Abstract
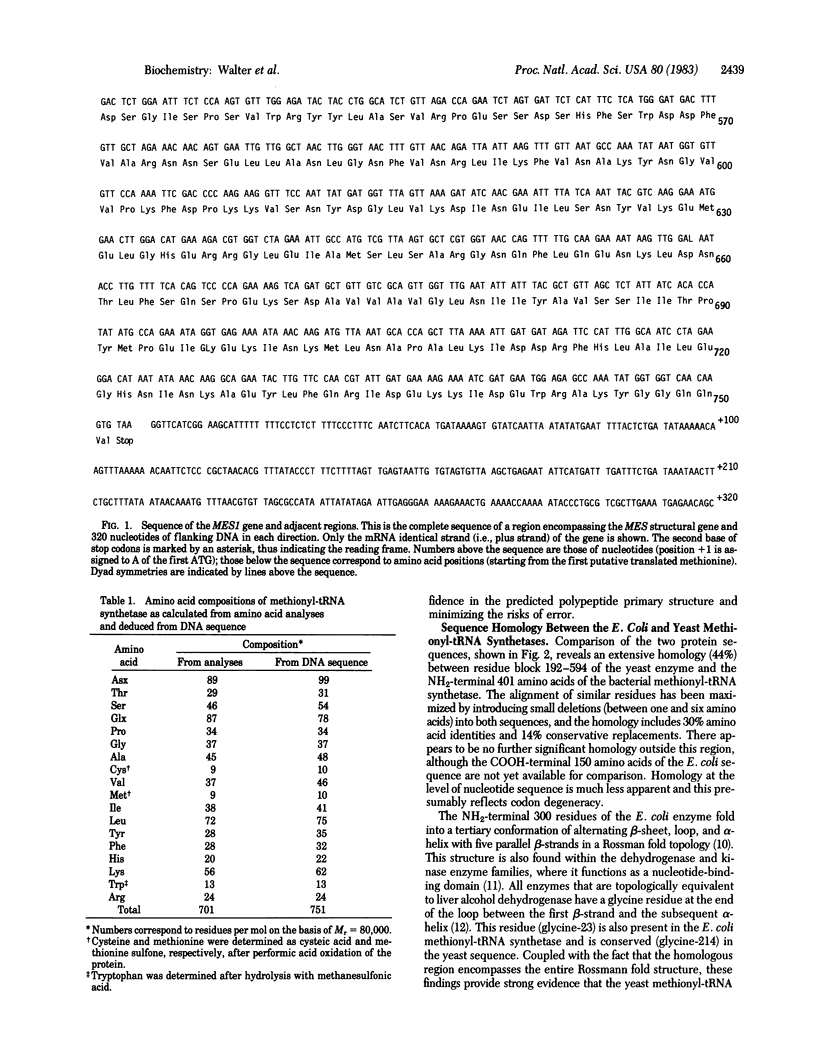
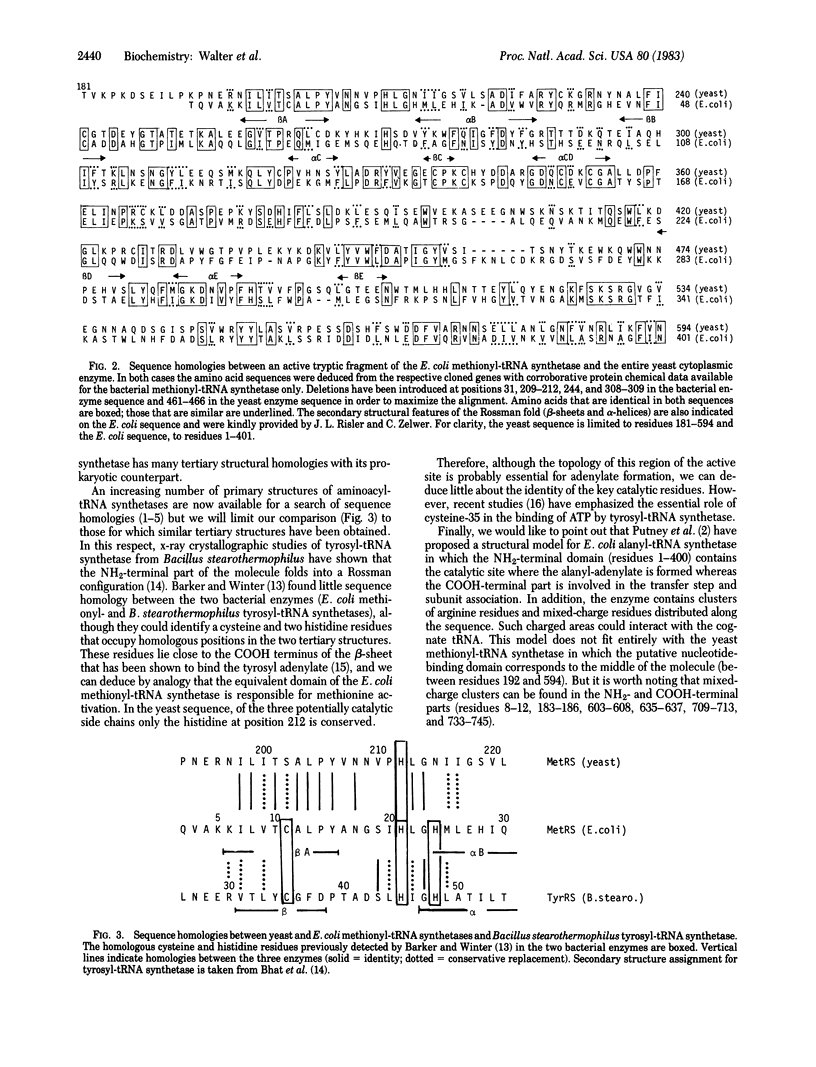
The sequence of a 5-kilobase DNA insert containing the structural gene for yeast cytoplasmic methionyl-tRNA synthetase has been determined and a unique open reading frame of 2,253 nucleotides encoding a polypeptide chain of 751 amino acids (Mr, 85,500) has been characterized. The data obtained on the purified enzyme (subunit size, amino acid composition, and COOH-terminal sequence) are consistent with the gene structure. The protein sequence deduced from the nucleotide sequence reveals no obvious internal repeats. This protein sequence shows a high degree of homology with that of Escherichia coli methionyl-tRNA synthetase within a region that forms the putative methionyl adenylate binding site. This strongly suggests that both proteins derive from a common ancestor.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D. G., Ebel J. P., Jakes R., Bruton C. J. Methionyl-tRNA synthetase from Escherichia coli. Primary structure of the active crystallised tryptic fragment. Eur J Biochem. 1982 Oct;127(3):449–457. [PubMed] [Google Scholar]
- Barker D. G., Winter G. Conserved cysteine and histidine residues in the structures of the tyrosyl and methionyl-tRNA synthetases. FEBS Lett. 1982 Aug 23;145(2):191–193. doi: 10.1016/0014-5793(82)80165-8. [DOI] [PubMed] [Google Scholar]
- Bhat T. N., Blow D. M., Brick P., Nyborg J. Tyrosyl-tRNA synthetase forms a mononucleotide-binding fold. J Mol Biol. 1982 Jul 15;158(4):699–709. doi: 10.1016/0022-2836(82)90255-8. [DOI] [PubMed] [Google Scholar]
- Fasiolo F., Bonnet J., Lacroute F. Cloning of the yeast methionyl-tRNA synthetase gene. J Biol Chem. 1981 Mar 10;256(5):2324–2328. [PubMed] [Google Scholar]
- Hall C. V., vanCleemput M., Muench K. H., Yanofsky C. The nucleotide sequence of the structural gene for Escherichia coli tryptophanyl-tRNA synthetase. J Biol Chem. 1982 Jun 10;257(11):6132–6136. [PubMed] [Google Scholar]
- Hoben P., Royal N., Cheung A., Yamao F., Biemann K., Söll D. Escherichia coli glutaminyl-tRNA synthetase. II. Characterization of the glnS gene product. J Biol Chem. 1982 Oct 10;257(19):11644–11650. [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney S. D., Royal N. J., Neuman de Vegvar H., Herlihy W. C., Biemann K., Schimmel P. Primary structure of a large aminoacyl-tRNA synthetase. Science. 1981 Sep 25;213(4515):1497–1501. doi: 10.1126/science.7025207. [DOI] [PubMed] [Google Scholar]
- Risler J. L., Zelwer C., Brunie S. Methionyl-tRNA synthetase shows the nucleotide binding fold observed in dehydrogenases. Nature. 1981 Jul 23;292(5821):384–386. doi: 10.1038/292384a0. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Argos P. The taxonomy of protein structure. J Mol Biol. 1977 Jan 5;109(1):99–129. doi: 10.1016/s0022-2836(77)80048-x. [DOI] [PubMed] [Google Scholar]
- Rubin J., Blow D. M. Amino acid activation in crystalline tyrosyl-tRNA synthetase from Bacillus stearothermophilus. J Mol Biol. 1981 Jan 25;145(3):489–500. doi: 10.1016/0022-2836(81)90541-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneller J. M., Schneider C., Stahl A. J. Distinct nuclear genes for yeast mitochondrial and cytoplasmic methionyl-tRNA synthetases. Biochem Biophys Res Commun. 1978 Dec 29;85(4):1392–1399. doi: 10.1016/0006-291x(78)91158-0. [DOI] [PubMed] [Google Scholar]
- Winter G. P., Hartley B. S. The amino acid sequence of tryptophanyl tRNA synthetase from Bacillus stearothermophilus. FEBS Lett. 1977 Aug 15;80(2):340–342. doi: 10.1016/0014-5793(77)80471-7. [DOI] [PubMed] [Google Scholar]
- Winter G., Fersht A. R., Wilkinson A. J., Zoller M., Smith M. Redesigning enzyme structure by site-directed mutagenesis: tyrosyl tRNA synthetase and ATP binding. Nature. 1982 Oct 21;299(5885):756–758. doi: 10.1038/299756a0. [DOI] [PubMed] [Google Scholar]



