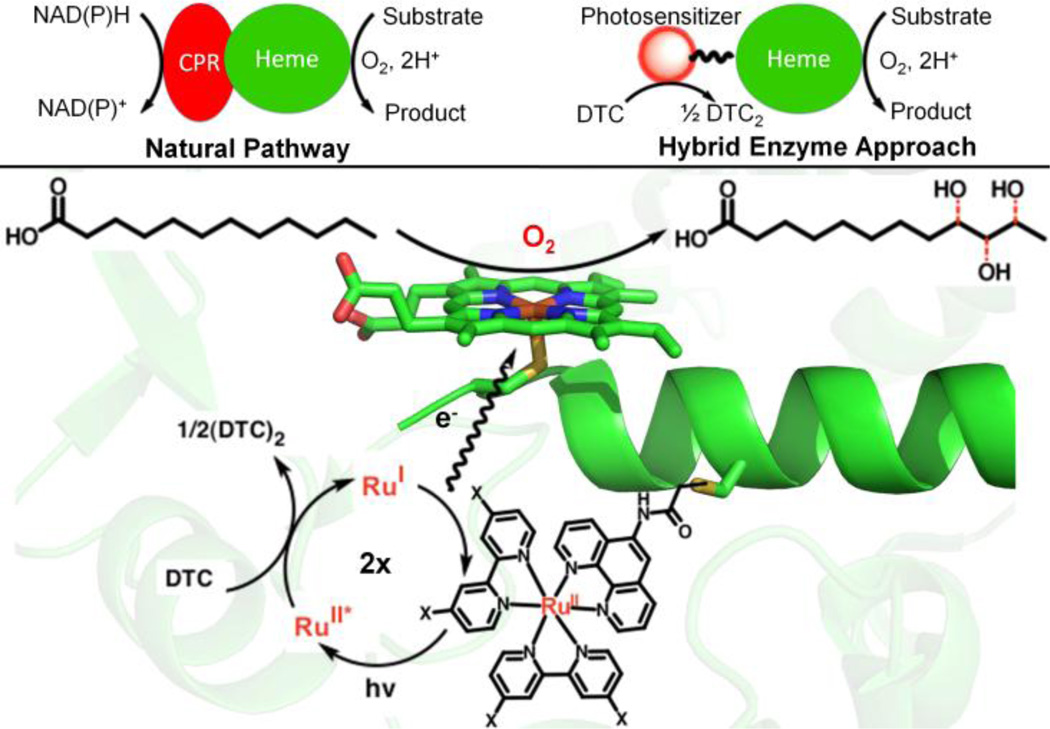
Our laboratory has focused on utilizing the photochemical properties of Ru(II)-diimine complexes14 covalently attached to P450 BM3 heme domain mutants to perform selective light-driven hydroxylation reactions (Figure 1).15 A light-activated system capable of harvesting the synthetic potential of P450 enzymes is of particular interest considering the current efforts dedicated to the utilization of light energy to drive chemical reactions. Herein, we report an optimized hybrid enzyme able to achieve, upon light activation, total turnover numbers of over 900 with initial reaction rate of 125 mol product / mol enzyme / min, among the highest values reported for alternative electron delivery approaches in the hydroxylation of lauric acid.9b, 11c–d, 13a Besides the use of light activation to drive P450 reactions, the hybrid enzyme approach circumvents the use of reductase and NAD(P)H cofactor (Figure 1).
Figure 1.
Schematic representation of the hybrid enzyme approach vs. the natural pathway involving the redox partner (CPR) and NAD(P)H (Top). (Bottom) Close up of the active site where the Ru(II) photosensitizer (X=H (1) or OMe (2)) is covalently attached to a non-native single cysteine (L407C) residue of P450 BM3 heme domain. Under flash quench reducing conditions (DTC: sodium diethyldithiocarbamate), the photogenerated Ru(I) species is able to provide electrons to the heme center in order to activate molecular dioxygen and carry out the hydroxylation of lauric acid, yielding three mono-hydroxylated products.
Briefly, the hybrid enzyme15 can be assembled by attaching Ru(II)-diimine complexes of general formula Ru(LL)2PhenA, (LL = bipyridine (bpy) and phenA = 5-acetamido-1,10-phenanthroline) to a strategically positioned non-native single cysteine residue (e.g. L407C) of P450 BM3 mutant heme domains. Under the photocatalytic cycle highlighted in Figure 1,16 the photogenerated Ru(I) is able to provide the necessary electrons to the heme domain in successive electron transfer steps and sustain photocatalytic activity. A series of hybrid P450 BM3 enzymes15 was generated by varying the nature and position of the Ru(II)-diimine photosensitizer to optimize the electron transfer rate according to the semi-classical Marcus theory.16,17 The most active tL407C-1 mutant having the Ru(bpy)2PhenA complex (1) covalently attached to the tL407C heme domain triple mutant (Table 1) showed modest total turnover numbers of 140. This L407C label attachment site has been the focus of our studies since, and changes in the nature of the photosensitizer as well as optimizations through protein engineering have led to a highly active P450 hybrid enzyme.
Table 1.
Mutations and half denaturation temperatures of the heme domains used in this study
| Enzymes | Mutations [a] | T50 (°C) [b] |
|---|---|---|
| hBM3 | Wild-type (C62, C156) | 58.1 ± 0.2 |
| sL407C | L407C | 56.4 ± 0.1 |
| tL407C | C62A, C156S, L407C | 51.8 ± 0.2 |
Residues in bold indicate position of photosensitizer covalent attachment;
T50 values are the mean ±SD of triplicate measurements.
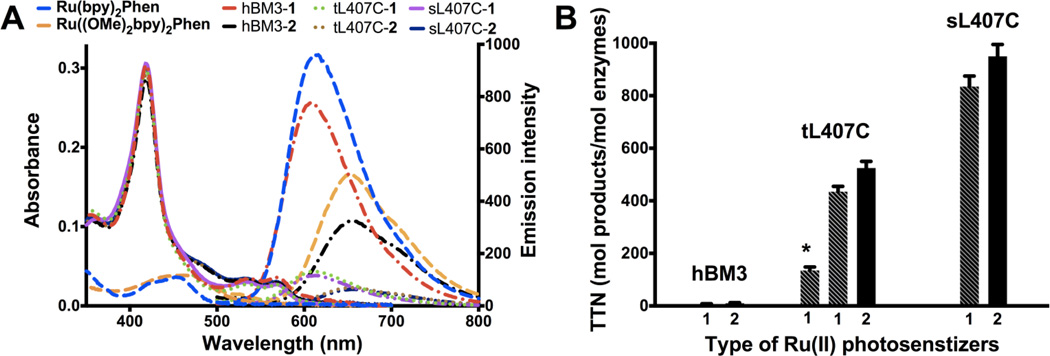
The nature of the Ru(II) photosensitizer was modified to increase the driving force of the electron transfer steps. The 4,4’-dimethoxy-2,2’-bipyridine ligand ((OMe)2bpy) containing the electron-donating methoxy groups was used instead of the bipyridine (used in 1). This substitution led to a 80 mV cathodic shift in the first reduction potential of Ru((OMe)2bpy)2Phen (E2+/1+= −1.27 V vs Ag/AgCl) compared to −1.19 V vs Ag/AgCl for Ru(bpy)2Phen (Figure S3). The labeled enzymes containing Ru((OMe)2bpy)2PhenA (2) also show a red-shifted metal-to-ligand charge transfer (MLCT) absorption and emission18 compared to their counterparts having the initial photosensitizer (1) as illustrated in Figure 2A.
Figure 2.
Photophysical properties and catalytic activities of the different hybrid enzymes. A) UV-vis and luminescence spectra of the different hybrid enzymes (hBM3: wild-type P450 BM3 heme domain, triply (tL407C) and singly (sL407C) mutated P450 BM3 heme domains; 1: Ru(bpy)2PhenA and 2: Ru((OMe)2bpy)2PhenA) and model complexes (Ru(bpy)2Phen and Ru((OMe)2bpy)2Phen) at the same concentration; B) Plot showing the total turnover numbers (TTN) as mol of hydroxylated products / mol of enzymes for the various hybrid enzymes in the light-activated hydroxylation of lauric acid. From left to right, the TTN range from 8 and 9 ± 1 for the hBM3-1 and -2, to 140 ± 5 (*,for previously reported tL407C-1), 438 ± 15 and 548 ± 22 for tL407C-1 and -2; and 856 ± 23, 935 ± 27 for sL407C-1 and -2. Error bars indicate standard deviations.
So far, P450 BM3 heme domain mutants with a single non-native cysteine residue were engineered in order to control the position of attachment of the Ru(II) photosensitizer.19 The library of triple mutants was originally generated where the two native cysteine residues, C62 and C156, were mutated to alanine and serine, respectively, and a non-native single cysteine residue was inserted by site-directed mutagenesis.19 Investigation into the thermostability of the enzymes revealed that the triply mutated heme domain, tL407C, has lower thermal stability (T50 = 52°C) compared to the wild-type P450 BM3 heme domain (hBM3, T50 = 58°C) (Table 1).20 This prompted us to reintroduce the two original cysteine residues in the L407C mutant in order to regain some of the stability and catalytic activity of the wild-type heme domain. The C62 residue appears to be involved in hydrogen bonding interactions with neighboring residues K391 and N395 (Figure S10) and likely plays a role in the enzyme stability. The resulting singly mutated sL407C has a T50 of 56°C, which is closer to the T50 observed for the hBM3 (Table 1).
The hBM3 and sL407C enzymes were then submitted to the same labeling reaction conditions. The hBM3 could be singly labeled in low yield (<10% after extended reaction time) at the more reactive C156 residue21 whereas the sL407C is singly labeled at the preferred L407C position in a much higher labeling yield (>95 %, similar to that observed with the tL407C mutant). The labeling of the heme domain mutants is verified by ESI-MS, UV-vis and luminescence measurements (Figure 2A and Supporting Information). The position of attachment of the photosensitizer was confirmed by Chymotrypsin digestion of the hybrid enzymes (Figure S7).
Significant differences in total turnover numbers (TTN) are observed (Figure 2B) when exposing a solution of the purified hybrid enzymes containing excess substrate and reductive quencher to visible light irradiation (with UV- and IR-cutoff filters). The TTN are determined as mol of hydroxylated products per mol of enzymes after 2 hours, after which no furthersignificant turnover was observed. The mono-hydroxylated products at subterminal positions (ω1, ω2, ω3) are identified and quantified, after derivatization, by gas chromatography-mass spectrometry (GC-MS) analysis. The ratio of hydroxylated products in the photocatalytic reaction is identical to that observed with the wild-type heme domain and values reported for NADPH/holoenzyme.6, 15 No catalytic turnover is observed in the absence of visible light irradiation or reductive quencher.15
Under optimal reaction conditions (1.5 µM hybrid enzymes, 1.5 mM lauric acid, and 100 mM sodium diethyldithiocarbamate), the labeled sL407C enzymes display a 100 fold increase in TTN (Figure 2B) in the photocatalytic hydroxylation of lauric acid compared to the labeled hBM3 enzymes (856 vs. 8 and 935 vs. 9 with labels 1 and 2, respectively), emphasizing the importance of the site of label attachment. The sL407C shows 7-fold higher TTN compared to the previously reported value for tL407C, which resulted from optimization of the protein expression conditions23 with a higher heme incorporation (Rz ratio,24 A418/A280, greater than 1.5) and greater thermostability of the heme domain achieved by reinserting the two native cysteines. In addition, ligand substitution in the photosensitizer resulted consistently in a 10 to 25% increase in TTN.
Initial reaction rates were measured as turnover numbers obtained after 1 min of visible light irradiation and range from 62 min−1 for the tL407C-2 to 125 min−1 for the sL407C-2 at 1.5 mM lauric acid. These catalytic rates are significantly improved relative to the apparent specific activity reported previously for the tL407C-1 (~1 min−1).15b The hybrid enzymes follow apparent Michaelis-Menten kinetics under the optimal flash–quench reductive conditions. The kcat and Km parameters were determined by measuring initial reaction rates at various substrate concentrations (Figure S9). The kcat for the tL407C-2 hybrid enzyme is 79 min−1 with a Km of 357 µM. For the sL407C-2, the kcat is 130 min−1 and the Km is 199 µM. The Km values are consistent with other values determined for the full-length wild-type enzyme with lauric acid.6
P450 BM3 is regarded as the benchmark P450 enzyme with its high catalytic rates and coupling efficiency largely due to its fused reductase. A number of alternative approaches have been developed to mimic such efficient system and artificially drive P450 BM3 and various other P450s.8–13 As shown in Table 2, the light activated sL407C-2 hybrid enzyme (entry 2) compares favorably in terms of TTN and reaction rate with the current P450 BM3 alternative approaches to hydroxylate lauric acid. NAD(P)H recycling systems8 and studies using other model substrates10, 11a–b have been omitted from the table for the sake of direct comparison. The entry 1 of the table is the full-length wild-type enzyme (holoBM3) with NADPH as source of reducing equivalents, showing initial reaction rates ranging from 81 to 4100 min−1. Such large discrepancies in the reaction rates have already been noted elsewhere.6 The other entries (from 3–10), listed in decreasing TTN order, include the electrochemical delivery of electrons via mediators (entries 3, 6, 7 and 9), the light-activated deazaflavin (entry 4) and the peroxide shunt (entries 5, 8). In general, low activity is usually observed using only the heme domain (entries 5, 7–9), and significant activity improvement is obtained using the full-length enzyme (holoBM3) with the fused reductase. The peroxide shunt (entry 8) typically lead to rapid protein degradation, which had been minimized through protein engineering in the more stable 21B3 heme domain. To the best of our knowledge, the observed total turnover numbers (935) and initial reaction rate (125 min−1) for the sL407C-2 are the highest among the existing alternative electron delivery approaches, even those taking advantage of the linked reductase.
Table 2.
Summary of the current data (TTN and reaction rates) on the alternative P450 BM3 strategies to perform the selective hydroxylation of lauric acid
| Entry | System | Enzyme[a] | Total turnover numbers (TTN) |
Initial reaction rate (mol product / mol enzyme / min) |
Reference |
| 1 | NADPH | holoBM3 | n/a[b] | 81.1± 2.7 – 4100 ± 150 | 6,22 |
| 2 | hν/Ru(II) photosensitizer | sL407C-2 | 935 ± 27 | 125 ± 4 | this work |
| 3 | Co(III) sepulchrate/electrode | holoBM3 | 835 ± 7 | 37.8 ± 0.3 | 11c |
| 4 | hν/deazaflavin/EDTA[c] | holoBM3 | 698 | 1.95 | 13a |
| 5 | H2O2 | 21B3 | 280 | 50 | 9 |
| 6 | Cobaltocene/electrode | holoBM3 | 224 ± 7 | 16.4 ± 0.6 | 11c |
| 7 | Co(III) sepulchrate/electrode | hBM3 | 76 ± 7 | 2.2 ± 0.1 | 11c |
| 8 | H2O2 | hBM3 | 70 | 10 | 9b |
| 9 | Cobaltocene/electrode | hBM3 | 58 ± 7 | 1.8 ± 0.5 | 11c |
| 10 | Pt electrode | holoBM3 | n/a[b] | 110 | 11d |
full-length wild-type enzyme (holoBM3), heme domain (hBM3) and 21B3 heme domain variant;
n/a : not available;
EDTA : ethylenediaminetetraacetic acid.
In comparison with other alternative P450 systems, the hybrid enzyme approach offers the advantage of achieving high catalytic activity upon light activation using only the heme domain, circumventing the need for redox partners and NAD(P)H cofactors. This approach has therefore the potential to be more generally applicable to other P450 isoforms, especially those lacking a fused reductase. In the hybrid enzyme, the high initial reaction rates and TTN indicate that oxygen activation is coupled to electron transfer from Ru(II) photosensitizers strategically positioned at the proximal heme side where the reductase is thought to interact (Figure S11).25 In conclusion, improvement in the stability of the hybrid enzymes with a more native-like structure and a change in nature of the Ru(II) photosensitizer have resulted in a sL407C-2 system able to efficiently catalyze the light-driven hydroxylation of lauric acid. The use of Ru(II) photosensitizer in the hybrid enzymes is becoming a valuable approach to perform P450 reactions upon light activation, expanding the scope of photocatalytic reactions carried out by enzymatic systems containing polypyridyl Ru(II) complexes.26
Supplementary Material
ACKNOWLEDGMENT
This work was financially supported by the National Institute of Health (GM095415), the Research Corporation for Science Advancement. L.C. thanks the National Science Foundation (MRI grant 0923573) and San José State University for the use of mass spectrometry facilities.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental section; photosensitizers and hybrid enzymes characterizations (1H NMR, ESI-MS, Chymotrypsin digest, GC-MS, cyclic voltammetry). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.(a) Yu J-Q, Shi Z, editors. Top. Curr. Chem. 2010;292:1. [PubMed] [Google Scholar]; (b) Crabtree RH. Chem. Rev. 2010;110:575. doi: 10.1021/cr900388d. and references therein. [DOI] [PubMed] [Google Scholar]; (c) Wencel-Delord J, Glorius F. Nat. Chem. 2013;5:369. doi: 10.1038/nchem.1607. [DOI] [PubMed] [Google Scholar]; (d) White MC. Science. 2012;335:807. doi: 10.1126/science.1207661. [DOI] [PubMed] [Google Scholar]
- 2.Prier CK, Rankic DA, MacMillan DWC. Chem. Rev. 2013;113:5322. doi: 10.1021/cr300503r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roduner E, Kaim W, Sarkar B, Urlacher VB, Pleiss J, Gläser R, Einicke W-D, Sprenger GA, Beifuß U, Klemm E, Liebner C, Hieronymus H, Hsu S-F, Plietker B, Laschat S. Chem-CatChem. 2013;5:82. [Google Scholar]
- 4.(a) Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem. Rev. 2005;105:2253. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]; (b) Urlacher VB, Girhard M. Trends Biotechnol. 2012;30:26. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]; (c) Fasan R. ACS Catal. 2012;2:647. [Google Scholar]; (d) Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. Trends Biochem. Sci. 2013;38:140. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]; (e) Coelho PS, Brustad EM, Kannan A, Arnold FH. Science. 2013;339:307. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- 5.Rittle J, Green MT. Science. 2010;330:933. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 6.Whitehouse CJC, Bell SG, Wong LL. Chem. Soc. Rev. 2012;41:1218. doi: 10.1039/c1cs15192d. [DOI] [PubMed] [Google Scholar]
- 7.O'Reilly E, Kohler V, Flitsch SL, Turner NJ. Chem. Commun. 2011;47:2490. doi: 10.1039/c0cc03165h. [DOI] [PubMed] [Google Scholar]
- 8.(a) Van der Donk WA, Zhao H. Curr. Opin. Biotechnol. 2003;14:421. doi: 10.1016/s0958-1669(03)00094-6. [DOI] [PubMed] [Google Scholar]; (b) Hollmann F, Hofstetter K, Schmid A. Trends Biotechnol. 2006;24:163. doi: 10.1016/j.tibtech.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 9.(a) Chen MM, Coelho PS, Arnold FH. Adv. Synth. Catal. 2012;354:964. [Google Scholar]; (b) Cirino PC, Arnold FH. Angew. Chem. Int. Ed. 2003;42:3299. doi: 10.1002/anie.200351434. [DOI] [PubMed] [Google Scholar]
- 10.Nazor J, Dannenmann S, Adjei RO, Fordjour YB, Ghampson IT, Blanusa M, Roccatano D, Schwaneberg U. Protein Eng. Des. Sel. 2008;21:29. doi: 10.1093/protein/gzm074. [DOI] [PubMed] [Google Scholar]
- 11.(a) Sadeghi SJ, Fantuzzi A, Gilardi G. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:237. doi: 10.1016/j.bbapap.2010.07.010. [DOI] [PubMed] [Google Scholar]; (b) Krishnan S, Wasalathanthri D, Zhao LL, Schenkman JB, Rusling JF. J. Am. Chem. Soc. 2011;133:1459. doi: 10.1021/ja108637s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Udit AK, Arnold FH, Gray HB. J. Inorg. Biochem. 2004;98:1547. doi: 10.1016/j.jinorgbio.2004.06.007. [DOI] [PubMed] [Google Scholar]; (d) Estabrook RW, Faulkner KM, Shet MS, Fisher CW. Methods Enzymol. 1996;272:44. doi: 10.1016/s0076-6879(96)72007-4. [DOI] [PubMed] [Google Scholar]; (e) Estavillo C, Lu Z, Jansson I, Schenkman JB, Rusling JF. Biophys. Chem. 2003;104:291. doi: 10.1016/s0301-4622(02)00383-6. [DOI] [PubMed] [Google Scholar]; (f) Rudakov YO, Shumyantseva VV, Bulko TV, Suprun EV, Kuznetsova GP, Samenkova NF, Archakov AI. J. Inorg. Biochem. 2008;102:2020. doi: 10.1016/j.jinorgbio.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Sadeghi SJ, Gilardi G. Biotechnol. Appl. Biochem. 2013;60:102. doi: 10.1002/bab.1086. [DOI] [PubMed] [Google Scholar]
- 13.(a) Zilly FE, Taglieber A, Schulz F, Hollmann F, Reetz MT. Chem. Commun. 2009;46:7152. doi: 10.1039/b913863c. [DOI] [PubMed] [Google Scholar]; (b) Jensen K, Jensen PE, Moller BL. ACS Chem. Biol. 2011;6:533. doi: 10.1021/cb100393j. [DOI] [PubMed] [Google Scholar]; (c) Girhard M, Kunigk E, Tihovsky S, Shumyantseva VV, Urlacher VB. Biotechnol. Appl. Biochem. 2013;60:111. doi: 10.1002/bab.1063. [DOI] [PubMed] [Google Scholar]; (d) Ipe BI, Niemeyer CM. Angew. Chem. Int. Ed. 2006;45:504. doi: 10.1002/anie.200503084. [DOI] [PubMed] [Google Scholar]
- 14.Balzani V, Bergamini G, Campagna S, Puntoriero F. Top. Curr. Chem. 2007;280:1. [Google Scholar]
- 15.(a) Tran NH, Huynh N, Bui T, Nguyen Y, Huynh P, Cooper ME, Cheruzel LE. Chem. Commun. 2011;47:11936. doi: 10.1039/c1cc15124j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tran NH, Huynh N, Chavez G, Nguyen A, Dwaraknath S, Nguyen TA, Nguyen M, Cheruzel L. J. Inorg. Biochem. 2012;115:50. doi: 10.1016/j.jinorgbio.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray HB, Winkler JR. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3534. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcus RA, Sutin N. Biochim. Biophys. Acta. 1985;811:265. [Google Scholar]
- 18.Anderson PA, Keene FR, Meyer TJ, Moss JA, Strouse GF, Treadway JA. Dalton Trans. 2002;20:3820. [Google Scholar]
- 19.Ener ME, Lee YT, Winkler JR, Gray HB, Cheruzel L. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18783. doi: 10.1073/pnas.1012381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar O, Cirino PC, Arnold FH. ChemBioChem. 2003;4:891. doi: 10.1002/cbic.200300660. [DOI] [PubMed] [Google Scholar]
- 21.Sevrioukova IF, Hazzard JT, Tollin G, Poulos TL. J. Biol. Chem. 1999;274:36097. doi: 10.1074/jbc.274.51.36097. [DOI] [PubMed] [Google Scholar]
- 22.(a) Cowart LA, Falck JR, Capdevila JH. Arch. Biochem. Biophys. 2001;387:117. doi: 10.1006/abbi.2000.2246. [DOI] [PubMed] [Google Scholar]; (b) Noble MA, Miles CS, Chapman SK, Lysek DA, Mackay AC, Reid GA, Hanzlik RP, Munro AW. Biochem. J. 1999;339:371. [PMC free article] [PubMed] [Google Scholar]
- 23.Pflug S, Richter SM, Urlacher VB. J. Biotechnol. 2007;129:481. doi: 10.1016/j.jbiotec.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 24.O’Keeffe DH, Ebel RE, Peterson JA. Methods Enzymol. 1978;52:151. doi: 10.1016/s0076-6879(78)52017-x. [DOI] [PubMed] [Google Scholar]
- 25.(a) Tripathi S, Li HY, Poulos TL. Science. 2013;340:1227. doi: 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]; (b) Sevrioukova IF, Li HY, Zhang H, Peterson JA, Poulos TL. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1863. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Roth LE, Nguyen JC, Tezcan FA. J. Am. Chem. Soc. 2010;132:13672. doi: 10.1021/ja1071866. [DOI] [PubMed] [Google Scholar]; (b) Roth LE, Tezcan FA. J. Am. Chem. Soc. 2012;134:8416. doi: 10.1021/ja303265m. [DOI] [PubMed] [Google Scholar]; (c) Reisner E, Powell DJ, Cavazza C, Fontecilla-Camps JC, Armstrong FA. J. Am. Chem. Soc. 2009;131:18458. doi: 10.1021/ja907923r. [DOI] [PubMed] [Google Scholar]; (d) Simaan AJ, Mekmouche Y, Herrero C, Moreno P, Aukauloo A, Delaire JA, Réglier M, Tron T. Chem. Eur. J. 2011;17:11743. doi: 10.1002/chem.201101282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.