Abstract
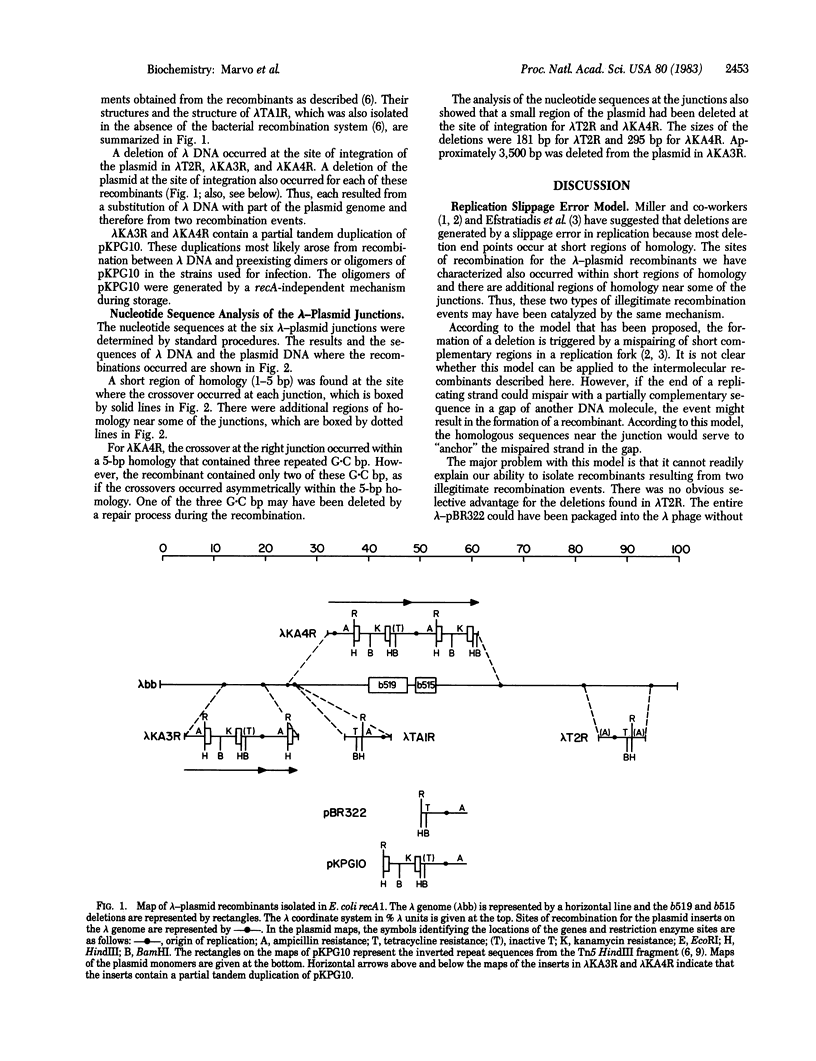
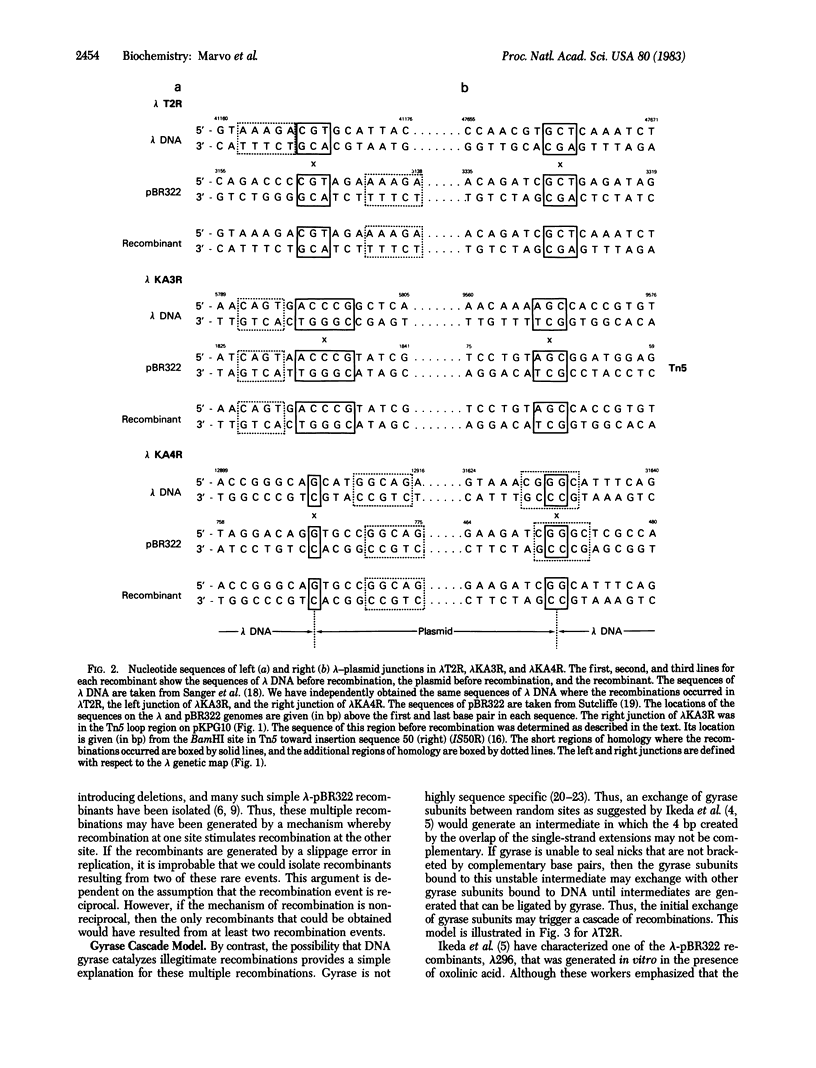
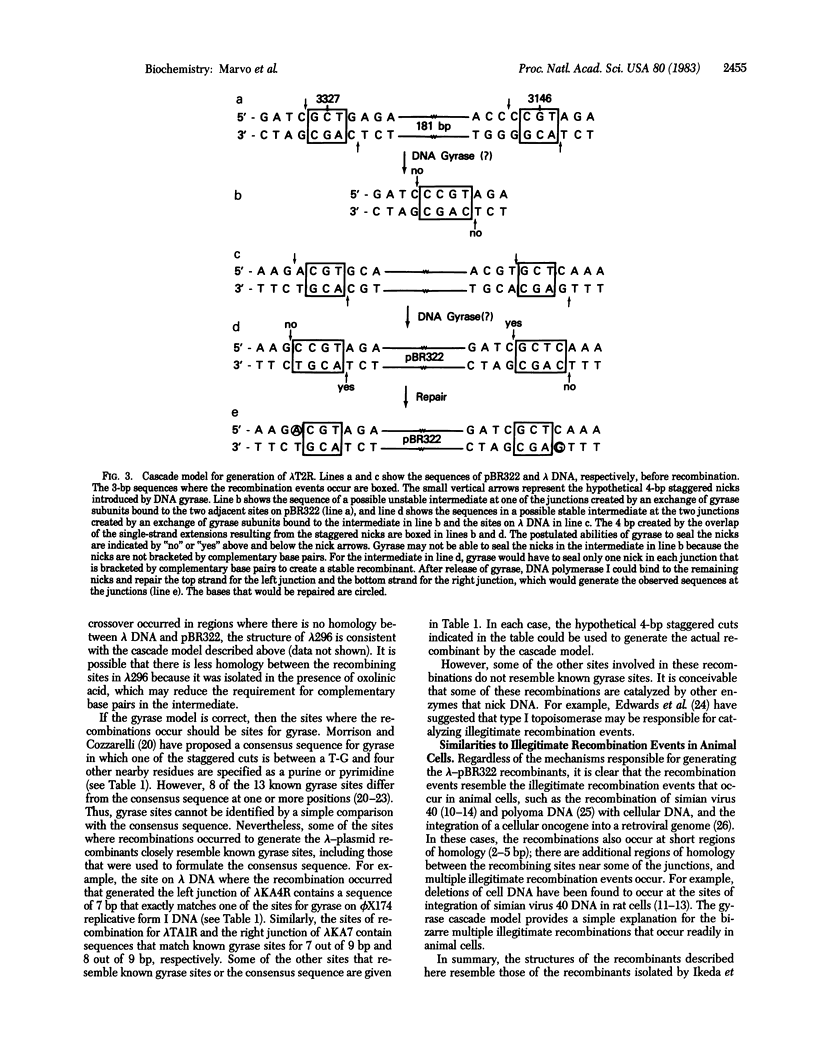
The structures of three recombinants between bacteriophage lambda DNA and plasmid pBR322 that were generated in a recA derivative of Escherichia coli are described. Each resulted from two illegitimate recombination events that resulted in the substitution of part of the lambda genome by part of the plasmid genome. The nucleotide sequences at the six lambda-plasmid junctions were determined and compared with the sequences of the lambda and plasmid genomes before recombination. Each recombination occurred at a short region of homology in the two genomes, and other short regions of homology were found near some of the junctions. The structures of these junctions are similar to those resulting from illegitimate recombination in animal cells. A model to explain how these multiple illegitimate recombination events could result from a cascade of DNA gyrase-catalyzed recombinations is discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Bourgaux P., Sylla B. S., Chartrand P. Excision of polyoma virus DNA from that of a transformed mouse cell: identification of a hybrid molecule with direct and inverted repeat sequences at the viral-cellular joints. Virology. 1982 Oct 15;122(1):84–97. doi: 10.1016/0042-6822(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Fisher L. M., Mizuuchi K., O'Dea M. H., Ohmori H., Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Aoki K., Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Moriya K., Matsumoto T. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- King S. R., Krolewski M. A., Marvo S. L., Lipson P. J., Pogue-Geile K. L., Chung J. H., Jaskunas S. R. Nucleotide sequence analysis of in vivo recombinants between bacteriophage lambda DNA and pBR322. Mol Gen Genet. 1982;186(4):548–557. doi: 10.1007/BF00337963. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981 Mar;23(3):721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979 May;17(1):175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- Pogue-Geile K. L., DasSarma S., King S. R., Jaskunas S. R. Recombination between bacteriophage lambda and plasmid pBR322 in Escherichia coli. J Bacteriol. 1980 Jun;142(3):992–1003. doi: 10.1128/jb.142.3.992-1003.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Yin J. C., Yong-di Z., Johnson R. C., Reznikoff W. S. Genetic organization of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):99–105. doi: 10.1101/sqb.1981.045.01.018. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Galleshaw J. A., Jonas V., Berns A. J., Doolittle R. F., Donoghue D. J., Verma I. M. Nucleotide sequence and formation of the transforming gene of a mouse sarcoma virus. Nature. 1981 Jan 22;289(5795):258–262. doi: 10.1038/289258a0. [DOI] [PubMed] [Google Scholar]


