Abstract
Mycobacterium tuberculosis (Mtb) secretes excess of a second messenger molecule, 3',5'-cyclic AMP (cAMP), which plays a critical role in the survival of Mtb in host macrophages. Although Mtb produces cAMP in abundance, its exact role in the physiology of mycobacteria is elusive. In this study we have analyzed the expression of 16 adenylate cyclases (ACs) and kinetics of intracellular cAMP levels in Mtb during in vitro growth under the regular culture conditions, and after exposure to different stress agents. We observed a distinct expression pattern of these ACs which is correlated with intracellular cAMP levels. Interestingly cAMP levels are significantly elevated in Mtb following heat stress, whereas other stress conditions such as oxidative, nitrosative or low pH do not affect intracellular cAMP pool in vitro. A significant increase in expression by >2-fold of five ACs namely Rv1647, Rv2212, Rv1625c, Rv2488c and Rv0386 after heat stress further suggested that cAMP plays an important role in controlling Mtb response to heat stress. In the light of these observations, effect of exogenous cAMP on global gene expression profile was examined by using microarrays. The microarray gene expression analysis demonstrated that cAMP regulates expression of a subset of heat stress-induced genes comprising of dnaK, grpE, dnaJ, and Rv2025c. Further we performed electrophoretic mobility shift assay by using cAMP-receptor protein of Mtb (CRPM), which demonstrated that CRPM specifically recognizes a sequence −301 AGCGACCGTCAGCACG −286 in 5'-untranslated region of dnaK.
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of disease tuberculosis (TB) has evolved a clever strategy of intoxicating host macrophages by secreting a signaling molecule, 3',5'-cyclic adenosine monophosphate (cAMP) [1], [2]. Cyclic AMP is continuously produced by Mtb during in vitro growth [3], probably due to presence of multiple adenylate cyclases (ACs). Genome sequence of Mtb reveals the presence of 16 ACs in Mtb H37Rv strain, 10 of which have been biochemically characterized in vitro [4]–[7]. Fusion of class III adenylyl cyclase catalytic region with different domains adds versatility to these multiple ACs. The first AC of Mtb which was characterized by in vitro biochemical assays is Cya which is encoded by a gene Rv1625c [8], [9]. Structurally each monomeric subunit of Cya homodimer contains six transmembrane regions and a catalytic domain, which corresponds to one half of the mammalian adenylyl cyclases [10]. Other functionally characterized ACs of Mtb are: pH-sensing Rv1264 which contains an autoinhibitory N-terminal domain [11]–[12]; Rv1318c, Rv1319c, Rv1320c and Rv3645 containing membrane anchored HAMP (present in Histidine kinases, Adenylate cyclases, Methyl accepting proteins and Phosphatases) region [13]; Rv0386 whose adenylyl cyclase catalytic domain is fused to an AAA-ATPase and a helix-turn-helix DNA-binding domain [14]; Rv1647 and Rv2212 which attain complete activity only in the presence of detergent and unsaturated fatty acids, respectively [15], [16]; and Rv1900c which forms asymmetric homodimers [17]. Cyclic AMP is also secreted into host cells during infection which perturbs signaling pathways and affects bacterial persistence and killing by host macrophages [1], [2].
In prokaryotes cAMP activates the function of a transcription factor known as cAMP-receptor protein or CRP which recognizes a specific sequence in the 5'-untranslated region (5'-UTR) and subsequently regulates the mRNA synthesis of candidate genes [4]. In silico analysis predicts 10 putative cNMP-binding proteins in Mtb [4]; two of these proteins encoded by Rv3676 (known as cAMP-receptor protein of M. tuberculosis, CRPM) and Rv1675c (annotated as Cmr, for cAMP and macrophage regulator) function as cAMP-responsive transcription factors that regulate expression of multiple genes by direct binding to their promoter regions [18]–[24]. In addition to regulating mycobacterial pathogenesis other important biological processes are also regulated by this signaling molecule. Exogenous cAMP stimulates the expression of galactokinase in the presence of glutamate and galactose in Mycobacterium smegmatis (Msm), which is otherwise not induced by galactose alone [25]. In M. bovis BCG cAMP regulates the expression of five proteins namely Rv1265, Rv2971, GroEL2, PE_PGRS6a, and malate dehydrogenase [26]. Very recently it is shown that cAMP plays a role in acetylation of stress proteins and acetyl-CoA synthetase [27]–[29], which suggests that cAMP is critical in functioning of central metabolic pathways of Mtb.
Though Mtb produces significant concentration of cAMP which is also secreted into extracellular environment [30], [31], direct role of cAMP in the physiology of Mtb is lacking. By using a systematic approach in this study we measured the intracellular cAMP levels and expression of ACs in Mtb during its in vitro growth in regular culture medium as well as under different stress conditions. By performing a whole genome microarray analysis, we studied the effect of cAMP on global gene expression profile of Mtb. Further, direct effect of cAMP on the expression of candidate genes was validated by performing electrophoretic mobility shift assay (EMSA). Our results demonstrate that in Mtb cAMP levels are significantly elevated after heat stress which in-turn regulates the expression of a subset of heat stress-induced genes encoding chaperones DnaK, GrpE and DnaJ respectively, by facilitating the direct binding of CRPM to the promoter region of dnaK operon.
Results
Analysis of intracellular cAMP levels in Mtb during in vitro growth
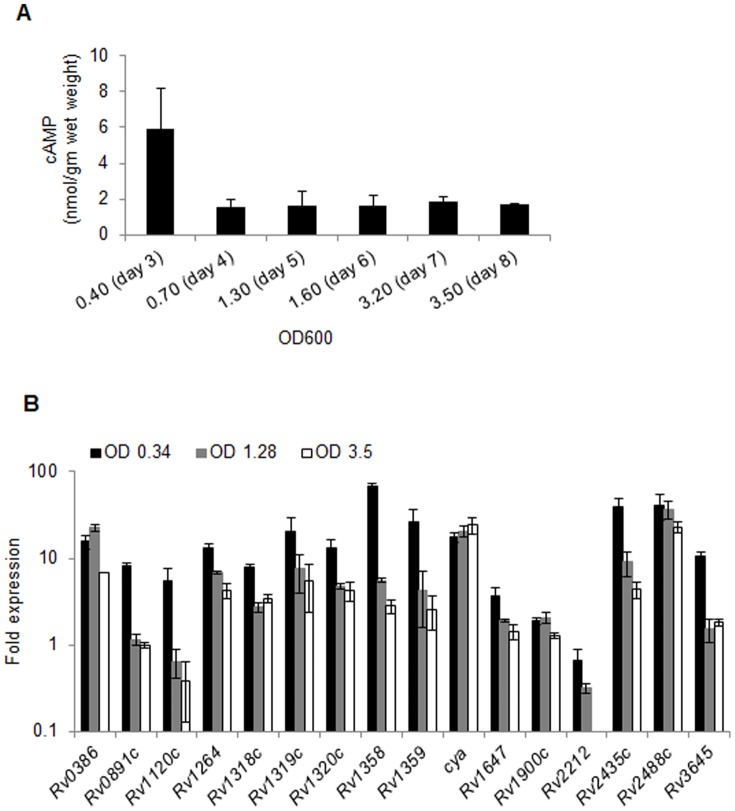
Cyclic AMP is known to exert an array of regulatory functions which advocates that the cellular concentration of cAMP must itself be subject to control by culture conditions. Here we estimated intracellular cAMP levels in pathogenic Mtb grown in 7H9 culture medium supplemented with 1x OADC (oleic acid, albumin, dextrose and catalase), 0.5% glycerol and 0.02% tween-80, at different growth stages. Lysates were prepared by boiling the bacterial pellets in 0.1 M HCl to avoid degradation of cyclic nucleotides during extraction by phosphodiesterase (PDE) of Mtb, and cAMP was measured by ELISA as described in materials and methods. Our results demonstrated that Mtb exhibited maximum intracellular cAMP at day3 post-inoculation when the optical density at 600 nm (OD600) of bacterial culture was 0.4. Subsequent growth on day 4 (OD600 of 0.70) resulted in sharp decline in the intracellular cAMP pool by ∼3.5 folds and this level remained constant for next four days of growth when cultures reached to stationary phase (Fig. 1A). By ELISA, it was estimated that cAMP concentrations were 5.9 nmol/gm wet weight on day 3, and 1.6–1.8 nmol/gm wet weight on days 4–8 respectively (Fig. 1A).
Figure 1. Kinetics of intracellular cAMP levels and expression of ACs in Mtb during in vitro growth.
A) Estimation of intracellular cAMP levels in Mtb. Wild-type Mtb CDC1551 was cultured in Middlebrook 7H9 broth supplemented with OADC, glycerol, and tween-80 at 37°C with shaking and growth was monitored at regular intervals by measuring optical density of the cultures at 600 nm (OD600). An aliquot of culture suspension was used at each time point up to 8 days for cAMP measurement, as described in materials and methods, and final cAMP concentration was calculated as nmol/gm wet weight. Data are the averages of two independent experiments and the mean values ± standard deviations are shown. B) Analysis of the expression of AC-encoding genes at different time points during in vitro growth of Mtb. Total RNA was isolated from Mtb CDC1551 cultures at regular intervals as shown in the graph. Quantitative RT-PCR (qRT-PCR) was performed, and the relative expression of each gene was calculated using values of sigA transcript for normalizing the RNA amounts and day-1 expression value as a control. The graph represents an average of two independent experiments and the mean values ± standard deviations are shown.
Expression analysis of Mtb ACs by real-time quantitative reverse-transcription PCR (qRT-PCR)
Cellular concentration of cAMP can be regulated at the level of expression and/or activity of AC and the PDE, or by a change in the rate of cAMP export [6], [32]–[34]. Although intracellular cAMP levels are significantly altered in Mtb, we observed that the extracellular cAMP pool remains constant over eight days of in vitro growth (data not shown). Since it is challenging to determine intracellular enzymatic activities of multiple ACs or PDEs, we focused on studying the expression profiles of mycobacterial ACs by qRT-PCR at various OD600 (Fig. 1B). Transcript levels of each of the 16 AC-encoding genes at designated OD600 were compared with their respective expression levels at day 1 post-inoculation in wild-type Mtb when the OD600 of culture was 0.1. Figure 1B shows that majority of AC-encoding genes except Rv1647, Rv1900c and Rv2212 were overexpressed by ≥5-fold when OD600 of culture reached to 0.34 on day 3. In contrast, transcripts of Rv1647 and Rv1900c were upregulated by 3.6 and 1.9-fold, respectively, whereas Rv2212 exhibited moderate reduction at OD600 of 0.34. Interestingly, at day 5 when OD600 of Mtb culture reached to 1.28, expression of most of the genes except Rv0386, cya, Rv1900c and Rv2488c was reduced by 2–12 folds compared to their respective expression levels at OD600 of 0.34 on day 3. Contrary to these, Rv0386, cya, Rv1900c and Rv2488c either maintained or displayed moderate increase in expression at OD600 of 1.28. Further growth of mycobacteria to OD600 of 3.5 at day 8 post-inoculation resulted in moderate decrease in expression of few genes that included Rv0386, Rv1358 and Rv2435c by 3.2-, 1.97- and 2.07-folds, respectively, while others were expressing at levels similar to OD600 1.28 (Fig. 1B). In contrast, Rv2212 transcript was not detected at OD600 of 3.5. These results are in line with the profile of cellular cAMP concentrations in Mtb which suggests that intracellular cAMP levels vary in proportion to the expression of ACs in Mtb (Fig. 1).
Effect of various stresses on intracellular cAMP levels and expression of Mtb ACs
Upon infection, virulent mycobacteria encounter stringent antimicrobial response within the host organism. However, tubercle bacilli are resistant to killing by host and persist for decades in the host tissues [35]. For their survival, mycobacterial pathogens sense and respond to exogenous stress conditions by modulating the expression of key genes. It is known that cAMP-associated transcription factors CRPM and Cmr regulate the expression of several genes in Mtb which play important roles in Mtb–host interactions [18]–[24]. Interestingly, intracellular cAMP levels in mycobacteria increase dramatically after infection to macrophages [33]. These observations led us to hypothesize that Mtb must be equipped to modulate its cAMP pool under different stress conditions imposed by the host. Therefore we monitored the intracellular cAMP levels in Mtb after its exposure to different stress conditions that bacteria encounter during host infection.
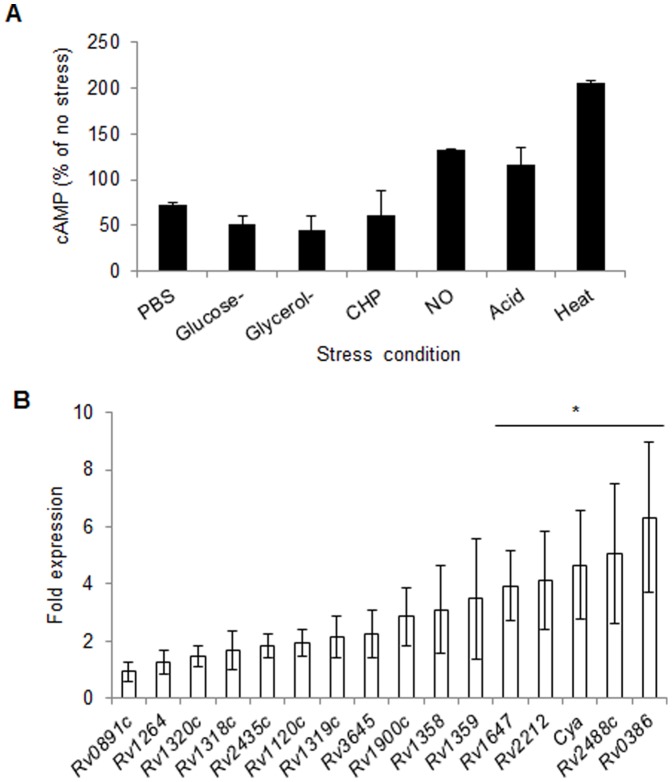
Wild-type Mtb cells were exposed to nutrient starvation (PBS), glucose deficiency, glycerol deficiency, oxidative stress (cumene hydroperoxide, CHP), nitosative stress (DETA/NO), acid stress (pH 4.5) and heat stress (42°C), as described earlier [36]. Subsequently, cells were lysed and cAMP was measured in the lysates by ELISA. As shown in figure 2A, a moderate decrease in intracellular cAMP levels was observed after nutrient starvation and oxidative stress, whereas deficiency of any of the two carbon sources resulted in ∼2-fold reduction in the intracellular cAMP concentration (Fig. 2A). In contrast, a mild increase in intracellular cAMP level was seen in the cells after nitrosative and acid stresses. Interestingly, of these stresses, maximum effect was observed after heat-stress, which caused >2-fold increase in intracellular cAMP level under these culture conditions.
Figure 2. Effect of various stresses on intracellular cAMP levels and expression of ACs in Mtb.
A) Estimation of cAMP in Mtb during in vitro growth under different stresses. Wild-type culture suspension in Middlebrook 7H9 broth at OD600 of 0.4 was exposed to different stresses as described earlier [36] and intracellular cAMP levels were determined by ELISA. Bar graph represents cAMP level in each of the stressed samples as percentage of un-treated control. Data are the averages of three independent experiments and the mean values ± standard deviations are shown. B) Expression analysis of the AC-encoding genes following thermal stress in Mtb. The fold-expression, as measured by qRT-PCR, indicates the ratio of sigA-normalized gene expression levels in Mtb exposed to 42 °C relative to those in the Mtb grown at 37°C. The graph represents an average of three experiments. Error bars indicate standard deviations and asterisk denotes p ≤0.05.
In order to assess whether increase in cAMP level after the heat-stress is related to the expression of ACs, we performed the qRT-PCR to measure mRNA levels of AC-encoding genes in Mtb following the heat-shock treatment. Expression level of individual gene was obtained after normalization with the level of a housekeeping gene sigA, which remained unaltered under these conditions. Figure 2B depicts that the mRNA levels of 5 out of 16 AC-encoding genes (Rv1647, Rv2212, cya, Rv2488c and Rv0386) are increased by >2-fold after heat-stress (p ≤0.05). Taken together, these observations indicate that Mtb responds to heat-shock exposure by inducing the expression of certain ACs which results in elevated cAMP-levels in the cell.
Identification of cAMP-regulon by microarray
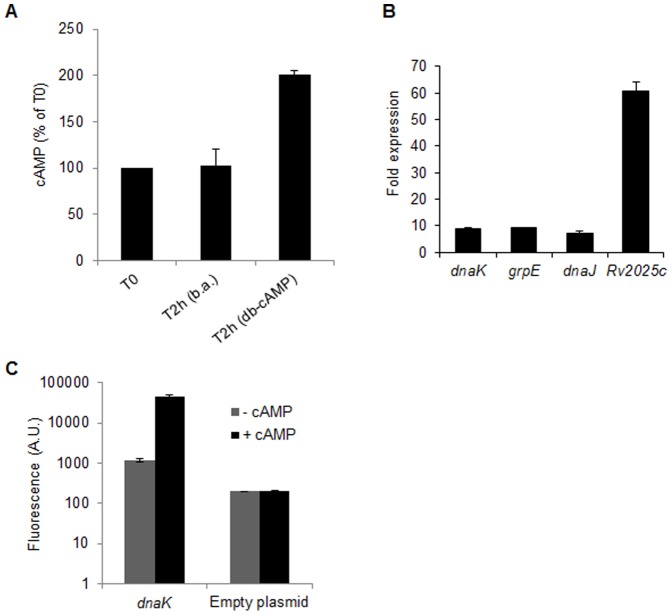
To identify cAMP-regulated genes of Mtb, wild-type Mtb CDC1551cultures were treated with di-butyryl cAMP (db-cAMP) and the expression of genes was compared with butyric acid-treated control samples by whole genome microarrays as described in materials and methods. Before proceeding to microarray experiments, intracellular cAMP levels were measured in both the samples by ELISA, which indicated a 2-fold increase in cAMP levels after 2 hrs of incubation with 20 mM db-cAMP (Fig. 3A). In contrast, there was no change in intracellular cAMP levels after incubation with 20 mM butyric acid for 2 hrs, which suggests that db-cAMP specifically elevates intracellular cAMP pool by 2-fold (Fig. 3A). A total of three hybridization experiments were performed with three independent RNA preparations, and the results were analyzed statistically. Expression level of individual gene was measured after normalization with the level of sigA, which was not changed under these conditions. Genes that were differentially expressed by >2.0-fold (p≤ 0.05) in db-cAMP-treated samples are shown in table 1. The microarray results demonstrated that a total of 7 genes were upregulated and 5 genes were downregulated by ≥2-fold after treatment with db-cAMP, in comparison to butyric acid-treated samples (Table 1). The most abundant transcripts were those of a subset of heat stress-induced genes [37] comprising of Rv2025c, dnaK, grpE and dnaJ. Further, differences in expression of these genes between db-cAMP-treated and butyric acid-treated samples by microarray were 9.8±4.4, 3.7±0.6, 2.9±0.9, and 3.2±0.7 respectively (Table 1). These differences were more prominent in qRT-PCR which exhibited 60.9±3.2, 8.9±0.4, 9.1±0.1, and 7.3±0.6-fold induction of Rv2025c, dnaK, grpE and dnaJ respectively after db-cAMP treatment (Fig. 3B). Moreover, expression levels of these genes were significantly higher than of cAMP-regulated whiB1 [20], which exhibited 1.4±0.18-fold induction under these conditions. Other genes that exhibited notable induction in expression levels were Rv0264c (2.9±1.6-fold), Rv1057 (2.3±0.2-fold), and Rv1330c (pncB1, 2±1-fold). In contrast, five genes that include Rv0146, mmpS5, fadD9, hupB, and Rv3830c were significantly downregulated by 5.9, 2.1, 5.1, 2.0 and 8.5-folds respectively, in db-cAMP-treated Mtb samples (Table 1).
Figure 3. Effect of cAMP on global gene expression profile of Mtb.
A) Effect of db-cAMP treatment on intracellular cAMP levels in Mtb. In vitro grown cultures of Mtb were treated with 20 mM butyric acid (b.a.) or db-cAMP for 2 hrs, followed by intracellular cAMP estimation by ELISA. Shown are the intracellular cAMP levels after 2 hrs of treatment (T2h) compared to that in untreated cells at zero hour time point (T0). B) Validation of differential expression of genes in microarray analysis by qRT-PCR. Genes upregulated in the db-cAMP-treated Mtb relative to the butyric acid-treated sample by microarray analysis were evaluated by qRT-PCR. The fold-expression indicates the ratio of sigA-normalized gene expression levels in the db-cAMP-treated strain relative to those in the butyric acid-treated Mtb. C) Effect of db-cAMP on in vivo activity of dnaK promoter. In vivo activity of dnaK promoter was assessed by measuring β-gal expression in Msm harboring pSD5B-dnaK (dnaK) using β-gal fluorescent substrate (C2FDG). Promoter activity was determined in mycobacterial cultures treated with 10 mM db-cAMP (+cAMP) or butyric acid (-cAMP) for 2 hrs. The empty plasmid pSD5B was used as a control. Data represent an average of three independent experiments and error bars indicate standard deviations.
Table 1. Differentially expressed genes in Mtb treated with db-cAMP relative to butyric acid treated bacteria.
| Gene productb | Locus | Change in expression (fold) | SD (fold) | P value |
| CHP | Rv0146 | –5.9 | 0.0 | 0.01 |
| CHP | Rv0264c | 2.9 | 1.6 | 0.02 |
| DnaK | Rv0350 | 3.7 | 0.6 | 0.02 |
| GrpE | Rv0351 | 2.9 | 0.9 | 0.05 |
| DnaJ | Rv0352 | 3.2 | 0.7 | 0.03 |
| MmpS5 | Rv0677c | –2.1 | 0.2 | 0.00 |
| CHP | Rv1057 | 2.3 | 0.2 | 0.04 |
| CHP | Rv1330c | 2.0 | 1.0 | 0.03 |
| CMP | Rv2025c | 9.8 | 4.4 | 0.02 |
| FadD9 | Rv2590 | –5.1 | 0.1 | 0.04 |
| HupB | Rv2986c | –2.0 | 0.0 | 0.02 |
| TRP | Rv3830c | –8.5 | 0.0 | 0.03 |
Expression of 3924 genes of Mtb was compared between the cells treated with db-cAMP and butyric acid. An average of data from three independent experiments (p≤ 0.05) is included in the table. Genes that exhibit ≥2-fold difference in expression levels by whole-genome microarray are shown. SD: standard deviation. bCHP, CMP and TRP denote conserved hypothetical protein, conserved membrane protein and transcription regulatory protein, respectively.
Next, we examined whether expression of these genes is altered in mycobacteria due to direct effect of cAMP on their promoter activity. A 450bp long DNA sequence corresponding to 5'-UTR of dnaK was cloned upstream to the lacZ in promoter probe plasmid pSD5B [20] and the recombinant plasmid pSD5B-dnaK was transformed into Msm. Expression of lacZ was subsequently monitored in the presence or absence of db-cAMP in Msm::pSD5B-dnaK. As shown in figure 3C, in the absence of db-cAMP (-cAMP), Msm::pSD5B-dnaK exhibited significant β-gal activity in comparison to Msm containing empty plasmid. Interestingly, addition of db-cAMP to bacterial cultures (+cAMP) resulted in ∼40-fold increase in activity of dnaK promoter, whereas there was no effect of db-cAMP on the basal expression levels of lacZ in empty plasmid containing control strain (Fig. 3C).
These results thus clearly demonstrate that cAMP acts as a regulator of the expression of multiple genes including a subset of heat stress-induced genes of Mtb.
Identification of CRPM-recognition sequence in 5'-UTR of cAMP-regulon
The CRP of E. coli binds to a 16bp sequence TGTGA-N6-TCACA in the 5'-UTR of cAMP-regulon. Similar to E. coli the CRP of Mtb (CRPM) was also shown to regulate expression of key Mtb genes that bear the similar sequences in their 5'-UTR [18]–[20], [22], [23]. In order to understand whether cAMP-mediated expression of genes in our study is due to direct binding of CRPM to their respective 5'-UTRs, we analyzed 350bp sequences upstream to the start codon of each of the 10 open reading frames (ORFs) by using the TubercuList database (http://genolist.pasteur.fr/TubercuList/) to identify the putative CRPM consensus sequences bearing two mismatches in either of the left (TGTGA) or right (TCACA) arms. Since dnaK is transcribed in operon with grpE and dnaJ [37], we omitted the upstream sequences to grpE and dnaJ in our analysis. As shown in table 2 except hupB, all other ORFs exhibited putative CRPM-recognition sequences in their 5'-UTR. These observations suggest that cAMP may regulate the expression of these genes by facilitating the binding of cAMP-activated CRPM to their promoter region.
Table 2. Analysis of putative CRP-recognition sequences in 5'-UTRs of cAMP-regulon of Mtb.
| Locus (Rv/annotation) | Sequencea | Position from Start codon |
| Rv0146 | tgtcgaggctttcacc | –325 |
| Rv0146 | tttcaccatgaacaca | –316 |
| Rv0146 | tgacaccggcatcacg | –45 |
| Rv0146 | agcgactcggtttaga | –266 |
| Rv0146 | tgtccaggcgttgacc | –244 |
| Rv0146 | cgagaccgtccgcacc | –103 |
| Rv0264c | gctgatctggatgacc | –300 |
| Rv0350/dnaK | agcgaccgtcagcacg | –301 |
| Rv0350/dnaK | cgttagcatgctcagt | –135 |
| Rv0677c/mmpS5 | tttcactgtactctga | –81 |
| Rv0677c/mmpS5 | tgttcgacgaattcca | –149 |
| Rv0677c/mmpS5 | tctgaaatctgtgacg | –70 |
| Rv1057 | cgtgacctaggtaaca | –248 |
| Rv1057 | tcagaatttggtcgct | –208 |
| Rv1330c | tgcggaccgcgtcgga | –21 |
| Rv1330c | agtcacgtagctcatc | –233 |
| Rv1330c | tggcatgcggctcgct | –276 |
| Rv2025c | tctgagcaagctcagc | –246 |
| Rv2025c | tgcgtatgaatgcaga | –82 |
| Rv2590/fadD9 | agtgaggggctggaca | –206 |
| Rv2590/fadD9 | cgtcatcattttgacc | –285 |
| Rv2590/fadD9 | cgtgccgcatctcaca | –133 |
| Rv3830c | tgcgcgcagtctcgcc | –72 |
| Rv3830c | tgtcaatgttgacaga | –42 |
| Rv3830c | tttgtcaatgttgaca | –44 |
Three hundred fifty base pair long DNA sequences upstream to the respective ORFs were analyzed to identify CRPM-recognition sequence. aThe underlined sequences represent putative CRPM-recognition sequences (TGTGA-N6-TCACA, with two mismatches in any of the arms) in the 5'-UTR of the respective genes.
EMSA to validate direct binding of CRPM- to 5'-UTR of dnaK operon
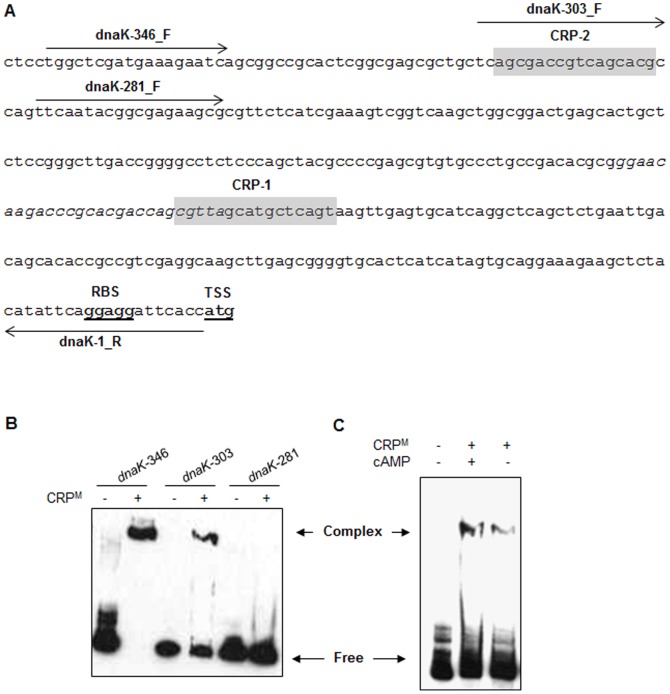
Analysis of 5'-UTR of dnaK operon indicates the presence of two putative CRPM-consensus sequences. The first sequence CRP-1 (5'- CGTTAGCATGCTCAGT-3') is located between positions -135 and -120 from translation start codon, whereas the second sequence CRP-2 (5'- AGCGACCGTCAGCACG-3') is situated further upstream between positions –301 and –286 from translation start codon (Fig. 4A). A careful observation demonstrates that CRP-2 is located 127bp upstream to SigH-binding site which is positioned between –158 to –130 from translation stop codon (5'- GGGAACAAGACCCGCACGACCAGCGTTA-3'), whereas CRP-1 overlaps with SigH-recognition sequence in 5'-UTR of dnaK (Fig. 4A). Since dnaK, grpE and dnaJ were amongst the most abundant transcripts in db-cAMP-treated Mtb, we sought to determine if their expression is controlled by direct binding of CRPM to the promoter sequence in the 5'-UTR. CRPM was purified as earlier [20] and subjected to binding with DNA fragment comprising of sequence between –1 to –346 from translation start codon of dnaK (dnaK-346). As shown in figure 4B, CRPM makes a specific complex with this sequence. Further deletion of 43bp from 5' end (dnaK-303) indicated that CRPM continues to bind this sequence, albeit with ∼2-fold reduced affinity as assessed by the intensity of CRPM-DNA complex. On the other hand deletion of additional 22bp (dnaK-281) which resulted in complete loss of CRP-2 site located between –301 and –286, completely abolished the complex formation (Fig. 4B). These results thus establish that: i) regulation of dnaK expression is governed by direct binding of CRPM to its 5'-UTR, ii) CRPM binds at CRP-2 site and not at CRP-1 site in the dnaK promoter, and iii) binding of CRPM to 5'-UTR of dnaK and its subsequent effect on mRNA expression is independent of SigH binding.
Figure 4. Identification of CRP-binding motif in promoter region of dnaK operon by EMSA.
A) Sequence of the 5'-UTR of dnaK operon. The putative ribosome-binding site (RBS), and the translation start site (TSS) are boldfaced and underlined. The putative CRP-binding sites are shaded, whereas the SigH-recognition sequence is shown in italics. Positions and directions of the primers that were used for PCR amplification of DNA fragments shown in (B) are marked by horizontal arrows. B) Interaction of CRPM with the 5'-UTR of dnaK operon. Biotin-labeled dnaK promoter fragments of various lengths, as shown, were incubated with CRPM in the presence of 1 mM cAMP before separation of DNA-protein complexes by gel electrophoresis. Reaction mixtures containing promoter fragments but lacking the CRPM were resolved in adjacent lanes as controls. C) Effect of cAMP on complex formation between CRPM and dnaK promoter. Purified CRPM pre-incubated with or without 1 mM cAMP for 30 min on ice, was subject to binding with full length dnaK promoter fragment (dnaK-346) for 15 min at 37°C before separation of CRPM–dnaK complexes by gel electrophoresis. Complex, CRPM–dnaK complex; Free, unbound DNA fragment.
Presence of cAMP enhances binding of CRPM to corresponding promoter DNA sequences [20], [23]. Hence, we analyzed if association of CRPM to dnaK promoter is also influenced by cAMP. Binding reactions were carried out in the absence (apo-CRPM) or the presence of 1 mM cAMP, and the CRPM-DNA complexes were resolved on polyacrylamide gel as described in materials and methods. Figure 4C shows that presence of cAMP enhances the complex formation between CRPM and dnaK-346 promoter fragment by 3-fold compared to that with apo-CRPM. These results thus clearly indicate that cAMP regulates the expression of dnaK operon by facilitating the binding of CRPM to its promoter.
Discussion
This study was designed to estimate the intracellular cAMP levels over a period of in vitro growth and its role in gene expression of Mtb. Cyclic AMP in bacteria was first reported half a century ago when the cyclic nucleotide was observed in the culture filtrate of Bravibacterium liquefaciens [38] and later in E. coli [39]. Subsequently occurrence of cAMP was reported in other bacteria including mycobacteria [3]. Later after discovery of cAMP in bacteria, it was studied that cAMP plays an important role in assimilation of sugar molecules by regulating a process called catabolite repression [40], [41]. Levels of cAMP keep changing as a function of glucose level in the cell [41]. Although in mycobacteria cAMP is not involved in carbon metabolism, variation in intracellular cAMP levels over a period of in vitro growth (Fig. 1A) indicates that cAMP levels are dynamic. A constant extracellular cAMP concentration rules out the prospect of differential export of cAMP to outside culture medium. Although effects of differential activities of ACs, PDEs and other associated factors on intracellular cAMP concentrations is not ruled out, a similar pattern of expression of 16 ACs and intracellular cAMP levels during in vitro growth of Mtb suggests that the relative abundance of ACs could be an important factor governing the intracellular cAMP levels in Mtb (Fig. 1A and B). While, the trigger(s) of sudden changes in expression of ACs and subsequent cellular cAMP levels at late growth stages of Mtb is yet to identify, the intracellular polyphosphates may be an important determinant of cellular cAMP levels [42]. Polyphosphates, generated by the activity of an enzyme known as polyphosphate kinase, is highly accumulated at the stationary phase as well as under different stress conditions in Mtb [43], and are known to inhibit the activity of ACs [42]. These information warrant further studies to analyze the effect of polyphosphates on intracellular cAMP levels.
Microbial pathogens including Mtb are adapted to survive under diverse stress conditions. Effects of cAMP on bacterial responses to cold-shock [44], expression of stress proteins [45] and RpoS, a late stationary sigma factor which also regulates oxidative stress response [46], [47] indicate that cAMP is an important determinant of the bacterial response to variety of stresses other than carbon metabolism in many pathogenic bacteria. Recently it was shown that cAMP regulates the response of uropathogenic E. coli to nitrosative, oxidative and acid stresses, and affects its virulence [48]. In contrast to these organisms, intracellular cAMP in Mtb was significantly stimulated after the heat stress whereas other stresses such as oxidative, nitrosative or acid exhibited a milder effect (Fig. 2A). Response to heat stress is an adaptive response to a sudden increase in ambient temperature, which involves a group of proteins such as chaperons, proteases, and regulatory factors commonly known as heat shock proteins [37]. Transcriptionally heat shock proteins are expressed in Mtb by involving an extracellular function sigma protein, SigH [49]. Upregulated expression of 5 ACs after the heat stress and presence of SigH-consensus sequences in the upstream promoter region of their respective ORFs (Fig. 2B and Table 3) together suggest that apparent induction of cAMP pool under these conditions may be due to differential transcription of mycobacterial AC-encoding genes by SigH. Parallel to these observations, induction of genes encoding primary heat shock proteins DnaK, GrpE, DnaJ1 and Rv2025c by exogenous cAMP indicates that a subset of heat shock proteins are expressed in Mtb under the effect of cAMP (Table 1 and Fig. 3B-C). Absence of CRP/Cmr-recognition sequences in sigH promoter and lack of its induction by cAMP together suggest that cAMP-driven expression of heat shock genes may not be directly controlled by SigH. Alternatively our results propose that cAMP regulates the expression of these genes by facilitating the binding of CRPM to their respective promoter sequences. This was further confirmed by the presence of CRP-consensus sequence in the upstream promoter regions of 9 out of 10 ORFs that were differentially expressed in Mtb after cAMP-treatment (Table 2), and by EMSA studies with CRPM which specifically recognized a 16-bp sequence -301 AGCGACCGTCAGCACG −286 in the 5'-UTR of dnaK operon, in a cAMP-dependent manner (Fig. 4).
Table 3. Identification of putative SigH-recognition sequences in the 5'-UTRs of AC-encoding genes of Mtb.
| Locus (Rv/annotation) | Sequencea | Position from Start codon |
| Rv0386 | cggaaatccaccgtccggtggcgtcgcttc | –98 |
| Rv1625c/cya | cgcaacatctcggccaggtccatgcggatg | –335 |
| Rv1647 | ggcaacgcggtgaccggcttcctgtttg | –311 |
| Rv2212 | cgaaactcgcacccagctcgcgatggcggtc | –114 |
| Rv2488c | cggcaccgcccacgatgcggtcgtgtcggttc | –152 |
Three hundred fifty base pair long DNA sequences upstream to the AC-encoding genes were analyzed to identify SigH-recognition sequences. aThe underlined sequences represent putative SigH-recognition sequences (c/gGGAAc-N17–21-c/gGTTc/g) in the 5'-UTR of the respective genes.
DnaK is a 70 kDa chaperone which is conserved in almost all living organisms. In Mtb DnaK is one of the most abundant proteins, which regulates Mtb virulence [50]. DnaK plays a crucial role in protecting bacteria from thermal or oxidative stress that cause partial unfolding and possible aggregation of cellular proteins. It has ability to bind the hydrophobic residues of partially unfolded proteins, exposed by stress which subsequently prevents them from aggregating, and allows them to refold. Cyclic AMP is universally present in mycobacteria and its levels are further elevated under stress conditions. Regulation of DnaK expression by cAMP indicates that this secondary messenger molecule is critical in regulating mycobacterial response to stress.
Materials and Methods
Bacterial strains, culture conditions and plasmid
For culturing of Mtb CDC1551, Middlebrook 7H9 broth supplemented with 1x OADC, 0.02% tween-80 and 0.5% glycerol, and Middlebrook 7H10 agar supplemented with 1x OADC and 0.5% glycerol were used. Msm mc2155 was cultured in 7H9 broth containing 0.02% tween-80 and 0.5% glycerol. Bacteria were grown at 37°C with (in 7H9 broth) or without (on 7H10-agar) shaking at 200 rotations per minute. For culturing in glucose-free medium, synthetic supplement containing oleic acid, albumin and saline equivalent to 1x concentration were added to 7H9 medium containing 0.02% tween-80 and 0.5% glycerol. Bacteria were treated with different stress agents as described earlier [36]. The promoter probe vector pSD5B [20] was kindly provided by Dr. Anil Tyagi, University of Delhi South Campus, New Delhi, India. To determine the in vivo activity of dnaK promoter in mycobacteria, the 450bp region upstream to translational start site of dnaK ORF was amplified using primer pairs dnak-450F (5'-GGGTCTAGAGCACCGTTGGCCCGTTCGATG-3') and dnak-1R (5'-CCCTCTAGAGGTGAATCCTCCTGAATATGTAG-3'), and cloned at Xba I site in pSD5B plasmid upstream to the lacZ resulting in pSD5B-dnaK. The pSD5B-dnaK plasmid harboring dnaK promoter in sense orientation was subsequently electroporated in Msm and the recombinant Msm::pSD5B-dnaK strains were selected on Middlebrook 7H10 agar containing 0.5% glycerol and 25 mg/L kanamycin.
Estimation of cAMP by ELISA
Intracellular cAMP determination in Mtb was performed with clarified cell lysate after heat lysis of cell pellets in 0.1 M HCl. Cyclic AMP was estimated by ELISAs using Enzyme Immunoassay Kits (Assay Designs Inc., Ann Arbor, MI) according to the manufacturer’s instructions. The intracellular cAMP levels in Mtb were estimated as nmol/gm wet weight.
RNA extraction
Total RNA was isolated from Mtb by using the TRIzol suspension according to manufacturer’s instructions (Invitrogen Corporation, Carlsbad, CA).
Real-time quantitative reverse transcription PCR
RNA isolated from the bacterial cultures was subjected to treatment with RNase-free DNase I (Ambion) to remove traces of contaminating DNA. Subsequently, absence of DNA in the RNA preparations was verified by 30 cycles of PCR followed by ethidium-bromide-stained agarose gel analysis before proceeding with reverse transcription of the RNAs. Complementary DNA (cDNA) synthesized from total RNA was subjected to real-time quantitative reverse transcription PCR typically as described earlier [36].
Microarray analysis
To analyze the effect of cAMP on the expression of mycobacterial genes, Mtb was cultured to OD600 of 0.6 and pelleted down by centrifugation at 6000 x g for 10min at 4°C. Culture pellets were washed twice with 1 x phosphate buffered saline, pH 7.4 (PBS) and suspended in 1/10th volume of 7H9 medium containing either of the 20 mM butyric acid (control) or di-butyryl cAMP (db-cAMP, test). After 2 hrs of incubation at 37°C in roller bottles, cells were pelleted down, washed 3 x with PBS and RNA was isolated as described above. For probe preparation, cDNA was synthesized from 5 µg of total RNA from the test and control strains of Mtb and labeled with Cy3 or Cy5 (GE Healthcare). Microarray slides were prepared by using 70-mer oligos encompassing the entire genome of Mtb (GEO accession number GSE54289) and hybridization of labeled cDNAs was performed typically as described earlier [36]. RNA samples were prepared from 3 biological replicates. Slide scanning and data analysis was performed as previously described [36].
β-Galatosidase assay
Promoter activity of dnaK was measured in Msm::pSD5B-dnaK by fluorescent based detection of lacZ expression, as described earlier [51]. Briefly, Msm::pSD5B-dnaK cultures were grown in 7H9 broth medium containing kanamycin (25 mg/L) to an OD600 of 1.0, washed twice with 7H9 medium and resuspended in 7H9 medium. The OD600 of culture suspension was adjusted to 1.0, and the cultures were incubated with either 10 mM db-cAMP or 10 mM butyric acid for 2 hrs. Hundred microliter of culture suspensions were taken into 3 separate wells of a 96-well black fluoroplate (Greiner Bio-One). Fluorescent β-gal substrate, 5-acetylamino-FDG (C2FDG) (Life Technologies) was subsequently added to a final concentration of 33 µM into each well and β-gal activity was estimated by measuring the fluorescence after 1 hr of incubation using a spectrofluorometer (Biotek) with an excitation of 485±20 nm and emission of 528±20 nm. A similar experiment was performed with Msm containing empty plasmid, pSD5B as control.
EMSA
DNA probes for EMSA analysis were synthesized by PCR amplification of the desired regions of the dnaK promoter, using biotin-labeled oligonucleotides as the primers (Fig. 4A). The amplicons were purified from agarose gels and used for gel-shift experiments. The EMSA experiments were performed typically as described earlier [20].
Funding Statement
Financial support by Department of Biotechnology, Government of India (grant no. BT/PR15063/GBD/27/296/2011) to NA is highly acknowledged. The support of NIH grants AI36973 and AI 37856, and HHMI to WB is gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Agarwal N, Lamichhane G, Gupta R, Nolan S, Bishai WR (2009) Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature 460: 98–102. [DOI] [PubMed] [Google Scholar]
- 2. Agarwal N, Bishai WR (2009) cAMP signaling in Mycobacterium tuberculosis. Indian J Exp Biol 47: 393–400. [PubMed] [Google Scholar]
- 3. Padh H, Venkitasubramanian TA (1977) Adenosine 3', 5'-monophosphate in mycobacteria. Life Sci 20: 1273–1280. [DOI] [PubMed] [Google Scholar]
- 4. McCue LA, McDonough KA, Lawrence CE (2000) Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res 10: 204–219. [DOI] [PubMed] [Google Scholar]
- 5. Shenoy AR, Sivakumar K, Krupa A, Srinivasan N, Visweswariah SS (2004) A survey of nucleotide cyclases in actinobacteria: unique domain organization and expansion of the class III cyclase family in Mycobacterium tuberculosis. Comp Funct Genomics 5: 17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shenoy AR, Visweswariah SS (2006) New messages from old messengers: cAMP and mycobacteria. Trends Microbiol 14: 543–550. [DOI] [PubMed] [Google Scholar]
- 7. Shenoy AR, Visweswariah SS (2006) Mycobacterial adenylyl cyclases: biochemical diversity and structural plasticity. FEBS Lett 580: 3344–3352. [DOI] [PubMed] [Google Scholar]
- 8. Guo YL, Seebacher T, Kurz U, Linder JU, Schultz JE (2001) Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. EMBO J 20: 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reddy SK, Kamireddi M, Dhanireddy K, Young L, Davis A, et al. (2001) Eukaryotic-like adenylyl cyclases in Mycobacterium tuberculosis H37Rv: cloning and characterization. J Biol Chem 276: 35141–35149. [DOI] [PubMed] [Google Scholar]
- 10. Ketkar AD, Shenoy AR, Ramagopal UA, Visweswariah SS, Suguna K (2006) A structural basis for the role of nucleotide specifying residues in regulating the oligomerization of the Rv1625c adenylyl cyclase from M. tuberculosis. J Mol Biol 356: 904–916. [DOI] [PubMed] [Google Scholar]
- 11. Linder JU, Schultz A, Schultz JE (2002) Adenylyl cyclase Rv1264 from Mycobacterium tuberculosis has an autoinhibitory N-terminal domain. J Biol Chem 277: 15271–15276. [DOI] [PubMed] [Google Scholar]
- 12. Tews I, Findeisen F, Sinning I, Schultz A, Schultz JE, et al. (2005) The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science 308: 1020–1023. [DOI] [PubMed] [Google Scholar]
- 13. Linder JU, Hammer A, Schultz JE (2004) The effect of HAMP domains on class IIIb adenylyl cyclases from Mycobacterium tuberculosis. Eur J Biochem 271: 2446–2451. [DOI] [PubMed] [Google Scholar]
- 14. Castro LI, Hermsen C, Schultz JE, Linder JU (2005) Adenylyl cyclase Rv0386 from Mycobacterium tuberculosis H37Rv uses a novel mode for substrate selection. Febs J 272: 3085–3092. [DOI] [PubMed] [Google Scholar]
- 15. Shenoy AR, Sreenath NP, Mahalingam M, Visweswariah SS (2005) Characterization of phylogenetically distant members of the adenylate cyclase family from mycobacteria: Rv1647 from Mycobacterium tuberculosis and its orthologue ML1399 from M. leprae. Biochem J 387: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abdel Motaal A, Tews I, Schultz JE, Linder JU (2006) Fatty acid regulation of adenylyl cyclase Rv2212 from Mycobacterium tuberculosis H37Rv. FEBS J 273: 4219–4228. [DOI] [PubMed] [Google Scholar]
- 17. Sinha SC, Wetterer M, Sprang SR, Schultz JE, Linder JU (2005) Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c. EMBO J 24: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bai G, McCue LA, McDonough KA (2005) Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J Bacteriol 187: 7795–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rickman L, Scott C, Hunt DM, Hutchinson T, Menendez MC, et al. (2005) A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol 56: 1274–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agarwal N, Raghunand TR, Bishai WR (2006) Regulation of the expression of whiB1 in Mycobacterium tuberculosis: role of cAMP receptor protein. Microbiology 152: 2749–2756. [DOI] [PubMed] [Google Scholar]
- 21. Akhter Y, Tundup S, Hasnain SE (2007) Novel biochemical properties of a CRP/FNR family transcription factor from Mycobacterium tuberculosis. Int J Med Microbiol 297: 451–457. [DOI] [PubMed] [Google Scholar]
- 22. Gazdik MA, Bai G, Wu Y, McDonough KA (2009) Rv1675c (cmr) regulates intramacrophage and cyclic AMP-induced gene expression in Mycobacterium tuberculosis-complex mycobacteria. Mol Microbiol 71: 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stapleton M, Haq I, Hunt DM, Arnvig KB, Artymiuk PJ, et al. (2010) Mycobacterium tuberculosis cAMP receptor protein (Rv3676) differs from the Escherichia coli paradigm in its cAMP binding and DNA binding properties and transcription activation properties. J Biol Chem 285: 7016–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bai G, Schaak DD, Smith EA, McDonough KA (2011) Dysregulation of serine biosynthesis contributes to the growth defect of a Mycobacterium tuberculosis crp mutant. Mol Microbiol 82: 180–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raychaudhuri S, Basu M, Mandal NC (1998) Glutamate and cyclic AMP regulate the expression of galactokinase in Mycobacterium smegmatis. Microbiology 144 ( Pt 8): 2131–2140. [DOI] [PubMed] [Google Scholar]
- 26. Gazdik MA, McDonough KA (2005) Identification of cyclic AMP-regulated genes in Mycobacterium tuberculosis complex bacteria under low-oxygen conditions. J Bacteriol 187: 2681–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nambi S, Basu N, Visweswariah SS (2010) cAMP-regulated protein lysine acetylases in mycobacteria. J Biol Chem 285: 24313–24323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu H, Hegde SS, Blanchard JS (2011) Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50: 5883–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nambi S, Gupta K, Bhattacharyya M, Ramakrishnan P, Ravikumar V, et al. (2013) Cyclic AMP-dependent Protein Lysine Acylation in Mycobacteria Regulates Fatty Acid and Propionate Metabolism. J Biol Chem 288: 14114–14124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee CH (1977) Identification of adenosine 3',5'-monophosphate in Mycobacterium smegmatis. J Bacteriol 132: 1031–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee CH (1979) Metabolism of cyclic AMP in non-pathogenic Mycobacterium smegmatis. Arch Microbiol 120: 35–37. [DOI] [PubMed] [Google Scholar]
- 32. Shenoy AR, Sreenath N, Podobnik M, Kovacevic M, Visweswariah SS (2005) The Rv0805 gene from Mycobacterium tuberculosis encodes a 3',5'-cyclic nucleotide phosphodiesterase: biochemical and mutational analysis. Biochemistry 44: 15695–15704. [DOI] [PubMed] [Google Scholar]
- 33. Bai G, Schaak DD, McDonough KA (2009) cAMP levels within Mycobacterium tuberculosis and Mycobacterium bovis BCG increase upon infection of macrophages. FEMS Immunol Med Microbiol 55: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bai G, Knapp GS, McDonough KA (2011) Cyclic AMP signalling in mycobacteria: redirecting the conversation with a common currency. Cell Microbiol 13: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, et al. (2013) Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4: 3–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agarwal N, Woolwine SC, Tyagi S, Bishai WR (2007) Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes. Infect Immun 75: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, et al. (2002) Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148: 3129–3138. [DOI] [PubMed] [Google Scholar]
- 38. Okabayashi T, Ide M, Yoshimoto A (1963) Excretion of adenosine-3',5'-phosphate in the culture broth of Brevibacterium liquefaciens. Arch Biochem Biophys 100: 158–159. [DOI] [PubMed] [Google Scholar]
- 39. Makman RS, Sutherland EW (1965) Adenosine 3',5'-Phosphate in Escherichia Coli. J Biol Chem 240: 1309–1314. [PubMed] [Google Scholar]
- 40. Perlman R, Pastan I (1968) Cyclic 3'5-AMP: stimulation of beta-galactosidase and tryptophanase induction in E. coli. Biochem Biophys Res Commun 30: 656–664. [DOI] [PubMed] [Google Scholar]
- 41. Ullmann A, Monod J (1968) Cyclic AMP as an antagonist of catabolite repression in Escherichia coli. FEBS Lett 2: 57–60. [DOI] [PubMed] [Google Scholar]
- 42. Guo YL, Mayer H, Vollmer W, Dittrich D, Sander P, et al. (2009) Polyphosphates from Mycobacterium bovis--potent inhibitors of class III adenylate cyclases. FEBS J 276: 1094–1103. [DOI] [PubMed] [Google Scholar]
- 43. Singh R, Singh M, Arora G, Kumar S, Tiwari P, et al. (2013) Polyphosphate Deficiency in Mycobacterium tuberculosis Is Associated with Enhanced Drug Susceptibility and Impaired Growth in Guinea Pigs. J Bacteriol 195: 2839–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohsaka Y, Ohgiya S, Hoshino T, Ishizaki K (2001) Cold-stimulated increase in a regulatory subunit of cAMP-dependent protein kinase in human hepatoblastoma cells. DNA Cell Biol 20: 667–673. [DOI] [PubMed] [Google Scholar]
- 45. Jeong KC, Baumler DJ, Kaspar CW (2006) dps expression in Escherichia coli O157:H7 requires an extended -10 region and is affected by the cAMP receptor protein. Biochim Biophys Acta 1759: 51–59. [DOI] [PubMed] [Google Scholar]
- 46. Barth E, Gora KV, Gebendorfer KM, Settele F, Jakob U, et al. (2009) Interplay of cellular cAMP levels, {sigma}S activity and oxidative stress resistance in Escherichia coli. Microbiology 155: 1680–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheng Y, Sun B (2009) Polyphosphate kinase affects oxidative stress response by modulating cAMP receptor protein and rpoS expression in Salmonella typhimurium. J Microbiol Biotechnol 19: 1527–1535. [PubMed] [Google Scholar]
- 48. Donovan GT, Norton JP, Bower JM, Mulvey MA (2013) Adenylate cyclase and the cyclic AMP receptor protein modulate stress resistance and virulence capacity of uropathogenic Escherichia coli. Infect Immun 81: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raman S, Song T, Puyang X, Bardarov S, Jacobs WR Jr, et al. (2001) The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol 183: 6119–6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stewart GR, Snewin VA, Walzl G, Hussell T, Tormay P, et al. (2001) Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat Med 7: 732–737. [DOI] [PubMed] [Google Scholar]
- 51. Rowland B, Purkayastha A, Monserrat C, Casart Y, Takiff H, et al. (1999) Fluorescence-based detection of lacZ reporter gene expression in intact and viable bacteria including Mycobacterium species. FEMS Microbiol Lett 179: 317–325. [DOI] [PubMed] [Google Scholar]