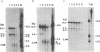Abstract
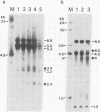
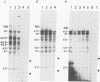
We have used DNase I as a probe to examine the chromatin structure of mouse immunoglobulin kappa light chain genes in rearranged and unrearranged chromosomes--i.e., in nuclei from myeloma cells and from brain and liver cells. Tissue-specific DNase I-hypersensitive sites are observed 0.7 and 1.7 kilobases upstream from the 5' end of the C kappa gene in the J kappa -C kappa intron region in myeloma nuclei but not in naked DNA or in brain or liver nuclei. In myeloma cells expressing one functional kappa light chain polypeptide, but with more than one rearranged allele, one DNase I-hypersensitive site is found 0.3 kilobases upstream from the start of the coding region of the V kappa sequence in both functionally rearranged and nonfunctionally rearranged alleles but not in cross-hybridizing V kappa genes in the germ line context. Thus, during the development of B lymphocytes, the commitment of immunoglobulin kappa light chain gene expression seems to be associated with the presence of DNase I hypersensitivities that reflect changes of chromosomal structure surrounding the single copy C kappa gene. In contrast, the germ line V kappa multigene family seems to be located in a chromosomal region that does not exhibit change of DNase I hypersensitivity in response to commitment of immunoglobulin expression; a V kappa gene acquires DNase I hypersensitivity only after it is translocated adjacent to the J kappa -C kappa intron region. The DNase I-hypersensitive site 5' to the V kappa sequence is similar in location to hypersensitive sites found for other eukaryotic genes and is probably associated with the promoter region. However, the DNase I-hypersensitive sites in the J kappa -C kappa intron region are not associated with any known promoters. In addition, the DNA sequences surrounding the C kappa-proximal DNase I-hypersensitive site have several stretches of homology with sequences within the 72-base-pair tandem repeat of simian virus 40, which has been shown to modulate the transcriptional activity of neighboring genes. This DNase I-hypersensitive site in the intron region may be significant for the differential expression of the translocated V kappa genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Farrell P. J., Rabbitts T. H. Unrearranged immunoglobulin variable region genes have a functional promoter. Nucleic Acids Res. 1982 Mar 25;10(6):1841–1856. doi: 10.1093/nar/10.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S. E., Botchan M. R. Isolation and characterization of human DNA fragments with nucleotide sequence homologies with the simian virus 40 regulatory region. Mol Cell Biol. 1982 Aug;2(8):949–965. doi: 10.1128/mcb.2.8.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan N. J., Secher D. S., Milstein C. Intracellular immunoglobulin chain synthesis in non-secreting variants of a mouse myeloma: detection of inactive light-chain messenger RNA. J Mol Biol. 1974 Dec 25;90(4):691–701. doi: 10.1016/0022-2836(74)90533-6. [DOI] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hieter P. A., Max E. E., Seidman J. G., Maizel J. V., Jr, Leder P. Cloned human and mouse kappa immunoglobulin constant and J region genes conserve homology in functional segments. Cell. 1980 Nov;22(1 Pt 1):197–207. doi: 10.1016/0092-8674(80)90168-3. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather E. L., Perry R. P. Transcriptional regulation of immunoglobulin V genes. Nucleic Acids Res. 1981 Dec 21;9(24):6855–6867. doi: 10.1093/nar/9.24.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Max E. E., Maizel J. V., Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Pech M., Höchtl J., Schnell H., Zachau H. G. Differences between germ-line and rearranged immunoglobulin V kappa coding sequences suggest a localized mutation mechanism. Nature. 1981 Jun 25;291(5817):668–670. doi: 10.1038/291668a0. [DOI] [PubMed] [Google Scholar]
- Sakano H., Hüppi K., Heinrich G., Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979 Jul 26;280(5720):288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Leder P. The arrangement and rearrangement of antibody genes. Nature. 1978 Dec 21;276(5690):790–795. doi: 10.1038/276790a0. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Max E. E., Leder P. A kappa-immunoglobulin gene is formed by site-specific recombination without further somatic mutation. Nature. 1979 Aug 2;280(5721):370–375. doi: 10.1038/280370a0. [DOI] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Storb U., Arp B., Wilson R. The switch region associated with immunoglobulin C mu genes is DNase I hypersensitive in T lymphocytes. Nature. 1981 Nov 5;294(5836):90–92. doi: 10.1038/294090a0. [DOI] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Strathearn M. D., Strathearn G. E., Akopiantz P., Liu A. Y., Paddock G. V., Salser W. Characterization of an immunoglobin cDNA clone containing the variable and constant regions for the MOPC 21 kappa light chain. Nucleic Acids Res. 1978 Sep;5(9):3101–3111. doi: 10.1093/nar/5.9.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness B. G., Weigert M., Coleclough C., Mather E. L., Kelley D. E., Perry R. P. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981 Dec;27(3 Pt 2):593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Glotov B. O., Schnell H., Zachau H. G. Differences in the nuclease sensitivity between the two alleles of the immunoglobulin kappa light chain genes in mouse liver and myeloma nuclei. Nucleic Acids Res. 1982 Jun 25;10(12):3627–3645. doi: 10.1093/nar/10.12.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Gilbert W. Tissue-specific exposure of chromatin structure at the 5' terminus of the rat preproinsulin II gene. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1577–1580. doi: 10.1073/pnas.78.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]