Abstract
Polycomb Group (PcG) proteins maintain the silent state of developmentally important genes. Recent evidence indicates that non-coding RNAs also play an important role in targeting PcG proteins to chromatin and PcG mediated chromatin organization, although the molecular basis for how PcG and RNA function in concert remains unclear. The Phe-Cys-Ser (FCS) domain, named for three consecutive residues conserved in this domain, is a 30 - 40 residue Zn2+ binding motif found in a number of PcG proteins. The FCS domain has been shown to bind RNA in a non-sequence specific manner, but how it does so is not known. Here, we present the three dimensional structure of the FCS domain from human Polyhomeotic homolog 1 (hPh1) determined using multi-dimensional NMR methods. Chemical shift perturbations upon the addition of RNA and DNA resulted in the identification of Lys 816 as a potentially important residue required for nucleic acid binding. The role played by this residue in Polyhomeotic function was demonstrated in a transcription assay carried out in Drosophila S2 cells. Mutation of the Arg residue to Ala in the Drosophila Polyhomeotic (Ph) protein, that is equivalent to hPh1 Lys 816, was unable to repress transcription of a reporter gene to the level of wild-type Ph. These results suggest that direct interaction between the Ph FCS domain and nucleic acids is required for Ph mediated repression.
The Polycomb Group (PcG) is a family of gene silencing proteins that repress important developmental regulator genes, including homeotic (HOX) genes. Once the expression of these genes is no longer required, the PcG proteins maintain the silent state over many cell divisions. In stem cells, the PcG represses genes that promote differentiation thus playing an important role in maintaining the pluripotency of these cells (1, 2).
The PcG functions at the level of chromatin, although the precise mechanism by which PcG complexes bind to chromatin has not been fully elucidated. While the presence of MBT and chromo domains within PcG proteins can allow direct association with methylated histones, DNA not bound to histones is also important for PcG function. For example, the Drosophila PcG protein Pleiohomeotic (Pho), the only PcG protein that houses a specific DNA binding domain, acts in concert with the multi-protein PcG complex called Polycomb Repression Complex 1 (PRC1) to associate with specific gene regulatory sequences called Polycomb response elements (PREs) (3, 4). Pho is also a component of a different PcG complex called PhoRC which includes the PcG protein dSfmbt (5). A single complex containing both a specific DNA binding protein (Pho) and ability to bind methylated histone through the MBT domains of dSfmbt (6) may utilize a combinatorial approach to bind specific chromatin sites that have both the Pho binding element and methylated histones.
RNA is also emerging as an important player in PcG-mediated repression. PRC2, a multi-protein PcG complex that catalyzes the tri-methylation of histone H3 K27, associates with a non-coding RNA (ncRNA) called Xist facilitating the inactivation of the X chromosome in female mammals (7). This interaction occurs between a region within Xist consisting of 7.5 repeats of a 28-nucleotide sequence (RepA) and the PRC2 component Ezh2, resulting in the proper targeting of PRC2 to the inactive X (Xi) chromosome (8). RING1B, a PRC1 component, is targeted to the Xi chromosome in a manner that is dependent on Xist but independent of PRC2 (9). Recent reports have revealed that many other ncRNAs can associate with PRC2. Long ncRNAs HOTAIR and Kcnq1oT1 are associated with PRC2 and, like Xist, play an important role in targeting the methyl transferase activity of PRC2 to specific chromatin locations (10, 11). Moreover, a global analysis of ncRNA interactions revealed that approximately 20% of all mammalian ncRNAs are associated with PRC2 (12).
RNAi related proteins have also shown to be involved in PcG mediated repression (13). The Drosphila RNAi proteins Dicer-2, PIWI and Argonaute were found to co-localize with PRC1 components Polycomb (Pc) and Polyhomeotic (Ph) and were required to maintain long range chromosomal interactions. Interestingly, small transcripts from an exogenous PRE were detected. These small RNAs appear to be required for maintaining the long-range interactions because mutant flies unable to make the transcripts could not mediate these interactions.
A domain found in a number of PcG proteins that is a candidate for directly binding RNA is the Phe-Cys-Ser (FCS) domain. In vitro, the FCS domain can bind both RNA and DNA independent of the sequence of the nucleic acids (14). The focus of that study was a C. elegans PcG protein called SOP-2. The RNA binding ability of SOP-2 allows its localization into large nuclear bodies often referred to as “PcG bodies” (13, 15). PcG bodies are thought to correspond to sites where clustering of PREs and gene silencing occur through long-range interactions between the PREs. SOP-2 does not contain an FCS domain but rather utilizes a different sequence of amino acids for RNA association. The conserved RNA binding function of the FCS domain and the RNA binding region of SOP-2 was demonstrated by a SOP-2 chimera that replaced its RNA binding regions with the mouse Ph homolog FCS domain which was able to partially restore the ability to localize into PcG bodies (14).
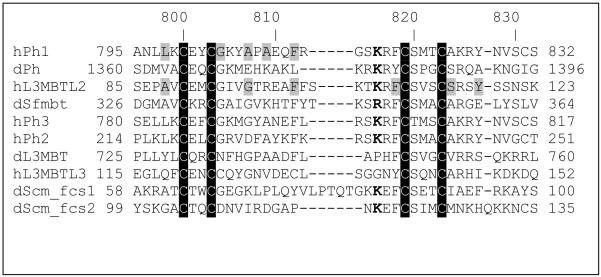
The FCS domain is a 30 - 40 residue sequence able to chelate a Zn2+ atom via four conserved Cys residues (Figure 1). A number of PcG proteins contain an FCS domain, including: all Ph orthologs; Drosophila Sex comb on midleg (Scm) where two FCS domains are present as a tandem repeat; Drosophila Sfmbt; and Drosophila lethal 3 malignant brain tumor (L3MBT) along with two of its related mammalian proteins L3MBT-like 2 (L3MBTL2) and L3MBTL3. A recent structure of the FCS domain from the human L3MBTL2 determined using multi-dimensional NMR methods revealed an architecture that is compatible with RNA binding (16). In this structure, three mostly conserved, positively charged residues are on one face of the structure and were suggested as potential functional residues that could bind the negatively charged backbone of RNA. Alternative to a nucleic acid binding role, a recent study has implicated the Scm and the dSfmbt FCS domains to be involved in protein-protein interactions (6).
Figure 1.
ClustalW (42) alignment of FCS sequences. The metal binding Cys residues are highlighted in black. The light shaded amino acids are buried residues (> 75% buried surface area) in the two FCS domain structures that have been determined. The “d” and “h” suffixes in the protein names indicate Drosophila melanogaster or human proteins, respectively. The two tandem FCS domains in the Drosophila Scm protein are indicated by fcs1 and fcs2.
In order to gain insight into the molecular basis of Ph, and also PcG function, we have both biochemically characterized and determined the solution structure of the FCS domain from human Ph homolog 1 (hPh1). Our results strongly suggest that hindering the ability of the Ph FCS domain to bind nucleic acids also hinders the ability of Ph to repress transcription.
Experimental Procedures
Protein preparation
The hPh1 FCS domain (residues 783-828) was cloned into a modified pET-3c (Novagen) plasmid containing an N-terminal hexahistidine tag followed by a TEV protease cleavage site. The hPh1 FCS domain was expressed in BL21-Gold (DE3) cells (Stratagene) that had been pre-transformed with the pRARE plasmid (Novagen). Bacterial cells from a 1 L culture were resuspended in 10 ml of 50 mM Tris pH 8.0, 100 mM NaCl, 25 mM imidazole pH 7.5, 1 mM PMSF, 10mM β-mercaptoethanol and 5% glycerol. Cells were lysed by sonication, and purified using Ni2+ affinity chromatography. The eluted protein was digested with TEV to remove the N-terminal sequence followed by a second Ni2+ affinity chromatography where the non-binding fractions were collected. The protein was further purified by ion exchange chromatography. The final polypeptide after TEV cleavage contained an N-terminal GTR sequence followed by the hPh1 residues.
UV/visible spectroscopy
The bacterially expressed and purified hPh1 FCS was stripped of metal by lowering the pH through the addition of trifluoroacetic acid and then purified by reversed-phase HPLC. In an anerobic environment, the lyophilized, metal-free hPh1 FCS domain was resuspended in 100 mM Hepes, 50 mM NaCl, pH 7.0 to a final concentration of 66.7 M. One equivalent of CoCl2 was added followed by the visible spectrum measurement. One equivalent of ZnCl2 was then added to the solution in order to observe any bleaching of the spectrum.
NMR spectroscopy: structure determination, dynamics and chemical shift perturbations
Stable isotope labeled protein samples (containing either 15N alone or both 15N and 13C) were expressed in minimal media containing isotopically labeled substrates, purified as described above and prepared to 1.5 mM in 10 mM NaPO4 pH 6.0, 50 mM NaCl. Fractionally labeled (10% 13C) samples were prepared in minimal media containing a ratio of 0.3:2.7 g/L of 13C labeled:unlabeled glucose. 2D 13C −1H HSQC spectra of biosynthetic fractionally [13C, 1H] labeled protein was used for stereospecific assignment of the side-chain methyl groups (valine, leucine) (17, 18) and the aromatic (Phe and Tyr) groups (19). NMR experiments were performed at 300 K using Bruker 600 MHz and 700 MHz spectrometers fitted with either conventional (700 MHz) or cryogenically cooled (600 MHz) 5 mm 1H probes equipped with 13C and 15N decoupler and pulsed field gradient coils. All spectra were processed and analyzed with NMRPipe (20) and NMRView (21). Structure calculations were performed with CNS 1.1 (22) using ARIA 1.2 (23), incorporating NOEs and TALOS (24) calculated dihedral angle restraints. The structure was refined using the 3JHNHα couplings, RDCs, hydrogen bonds, and the tetrahedral Zn2+ geometry restraints (Table 1). Backbone amide HSQC based longitudinal (T1) and transverse (T ) relaxation times and heteronuclear Overhauser effects ({1 2 H}-15N NOE) were recorded using standard Bruker pulse programs. Time scale estimations of the hPh1 FCS internal conformational dynamics were determined by analyzing the 15N relaxation parameters using the Lipari-Szabo model-free formalism (25, 26) in the program ModelFree 4.0 (http://cpmcnet.columbia.edu/dept/gsas/biochem/labs/palmer/software/modelfree.html). The chemical shift assignments have been deposited to the BMRB (accession number is 17396) and the RCSB PDB (RCSB ID: rcsb102085). The coordinates have been deposited to RCSB PDB (PDB ID: 2L8E).
Table 1.
Structural restraints and statistics for 10 lowest energy structure of hPh1 FCS domain
| Structural Restraints | |
| Total NOE distance restraints | 1024 |
| NOE distance restraints | |
| Intra residual (∣i-j∣=0 ) | 374 |
| Sequential (∣i-j∣=1 ) | 285 |
| Short range (2<∣i-j∣<5) | 146 |
| Long range (∣i-j∣>5) | 220 |
| Dihedral restraints (extracted from TALOS) (24) | |
| φ | 26 |
| ψ | 26 |
| RDC restraints | |
| DNH | 28 |
| DCH | 28 |
| Coupling restraints | |
| 3JHNHα | 32 |
| Hydrogen bond restraints | 9 |
| Zinc coordinate restrains | 6 |
| Structural Statistic | |
| RMS deviations from the ideal geometry (±SD) | |
| Bond lengths | 0.0046 ± 0.00024 |
| Bond angles | 0.8473 ± 0.039 |
| Improper angels | 2.125 ± 0.019 |
| Average atomic RMSD from mean structure (±SD) | |
| Residues (795-813, 815-828) (backbone) | 0.46 |
| Residues (795-813, 815-828) (heavy atoms) | 1.05 |
| Structure evaluation (Ramachandran plot) | |
| (residues 783-828) | |
| Residues in most favored regions | 62.4% |
| Residues in additional allowed regions | 30.4% |
| Residues in generally allowed regions | 5.2% |
| Residue in disallowed regions (Asp793) | 2.0% |
For the measurement of the chemical shift perturbations to the hPh1 FCS domain in the presence of nucleic acids, the following oligonucleotides were used: 34nt RNA: 5′-GGGCACGCGUAUUGCCCUAGUGGCCGGCGUGCCC-3′ 14bp dsDNA: 5′-CAGCCATATGGCTG-3′ with its reverse complement. All oligonucleotides were placed into the same buffer as the hPh1 FCS domain prior to the titration experiment. The hPh1 FCS peptide was titrated with increasing amounts of nucleic acids. A {1H}-15N HSQC spectrum was collected after each titration point. The weighted average chemical shift changes of the assigned residues were calculated using the equation Δav = sqrt ((Δδ2NH + Δδ2N/25)/2).
The dissociation constant (Kd) was estimated by globally fitting the shifts for different residues to a common Kd value using the following equation as described by Lian and Roberts (27):
Δδobs is the observed difference in weighted average chemical shifts between the free state and that observed at each titration point; ( b - f) is the total weighted average chemical shift difference between the bound and free states; PT and LT are the total concentrations of protein and ligand (RNA), respectively. Pro Fit software (QuantumSoft) was used for the nonlinear least-squares fitting.
Transcription assay
On day one of the assay, the following plasmids were transfected into 1 × 105 Drosophila S2 cells using the Fugene HD transfection reagent (Roche Applied Science): 1) 100 ng of the Drosophila Ph expression plasmid under control of a constitutive actin 5c promoter (kind gift from Dr. Albert J. Courey); 2) 7.5 ng of a lacZ gene expression plasmid, also under control of the actin 5c promoter (kind gift from Dr. Yuzuru Shiio); and 3) 7.5 ng of pGL2-Basic with three tandem repeats of the zif268 DNA binding sites cloned immediately upstream of a metallothionine promoter (MTp) that controls expression of the luciferase gene. On day three, the expression of the luciferase gene was induced by adding CuSO4 (100 M). On day four, cells were harvested and lysed using 100 mM potassium phosphate buffer pH 7.8, 0.2% Triton X-100 and 0.5 mM DTT. For all individual transfections, equal volumes of lysate were used for the Dual-Light Combine Reporter Gene Assay System (Applied Biosystems) to measure both luciferase and -galactosidase activities. The data are presented as the ratio of the two enzyme activities.
Results
Tetrahedral coordination of metal by the hPh1 FCS domain
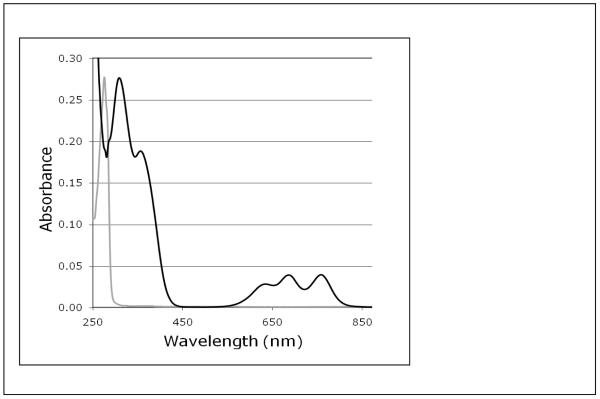
The ability of the FCS domain to bind Zn2+ was originally suggested based on the presence of four conserved Cys residues whose sulfide groups were predicted to be the chelating residues for the zinc metal. If so, the four thiolates would be arranged in a tetrahedral geometry around the Zn2+. In order to utilize the tetrahedral geometric constraints in our structure calculations, we needed to first confirm that the FCS domain does indeed bind Zn2+ with tetrahedral geometry. We measured the UV/visible spectrum of hPh1 FCS domain (residues 783 - 828 of hPh1) fully substituted with Co2+. The d-d electron transitions of the Co2+ that is ligated through four ligands arranged tetrahedrally produces a distinct spectrum while the Zn2+ bound FCS domain is spectroscopically silent. The solution of the Co2+ bound hPh1 FCS domain displays a clear blue color with three distinct d-d transition peaks at 635, 686, and 759 nm in the visible spectrum (Figure 2). In addition, the Cys-S to Co2+ charge transfer bands are observed between 275 - 450 nm. This spectrum resembles the UV/visible spectrum of other Cys4-coordinated Zn2+ binding peptides when bound to Co2+(28) including a treble-clef zinc finger peptide that is structurally similar to the L3MBTL2 FCS domain (29). Our results are consistent with a structure in which the hPh1 FCS domain does indeed bind Zn2+ with tetrahedral geometry through four cysteine bonds. Upon addition of Zn2+, all cobalt transitions were bleached as a consequence of Zn2+ substitution for Co2+.
Figure 2.
UV/Visible spectrum of the hPh1 FCS domain bound to Co2+. Black spectrum is the spectrum with Co2+ bound and gray is the spectrum after the addition of ZnCl2.
NMR structure determination of the hPh1 FCS domain
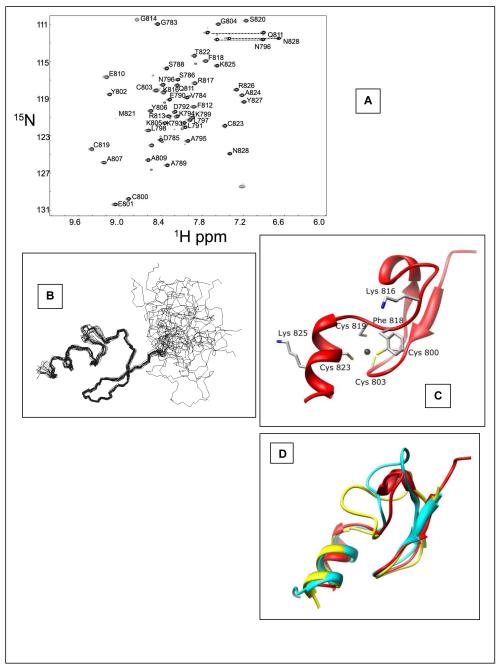
Our initial {1H}-15N HSQC measurement of the hPh1 FCS domain showed a spectrum with well-dispersed pattern of backbone amide signals whose total number was nearly equal to the anticipated value (Figure 3A). We, thus, proceeded to prepare a 13C and 15N labeled hPh1 FCS sample in order to assign the backbone and side-chain resonances and to determine the three-dimensional structure. The structural restraints used and statistics are summarized in Table 1. The ensemble of the 10 calculated lowest energy structures consistent with the NMR structural restraints is shown in Figure 3B. The hPh1 residues 783 - 795 of our construct appear disordered as this region lacks sufficient structural restraints required to define a consistent structure. The ordered region of the hPh1 FCS structure (Asn 797 - Asn 828; Figure 3C) consists of an anti-parallel beta sheet (Asn 797 - Pro 808), followed by a loop region (Ala 809 - Ser 820), then an alpha helix (Met 821 - Asn 828). The Zn2+ binding residues Cys 800 and Cys 803 are in the beta sheet region, Cys 819 is in the loop, and Cys 823 in the alpha helix. As with other similar zinc binding peptides (29), zinc binding appears to be critical for maintaining the fold of the FCS domain as the Zn2+ chelation is responsible for determining the orientation of the hPh1 FCS alpha helix with respect to the beta sheet. The structure of the hPh1 FCS domain closely resembles that of the FCS domain of L3MBTL2 (16) (r.m.s.d. 1.7 Å2 over 28 C atoms) and the E. coli YacG zinc binding domain (30) (r.m.s.d 2.3 Å2 over 29 Ca atoms), with the greatest variation in structures occurring in the loop region of the structures (Figure 3D).
Figure 3.
Solution structure determination of the hPh1 FCS domain. A. hPh1 FCS {1H}-15N HSQC. The chemical shift assignment for each residue is indicated. B. Ensemble of the 20 lowest energy structures of the hPh1 FCS domain. The RMSD of secondary structure backbone is 0.46. C. Ribbon diagram of the hPh1 FCS domain. The four metal binding residues are highlighted along with other residues which were mutated for the transcription assay. D. Overlay of hPh1 FCS (red), L3MBTL2 FCS (yellow, PDBID: 2W0T) (16), and the bacterial YacG peptide (cyan, PDBID: 1LV3) (30). The overlay shows the greatest variability of this fold occurs in the loop region.
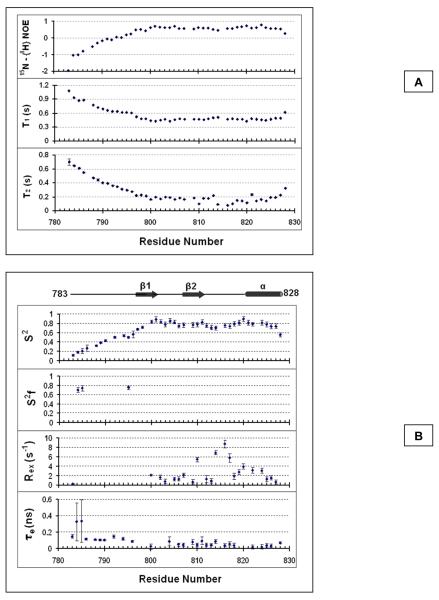
hPh1 FCS dynamics indicates an ordered loop region
Aside from the disordered N-terminal region of our construct, the central loop region (Ala 809 - Ser 820) within the FCS domain exhibited increased RMSDs among the ensemble of low energy structures as compared to the regions of defined secondary structure (Figure 3B). As the restraint density is lower in this region compared to the regions of regular secondary structure (18 restraints per residue as compared to 27 in the beta sheet and alpha helix), it is unclear whether the increased RMSD is due to this or intrinsic flexibility of the loop. We investigated this in greater detail by measuring the T1, T2, and {1H}-15N NOE (HNNOE) relaxation parameters (Figure 4A). The long T1 and T2 times along with the low HNNOE values for hPh1 FCS N-terminal residues 783 - 797 indicate significant flexibility in this region on the ps – ns time scale, consistent with disorder of this region. We conducted a more detailed analysis of the molecular motions of the FCS domain using the Lipari-Szabo extended model-free formalism (25, 26) in order to distinguish motions on different time scales (Figure 4B). hPh1 residues 783 – 797, as expected, have low S2 and high e values indicating that this region is much more flexible on the ps – ns time scale than the rest of the structure. The loop, residues 809 – 820, appears to be rigid on the ps – ns time scale as it exhibits comparably high S2 values. However, several residues in the loop, including Glu 810, Gly 814, Lys 816, and Arg 817, exhibit elevated Rex values indicating greater mobility of these residues on the s – ms time scale. Interestingly, the residue with the highest Rex and, thus, greatest motion in the s – ms time scale is Lys 816, a residue that was identified as playing an important role in nucleic acid recognition (see below). We conclude from these results that the loop is rigid on the ps - ns timescale, but flexible on the s - ms timescale.
Figure 4.
Backbone conformational dynamics of the hPh1 FCS domain. A. The longitudinal (T1), transverse (T2), and {1H}-15N NOEs. The errors in individual T1 and T2 measurements were estimated by Monte Carlo simulations. B. Lipari-Szabo model-free results calculated from 15N T1, 15N T2 and {1H}-15N NOE data. The modeling was performed at τ = 2.85 ns. The calculated Lipari-Szabo S2, S2f, τe and Rex parameters are shown. Residues that could not be fit to a specific motional model were not assigned S2f, τe and Rex values.
Identification of nucleic acid binding residues of hPh1 FCS domain
The Ph FCS domain can bind nucleic acids in a non-sequence specific manner (14). In the hPh1 FCS domain used in this study, there are several positively charged residues within the ordered hPh1 FCS domain as well as two in the disordered N-terminus. These residues could contact the negatively charged phosphate backbone of nucleic acids. Moreover, there are three mostly conserved basic residues (Lys 816, Arg 817, and Lys 825) that are clustered together on both the hPh1 FCS and the L3MBTL2 FCS structures (16), suggesting a potential conserved role for these residues. If indeed the function of the FCS domain is to bind nucleic acids, albeit independent of the nucleic acid sequence, then it can be predicted that a particular set of FCS domain residues, and not a random collection of positively charged residues, would be more inclined to perform this function.
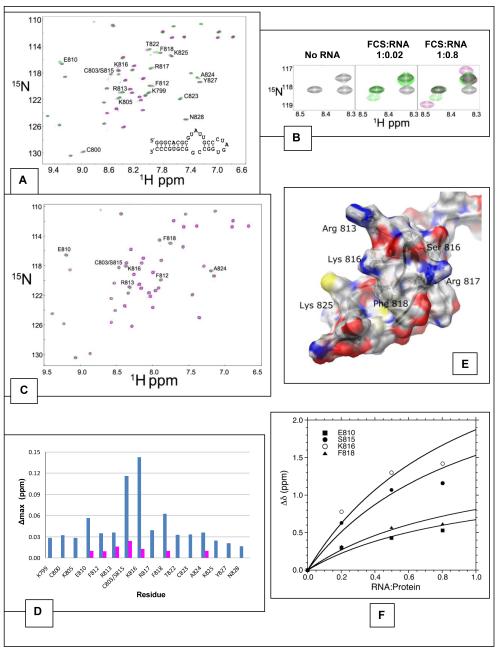
To identify the residues that contact nucleic acids, we measured the perturbations of the hPh1 FCS backbone amide resonances upon addition of RNA and DNA. Because the FCS domain can bind RNA and DNA with no sequence specificity, we arbitrarily chose a 34nt RNA predicted to form a stem-bulge-loop structure (Figure 5A, inset) and a 14-base pair double stranded DNA. A {1H}-15N HSQC spectrum was acquired at several titration points for each of the nucleic acid titration experiments (Figure 5). The hPh1 FCS domain titration with the 34nt RNA showed a number of residues whose HSQC signals were altered, but which were not appreciably broadened and thus falling under the fast exchange regime (Figure 5A). Most notable were the Lys 816, Phe 818 and either Cys 803 or Ser 815 (the identity of this backbone amide signal could not be discerned because these two residues are degenerate) (Figure 5B). Titration with the 14 bp double-stranded DNA was less remarkable than titration with the 34nt RNA (Figure 5C). However, all the residues whose signals were altered in the presence of the DNA are also perturbed with RNA (Glu 810, Phe 812, Arg 813, Lys 816, Phe 818). All these residues are in the loop region of the hPh1 FCS domain. The results of these titration experiments are analyzed by the backbone N-H maximum weighted average shift chemical differences (Figure 5D). A charged surface representation of the FCS structure (Figure 5E) clearly shows the close proximity of the loop residues whose chemical shifts show the greatest perturbation. Lys 825, which was previously pointed out as being part of a potential nucleic acid binding surface on the L3MBTL2 FCS structure (16) but does not appear to be involved in binding, is quite distant from the loop. From these experiments we conclude the FCS domain does indeed use a specific set of residues to contact nucleic acids and that the loop region appears to be especially important for binding.
Figure 5.
Nucleic acid titration of the hPh1 FCS domain. A. Overlay of the HSQC spectra for hPh1 FCS titrated with the 34nt RNA. The black, green and purple HSQC spectra correspond to the addition of 0, 0.2, 0.8 molar equivalents, respectively, of the 34nt RNA to the 15N labeled hPh1 FCS domain. Inset: predicted secondary structure of the 34nt RNA used in the titration. B. The expanded view of the section of HSQC that includes the backbone amide signals for Cys 803/Ser 815 and Lys 816. These two signals show the largest perturbation in the presence of the 34nt RNA. Unfortunately, the signals for Cys 803 and Ser 815 could not be discerned. C. Overlay of the spectra for hPh1 FCS titrated with 0, 0.5 and 2 molar equivalents (black, green and purple, respectively) of the 14 bp dsDNA. Residues that show perturbation in the presence of the DNA are highlighted. D. Backbone N-H weighted average chemical shift differences of the hPh1 FCS domain upon binding the 34nt RNA (blue) and the 14bp dsDNA (purple). The maximum weighted average chemical shift difference, Δmax, was calculated from chemical shift values obtained from the {1H}-15N HSQC spectra collected in the absence of nucleic acids and the highest, saturated, concentration. E. Charged surface representation of the hPh1 FCS. The structure is oriented similarly to the view in Figure 3C. Several of the loop residues as well as Lys 825 are highlighted. F. hPh1 FCS/34nt RNA binding curves of the four residues that show the largest chemical shift perturbation in the titration experiment. The data were fit to the equation described by Lian and Roberts (27) (see methods).
The affinity between the hPh1 FCS and the 34nt RNA was calculated using the equation described by Lian and Roberts (27) (see methods for equation). This equation relates the observed chemical shift difference (Δδobs) to the dissociation constant (Kd) and the total concentrations of the protein and RNA (ligand), PT and LT, respectively. The maximum weighted average chemical shift changes of four residues with the greatest changes to their chemical shifts (Glu 810, Ser 815, Lys 816 and Phe 818) were plotted as a function of the total concentrations of protein and RNA concentrations resulting in an estimated Kd of 3.0 ± 1.1 mM (Figure 5F). The changes in chemical shifts were insufficient to estimate a similar Kd with the 14bp DNA. This likely indicates a weaker affinity for the 14bp DNA due to insufficient amounts of the intact FCS/DNA complex allowable for detection of perturbed chemical shifts.
Mutation of the nucleic acid binding residues hinders Ph transcription repression ability
We next determined what the consequence was on gene silencing of disturbing the potential Ph FCS domain interaction with nucleic acids. For this purpose, we utilized a transcription assay carried out in Drosophila S2 cells using the full-length Drosophila Ph (dPh) protein. There is abundant evidence showing Drosophila PcG proteins function in a very similar manner as their mammalian counterparts. Most compellingly, mammalian PcG proteins have been used to rescue Drosophila phenotypes resulting from mutant orthologs (31, 32, 33) including dPh (34). In our transcription assay, we fused the DNA binding domain of human zif268 to the N-terminus of a variety of dPh proteins in order to target the zif268 fused chimeric proteins to three zif268 binding sites immediately upstream of the metallothionine promoter (MTp) that controls expression of the luciferase gene. Based on the results of our in vitro binding studies with the hPh1 FCS domain, we introduced four different mutations into the dPh FCS domain and determined the ability of these mutant dPh proteins, compared to wild-type dPh, to repress expression of the luciferase gene. The four individual mutations we introduced into dPh were Lys1380Ala (equivalent to hPh1 Lys 816), Tyr1382Ala (equivalent to hPh1 Phe 818), Cys1383Ala (equivalent to hPh1 Cys 1389), and Arg1389Ala (equivalent to hPh1 Lys 825). Cys 1383 is a metal binding residue and would be expected to disrupt the structure of the FCS domain. Arg 1389 is equivalent to hPh1 Lys 825 and is mostly conserved as a positively charged residue in the sequence alignment (Figure 1). However, unlike the other conserved positively charged residue hPh1 Lys 816, Lys 825 displays minimal HSQC signal perturbation upon addition of RNA and DNA (Figure 5). This residue was, therefore, chosen as a control for the mutation experiments, anticipating that if nucleic acid binding is required for Ph mediated repression, then a Lys825Ala mutant would not be defective in its repressive abilities.
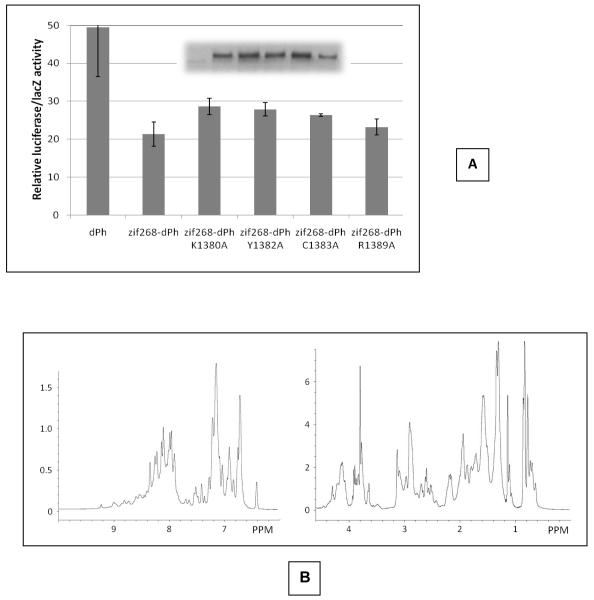
The results of the transcription assay are shown in Figure 6. Zif268 fused wild-type dPh and mutant dPh proteins exhibit substantial repression activity compared to dPh not fused to zif268 and thus not targeted to the MTp, indicating that dPh can repress transcription with a non-functional FCS domain. However, some of the mutants showed a slight but significant reduction in their repressive abilities. Lys1380Ala, Tyr1382Ala and Cys1383Ala, mutations expected to disrupt the structure or nucleic acid binding ability of the FCS domain, showed diminished ability to repress transcription compared to the levels of wild-type. Intriguingly, dPh Cys1383Ala, which we expected to fully destabilize the FCS domain, was slightly better at repressing transcription than dPh Lys1380Ala, though it is within the standard deviation. One possibility for this outcome is that the mutation does not fully unfold the FCS structure. To determine whether this was the case, we introduced the equivalent mutation onto the hPh1 FCS domain, Cys819Ala, and measured its 1D 1H NMR spectrum. To our surprise, hPh1 FCS Cys819Ala does appear to possess some tertiary structural features as evidenced by the presence of both up and downfield chemical shifts (Figure 6B). Additionally, the stability of the FCS domain may increase further inside the nucleus upon binding nucleic acids. The Arg1383Ala mutant, whose hPh1 Lys 825 counterpart appears to play a lesser role in nucleic acid binding (Figure 5), showed no difference in repression ability compared to wild-type. Although Lys 825 is not directly binding nucleic acid, it may simply contribute by providing a more positive charge environment for nucleic acid binding. These results, which nicely correlate our in vitro nucleic acid binding studies with the transcription repression assay carried out in S2 cells, further confirm the important role played by Lys 1380 (or hPh1 Lys 816) in nucleic acid binding which is required for full repression mediated by dPh.
Figure 6.
Transcription repression assay. A. Error bars show the standard deviation of the results from three independent transfections. The inset shows an immunoblot against the Flag epitope tagged Ph proteins demonstrating relative equal amounts of the zif368 fused Ph proteins expressed in the S2 cells. This experiment has been repeated several times and in every case, the results have been consistent with what is shown. B. 1D 1H NMR spectrum of the hPh1 FCS Cys819Ala mutant.
Discussion
The key accomplishments of our experiments are the following: 1) determination of the three-dimensional structure of the hPh1 FCS domain using multi-dimensional NMR methods; 2) identification of the FCS domain loop region, in particular hPh1 Lys 816, as being important for binding RNA; and 3) demonstrating that the in vitro nucleic acid binding ability of the Ph FCS domain correlates with the ability of Ph to repress transcription.
Two different NMR experiments indicate that the loop region of the FCS domain is important for nucleic acid binding. The perturbations of the hPh1 FCS HSQC signal in the presence of RNA are greatest for the loop residues. hPh1 Lys 816, along with the conserved Phe 818, shows substantial movement of their resonances in the presence of the 34nt RNA. The signals of other nearby loop residues (Phe 812, Arg 813 and Arg 816) also showed perturbations in the presence of nucleic acids (Figure 5). In addition to the movement of the backbone amide signals in the HSQC spectra, it is also interesting that our backbone conformational dynamics study indicated that residues in the loop exhibit higher mobility on the s – ms time scale compared to the beta sheet or alpha helix regions of the structure (Figure 4B). Such flexibility within the loop of the FCS domain could be utilized to bind to specific targets via conformational selection (35). If so, the flexible loop region pre-existing in a number of different conformations may allow the FCS domain to bind to different target nucleic acids with high affinity. A recent study has revealed that the more flexible regions within the ubiquitin structure are correlated with the regions that are involved in protein-protein interactions due to conformational selection (36). Therefore, the greater flexibility of the loop region is likely important for nucleic acid recognition by the FCS domain. The loop region is also the most variable section of the FCS domain, both in sequence (Figure 1) and structure as it is the section within the FCS domains from hPh1 and L3MBTL2 where the difference is the greatest in the overlay of the two structures (Figure 3D). Because the loop appears to be important for nucleic acid recognition, it may be possible that the different loop structures among the FCS domains allows recognition of different nucleic acid sequences and/or structures, thereby providing additional specificity determinants.
An additional factor that could influence specificity and affinity to the binding reactions involving Ph FCS domain is its oligomeric state. All Ph orthologs contain a C-terminal sterile alpha motif (SAM) domain while dSfmbt, Scm and the L3MBT proteins contain MBT domains. In vitro, the Drosophila Ph SAM domain can self-associate into a helical polymer architecture (37); the dSfmbt and Scm MBT domains may also form higher order stoichiometries (6, 38, 39). Ph oligomerization via the SAM domain would result in the presence of multiple FCS domains that would potentially contribute to the specificity and affinity of the nucleic acid that bind to the FCS domain. The presence of the FCS, MBT and SAM domains in the same PcG protein is common. Drosophila Scm not only contains two adjacent FCS domains but also a polymeric SAM domain and an MBT domain. The Drosophila Sfmbt and L3MBT as well as the mammalian L3MBTL3 have both FCS and SAM domains as well as an MBT domain. It will be of interest to determine whether oligomerization related to the presence of the SAM or MBT domains influences the function of the FCS domain.
If multimerization of the FCS occurs via the protein-protein interactions with the other parts of the protein, it might be expected that the affinity between a single isolated FCS domain would be relatively weak. Consistent with this, the chemical shifts in the HSQC do not broaden and remain in fast exchange during the titration with nucleic acids. In addition, the Kd of the hPh1 FCS/34nt RNA was estimated to be 3 mM and even weaker with DNA. It is important to point out that there is currently no RNA molecule or DNA sequence that is known to be a specific binding partner to any of FCS domain. While the affinity to a specific binding partner would obviously increase, we speculate that the ability of the FCS domain to bind weakly and non-specifically to many different sequences of DNA and structures of RNA may be how it precisely evolved to function in order to organize different nucleic acid elements around a higher oligomeric display of FCS domains.
The results presented here show that Ph that contain mutations that hinder FCS - nucleic acid interactions exhibit decreased ability to repress transcription. The molecular role of the FCS domain in Ph mediated repression remains to be determined. However, in view of the recently-recognized participation of ncRNA in PcG-mediated represssion, it is possible that the FCS domain binds ncRNAs which in turn would provide a targeting function, allowing Ph and other FCS containing proteins to associate at specific chromatin locations. Alternatively, the FCS domain may function to bind DNA directly. If so, the FCS domain would need to bind to DNA that is free of histones. PREs are susceptible to histone replacement and have a reduced level of histones in chromatin immunoprecipitation experiments (40, 41). Furthermore, PRC1 can form a complex with PRE DNA in a manner that would only be possible in the absence of histones (4). These results point to the ability of PRC1 to form a complex with naked DNA. Binding of the FCS domain to DNA may help facilitate the formation of such complexes. Future studies of the FCS domain will help provide insights into these possibilities.
Acknowledgments
We would like to thank Drs. P. John Hart and Susan T. Weintraub for comments on the manuscript. We thank Dr. Jens Wöhnert for providing the RNA used in this study.
This work was supported by the American Heart Association (0830111N), the American Cancer Society (RSG-08-285-01-GMC) and the Department of Defense Breast Cancer Research Program (BC075278) (CAK). Support for the NMR facility was provided by UTHSCSA and NIH-NCI P30 CA54174 (CTRC at UTHSCSA).
Abbreviations and Textual Footnotes
- FCS domain
Phenylalanine-Cysteine-Serine domain
- PcG
Polycomb Group
- PRC1 & 2
Polycomb repression complex 1 & 2
- PRE
Polycomb response element
- Ezh2
Enhancer of zeste homolog 2
- Ph
Polyhomeotic
- hPh1
Human Polyhomeotic homolog 1
- Scm
Sex comb on midleg
- L3MBTL2
lethal 3 malignant brain tumor like 2
- Sfmbt
Scm-like with four MBT domains
- NMR
Nuclear magnetic resonance
- HSQC
heteronuclear single quantum coherence
- HNNOE
{1H}-15N NOE
- RDC
residual dipolar coupling
- HOX
Homeotic
- ncRNA
non-coding RNA
- Xi
inactivated X chromosome
- SAM
sterile alpha motif
- TEV
tobacco etch virus
- MBT domain
Malignant brain tumor domain
References
- 1.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb Complexes Repress Developmental Regulators in Murine Embryonic Stem Cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 2.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of Developmental Regulators by Polycomb in Human Embryonic Stem Cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic Recognition of an Epigenetic DNA Element by Pleiohomeotic and a Polycomb Core Complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a Polycomb Nucleoprotein Complex. Mol. Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb Group Protein Complex with Sequence-Specific DNA-Binding and Selective Methyl-Lysine-Binding Activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Muller J, Muller CW. Molecular Recognition of Histone Lysine Methylation by the Polycomb Group Repressor dSfmbt. EMBO J. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of Histone H3 Lysine 27 Methylation in X Inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 Function at the Initiation of X Inactivation Independent of PRC2 and Silencing. EMBO J. 2006 doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 12.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi Components are Required for Nuclear Clustering of Polycomb Group Response Elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Christoforou A, Aravind L, Emmons SW, van den Heuvel S, Haber DA. The C. Elegans Polycomb Gene SOP-2 Encodes an RNA Binding Protein. Mol. Cell. 2004;14:841–847. doi: 10.1016/j.molcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. The Human Polycomb Group Complex Associates with Pericentromeric Heterochromatin to Form a Novel Nuclear Domain. J. Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechtenberg BC, Allen MD, Rutherford TJ, Freund SM, Bycroft M. Solution Structure of the FCS Zinc Finger Domain of the Human Polycomb Group Protein L(3)Mbt-Like 2. Protein Sci. 2009;18:657–661. doi: 10.1002/pro.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific Nuclear Magnetic Resonance Assignments of the Methyl Groups of Valine and Leucine in the DNA-Binding Domain of the 434 Repressor by Biosynthetically Directed Fractional 13C Labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 18.Senn H, Werner B, Messerle BA, Weber C, Traber R, Wüthrich K. Stereospecific Assignment of the Methyl 1H NMR Lines of Valine and Leucine in Polypeptides by Nonrandom 13C Labelling. FEBS Lett. 1989;249:113–118. [Google Scholar]
- 19.Jacob J, Louis JM, Nesheiwat I, Torchia DA. Biosynthetically Directed Fractional 13C Labeling Facilitates Identification of Phe and Tyr Aromatic Signals in Proteins. J. Biomol. NMR. 2002;24:231–235. doi: 10.1023/a:1021662423490. [DOI] [PubMed] [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Johnson B,A, Blevins R,A. NMR View: A Computer Program for the Visualization and Analysis of NMR Data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Linge JP, O’Donoghue SI, Nilges M. Automated Assignment of Ambiguous Nuclear Overhauser Effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 24.Cornilescu G, Delaglio F, Bax A. Protein Backbone Angle Restraints from Searching a Database for Chemical Shift and Sequence Homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 25.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 1. Theory and Range of Validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 26.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 2. Analysis of Experimental Results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 27.Lian LY, Roberts GCK. In: NMR of Macromolecules: A Practical Approach. Roberts GCK, editor. Oxford Univ. Press; New York: 1993. p. 153. [Google Scholar]
- 28.Ghering AB, Shokes JE, Scott RA, Omichinski JG, Godwin HA. Spectroscopic Determination of the Thermodynamics of Cobalt and Zinc Binding to GATA Proteins. Biochemistry. 2004;43:8346–8355. doi: 10.1021/bi035673j. [DOI] [PubMed] [Google Scholar]
- 29.Seneque O, Bonnet E, Joumas FL, Latour JM. Cooperative Metal Binding and Helical Folding in Model Peptides of Treble-Clef Zinc Fingers. Chemistry. 2009;15:4798–4810. doi: 10.1002/chem.200900147. [DOI] [PubMed] [Google Scholar]
- 30.Ramelot TA, Cort JR, Yee AA, Semesi A, Edwards AM, Arrowsmith CH, Kennedy MA. NMR Structure of the Escherichia Coli Protein YacG: A Novel Sequence Motif in the Zinc-Finger Family of Proteins. Proteins. 2002;49:289–293. doi: 10.1002/prot.10214. [DOI] [PubMed] [Google Scholar]
- 31.Muller J, Gaunt S, Lawrence PA. Function of the Polycomb Protein is Conserved in Mice and Flies. Development. 1995;121:2847–2852. doi: 10.1242/dev.121.9.2847. [DOI] [PubMed] [Google Scholar]
- 32.Gorfinkiel N, Fanti L, Melgar T, Garcia E, Pimpinelli S, Guerrero I, Vidal M. The Drosophila Polycomb Group Gene Sex Combs Extra Encodes the Ortholog of Mammalian Ring1 Proteins. Mech. Dev. 2004;121:449–462. doi: 10.1016/j.mod.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription Factor YY1 Functions as a PcG Protein in Vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringrose L, Paro R. Epigenetic Regulation of Cellular Memory by the Polycomb and Trithorax Group Proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 35.Boehr DD, Nussinov R, Wright PE. The Role of Dynamic Conformational Ensembles in Biomolecular Recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition Dynamics Up to Microseconds Revealed from an RDC-Derived Ubiquitin Ensemble in Solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 37.Kim CA, Gingery M, Pilpa RM, Bowie JU. The SAM Domain of Polyhomeotic Forms a Helical Polymer. Nat. Struct. Biol. 2002;9:453–457. doi: 10.1038/nsb802. [DOI] [PubMed] [Google Scholar]
- 38.Nagem RA, Dauter Z, Polikarpov I. Protein Crystal Structure Solution by Fast Incorporation of Negatively and Positively Charged Anomalous Scatterers. Acta Crystallogr. D Biol. Crystallogr. 2001;57:996–1002. doi: 10.1107/s0907444901007260. [DOI] [PubMed] [Google Scholar]
- 39.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a Unifying Role in Cellular Lifespan. Nat. Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 40.Papp B, Muller J. Histone Trimethylation and the Maintenance of Transcriptional ON and OFF States by trxG and PcG Proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mito Y, Henikoff JG, Henikoff S. Histone Replacement Marks the Boundaries of Cis-Regulatory Domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 42.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple Sequence Alignment with the Clustal Series of Programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]