Abstract
The transcriptome profiles were compared for buffalo embryos with normal growth and embryos with retarded growth on Day 25 after mating. Embryos with retarded growth on Day 25 after mating have a reduced likelihood of undergoing attachment to the uterine endometrium and establishing a pregnancy. Italian Mediterranean buffaloes were mated by AI and on Day 25 underwent trans-rectal ultrasonography to ascertain embryo development. Embryos with an embryonic width (EW)>2.7 mm were classed as normal embryos and embryos with an EW<2.7 mm were classed as retarded embryos. Three buffaloes with embryos of the largest EW (3.7, 3.7 and 3.9 mm) and three buffaloes with embryos of the smallest EW (1.5, 1.6 and 1.9 mm) were slaughtered on Day 27 to recover embryos for transcriptome analysis using a bovine custom designed oligo array. A total of 1,047 transcripts were differentially expressed between embryos with normal growth and embryos with retarded growth. Retarded embryos showed 773/1,047 (74%) transcripts that were down-regulated and 274/1,047 (26%) transcripts that were up-regulated relative to normal embryos; in silico analyses focused on 680/1,047 (65%) of the differentially expressed transcripts. The most altered transcripts observed in retarded embryos were associated with membrane structure and function and with metabolic and homeostasis maintenance functions. Other notable functions altered in retarded embryos were developmental processes and in particular nervous system differentiation and function. Specific biochemical pathways such as the complement cascade and coagulation were also altered in retarded embryos. It was concluded from the findings that buffalo embryos with retarded growth on Day 25 after mating show altered gene expression compared with normal embryos, and some de-regulated functions are associated with attachment to the uterine endometrium.
Introduction
The water buffalo (Bubalus bubalis) is a short-day breeder and at higher latitudes females show distinct seasonal changes in reproductive function. Optimal fertility occurs during decreasing day length in autumn and fertility to both natural mating and AI declines appreciably during increasing day length from late-winter to early-spring [1], [2]. The latter period is recognized as the transition phase from the breeding to non-breeding season [1], [2]. Notable features of ovarian function during the transition phase are the reduced size of the corpus luteum, decreased vascularization, and relatively low concentrations of circulating progesterone (P4) [3], [4]. The latter was proposed to be a major factor in the increased incidence of embryonic mortality, which is the primary cause of reduced fertility in buffaloes during the transition phase [5]. Progesterone has been shown to have a fundamental role in embryonic development and implantation in buffalo [2] and also cattle [6]–[8].
A notable feature of embryonic mortality in buffaloes is that the highest incidence occurs between Days 25 and 45 after mating [1], [5], [9], [10]. This represents late embryonic mortality and differs from cattle that typically undergo embryonic mortality between Days 8 and 17 after mating [11],[12]. The latter covers the period during which there is maternal recognition of pregnancy. It was proposed that late embryonic mortality in buffaloes results from the failure of embryonic attachment [2], [13] which is a consequence, at least in part, of reduced concentrations of P4 in circulation from around Day 10 after mating [3].
Buffalo embryos destined to undergo late embryonic mortality show retarded growth at Day 25 after mating compared with embryos that maintain pregnancy [14]. Balestrieri et al. [13] recently reported that the chorioamnion of embryos with retarded growth (embryo width<2.7 mm) had a different proteomic profile to that of embryos with normal growth (embryo width>2.7 mm). Differences in proteome profiles were also reported for the caruncles adjacent to the chorioamnions of embryos with retarded and normal growth. Differentially expressed proteins were related to antioxidant protection, protease inhibition and protein folding [13]. These findings supported other evidence that a disruption of conceptus-uterus interaction, as a result of reduced progesterone, is the major cause of late embryonic mortality in buffaloes [1], [2].
Global transcriptome profiling has been used in cattle to gain insight into the molecular features of embryonic development [7], [15], [16] and embryonic attachment to the uterine endometrium [6], [17]–[19]. As noted above, embryonic mortality in cattle is most likely to occur before the window for maternal recognition of pregnancy closes, which is around Days 16–17. Embryo and uterine transcriptome profiling in cattle has therefore mainly been studied up to this period. In a study carried out on Day 16, 46 transcripts were conceptus-specific and 34 transcripts were endometrium-specific [19]. It was proposed that the differentially expressed transcripts were involved in the maternal recognition of pregnancy. In a second study in cattle, changes were observed from Day 21 to Day 28 in the gene expression profile of conceptus trophoblast membranes and it was similarly proposed that the changes in expression were related to embryonic attachment [20].
The decline in fertility in buffaloes during the transition phase to the non-breeding season has a significant impact on the rate of genetic improvement and distribution of germplasm, which reduces the efficiency of production. An improved understanding of the molecular basis for late embryonic mortality in buffaloes would contribute to fundamental comparative reproductive biology and also lead to strategies to enhance fertility during the transition phase. In the present study, global transcriptome profiling was undertaken for the first time in buffalo embryos that showed either normal or retarded growth at Day 25 after mating.
Materials and Methods
Embryo collection
Embryos were obtained from multiparous Italian Mediterranean buffaloes (Bubalus bubalis) that had undergone synchronization of ovulation and artificial insemination (AI) during the transition phase to the non-breeding season as described by Balestrieri et al. [13]. The “Ethical Animal Care and Use Committee” of the University of Naples Federico II approved the experimental design and animal treatments (Permit Number: 2013/010858).
Buffaloes underwent ultrasonography on day 25 after AI and were divided in two groups according to the embryo width (> or<than 2.7 mm). Three buffaloes that had normal embryos, i.e. with the largest embryo width and three buffaloes that had retarded embryos, i.e. with the smallest embryo width on Day 25 were slaughtered on Day 27 to collect embryos, as previously described [13]. At slaughter (Day 27) the embryos with the largest EW (3.7, 3.7 and 3.9 mm) had a respective EL of 9.8, 7.5 and 9.8 mm and embryos with the smallest EW (1.5, 1.6 and 1.9 mm) had a respective EL of 5.0, 5.5 and 5.0 mm.
RNA extraction
After removal from the uterus embryos were measured and placed in a test tube containing RNAlater solution. Total RNA was extracted from each embryo according to the Trizol protocol (Invitrogen). The concentration of RNA was determined with a Nanodrop (NanoDrop,) spectrophotometer and RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies).
Microarray hybridization
A comparative genomic approach was adopted [21] which utilized a heterologous hybridization technique [22], [23]. The EmbryoGENE bovine transcriptome microarray [24] was chosen for genome-wide analysis. This was a custom designed oligo array, constructed from global transcriptome RNA sequencing (RNA-Seq) experiments and enriched for embryo-specific transcripts and 3′UTR variants.
For each sample (embryo), 200 ng of total RNA were synthesized to Cy3-labeled, linearly amplified cRNA using the One-Color LowInput Quick Amp Labeling kit (Agilent Technologies). The concentration and quality of cRNA was assessed using a Nanodrop (NanoDrop) spectrophotometer. For each of the normal and retarded embryos, three assay replicates were produced and 1.65 µg of Cy3-labeled linearly amplified cRNA was fragmented and hybridized for 17 h to the Agilent bovine (Bos taurus) 4×44 k oligonucleotide microarray according to the protocol provided by the manufacturer (Agilent Technologies).
Array scanning and data analysis
Agilent Feature Extraction (AFE) 11.0.1.1 image analysis software was used to extract intensity from scanned images (file.tiff) obtained after microarray slides reading. The intensity files were loaded into GeneSpring software version 11.5 (Agilent Technologies) for quality control and gene expression analysis. First, the percentile shift normalizing algorithm, shifting to 75 percentile, was applied on the dataset to correct systematic errors. For differential expression analysis, assay replicates and embryo replicates were grouped together, and only genes detected in 100% of assay replicates for each embryo, were considered. The Unpaired t-Test with FDR correction was used to calculate a P-value for each differentially expressed gene, whilst fold-change was used to calculate differential expression between the two conditions. Only genes with adjusted P values≤0.05, and fold-change≥1.5 and ≤-1.5, were regarded as differentially expressed. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2064.
Quantitative RT-PCR analysis
The transcripts to analyze were chosen according to three main criteria: fold change degree, biochemical and functional roles, the availability of working primers, as the RT-PCR primers were designed according to bovine sequences in GenBank. The six up-regulated transcripts analyzed were: annexin A3 (ANXA3), serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (SERPINE1), keratin 18 (KRT18), syntaxin 7 (STX7), tetraspanin 8 (TSPAN8) and ectonucleotide pyrophosphatase/phosphodiesterase 3 (ENPP3). The four down-regulated transcripts were: the complement component C5 (C5), D-aspartate oxidase (DDO), dihydropyrimidinase (DPYS) and myosin X (MYO10). Specific primers were designed through the Primer-Blast algorithm (http://www.ncbi.nlm.nih.gov/tools/primer-blast) referring to the corresponding bovine gene sequences available in GeneBank (www.ncbi.nlm.nih.gov/genbank) and through the Blast-primer algorithm.
The primers used for amplification of GAPDH, the reference gene, were designed according to the Bubalus-specific sequence. Primer sequences and melting temperature (Tm), amplicons length, and the accession numbers for the bovine and buffalo sequences are shown in Table 1. Each RT-PCR was performed in triplicate using the CFX96 Real-Time PCR (Bio-Rad Laboratories). The reactions were performed in 20 µl final volume with the 2× Master mix Bio-Rad iQ SYBR Green Supermix, 125 µM of each primer and 1 µl of a 1/10 dilution of cDNA obtained through reverse transcription starting from 1 µg of total RNA, DNA free, in 20 µl of reaction volume. The RT-PCR protocol includes 35 amplification cycles. The expression level of each transcript was normalized to GAPDH transcript levels. The relative expression levels in the analyzed tissues were obtained using the 2−ΔΔ Ct method [25].
Table 1. Accession number (a), Gene Symbol (b), primers sequence (c), amplicon length (d) and Tm (e) of transcripts validated in qPCR experiments.
| a | b | c | d | e |
| NM_001035325 | ANXA3 | F 5′ GACCCCACCGGCAGTGTTCG 3′ R 5′ GGCATGGCCGATCTCCTGCAT 3′ | 130 bp | 64° |
| NM_174137 | SERPINE1 | F 5′ GTTTAGGCCGAGCCAGGCGG 3′ R 5′ TGGGATTGTGCCGCACCACG 3′ | 206 bp | 64° |
| NM_001079629 | TSPAN8 | F 5′ CCTGGGATGCTGTGGTGCCAT 3′ R 5′ ACGATACCTGCCGCCACCTGC 3′ | 102 bp | 64° |
| NM_001192095 | KRT18 | F 5′ GCAGACCGCTGAGATAGGAG 3′ R 5′ CAAGCTGGCCTTCAGATTTC 3′ | 109 bp | 60° |
| NM_001077864 | STX7 | F 5′ ACAGAGGACGACCTGCGCCTT 3′ R 5′ TGCGCTGATAGTCTGCCGCC 3′ | 217 bp | 64° |
| NM_173908 | DDO | F5′TGTGAAGGCCCTGCCTACCTCC 3′ R5′ CGGGTGAAGCTCCCACAGGTC 3′ | 102 bp | 64° |
| NM_001101065 | MIOX | F 5′ CTACACGTCTGGCCCGCTCCT 3′ R 5′ CCCCGAACTGGGCATGCTTCC 3′ | 104 bp | 64° |
| NM_001166616 | C5 | F 5′CGCAAACGCAGATGACACCCG 3′ R 5′ TGGGGCCTGCCTGAATCCGA 3′ | 200 bp | 62° |
| NM_001075923 | ENPP3 | F 5′ TGTTGGTGGCTGTGTCACTT 3′ R 5′GGCAGCCCTCTAGTCCTCTA 3′ | 117 bp | 60° |
| NM_001192214 | DPYS | F 5′ CCTTCAACGCCTGACTTCCT 3′ R 5′ AGAGCTTTCTGGCAGCTGTT 3′ | 98 bp | 58° |
| GU324291 | GAPDH | F 5′ TCACTGGCATGGCCTTCCGC3′ R 5′ GCCCTCTGACGCCTGCTTCAC 3′ | 122 bp | 60° |
Bioinformatic tools and databases
The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) [26] was used to ascertain major biochemical and functional pathways. The full list of differentially expressed transcripts and the list of the up- and down-regulated genes, respectively, were separately analyzed and compared. The molecular features taken into account for analysis were selected according to a statistical cut off P-value of 0.05, after the Benjamini method correction, and the percentage of genes included in each biological group.
The visualization of the main gene networks involved was obtained through Ingenuity Pathways Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com).
Results
Gene expression profiling
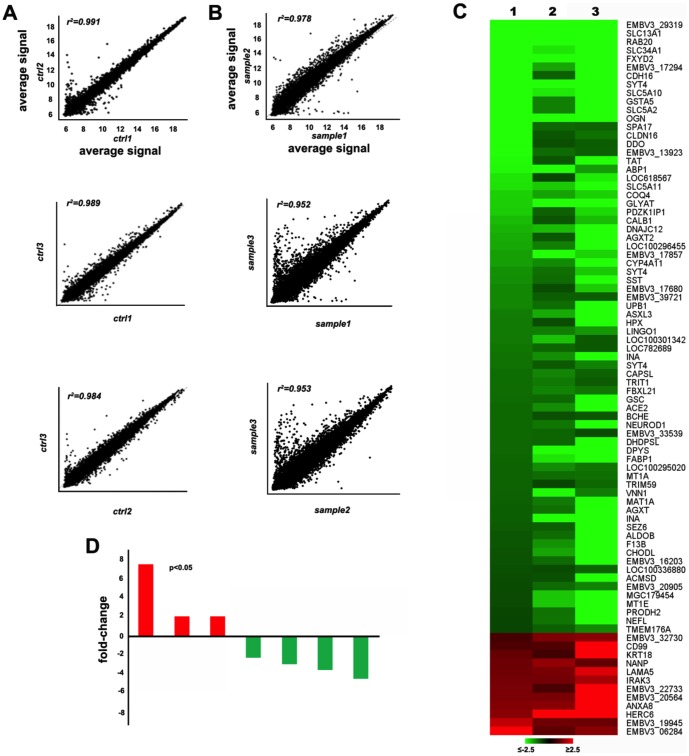
The heterologous hybridization of the bovine array showed that, a total of 1,047 transcripts were differentially expressed between embryos with normal growth and embryos with retarded growth (Figure 1). In panel A and B the correlation of the results among the samples of each group (respectively normal and retarded embryo) is shown. Retarded embryos showed 773/1,047 (74%) transcripts that were down-regulated and 274/1,047 (26%) transcripts that were up-regulated relative to normal embryos. A heat map of representative differentially expressed transcripts and a graphic representation are shown in panels C and D of Figure 1.
Figure 1. Microarray hybridization experiments.
Panel A and panel B show the correlation of the results among the three samples of each group (respectively ctrl = normal embryos and sample = retarded embryos); Panel C depicts the heat map corresponding to some differential expressed transcripts. The number 1, 2, 3 refer to retarded embryo samples; Panel D shows the relative fold change of some differential expressed transcripts.
Quantitative RT-PCR
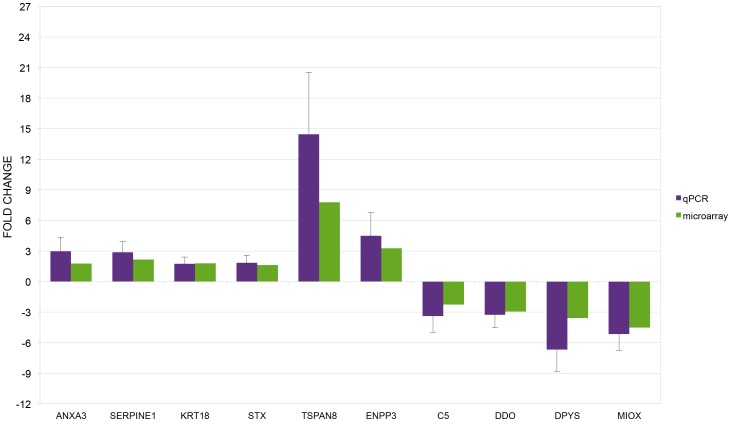
The microarray hybridization results were corroborated by the quantitative RT-PCR analysis of ten transcripts. In Figure 2 are shown the results of qPCR (retarded embryos versus normal embryos) compared to the corresponding results in microarray experiment. There was agreement for all compared transcripts. The direction of the expression change (up- or down-regulation) was confirmed for all transcripts.
Figure 2. qPCR microarray data validation.
Graph of qPCR data for selected genes compared to corresponding microarray data. The results are expressed as the mean + standard deviation of the relative expression of each transcript.
In silico analysis of differentially expressed transcripts
The in silico analyses focused on about 680/1,047 (65%) of the differentially expressed transcripts. Of the transcripts not included, about 15% were described as “Novel Transcribed: embryo expressed sequence tags (ESTs)” that at present lack a GenBank annotation; about 10% were only predicted in the bovine genome and lack any functional annotations; and about 7% were alternate forms of the same locus or polymorphic alleles. The Database for Annotation, Visualization and Integrated Discovery (DAVID), and the Ingenuity Pathway Analysis (IPA) tools were used to explore and enlighten the main molecular features and the biochemical networks that were altered in retarded versus normal embryos.
Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis
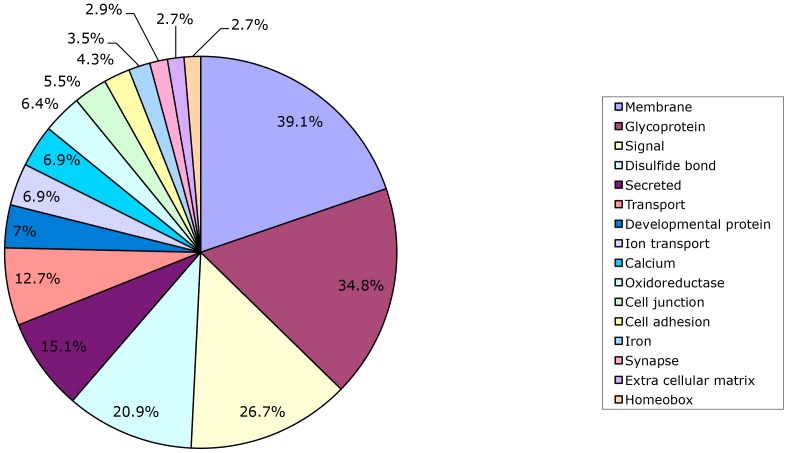
A scheme of the main classes of putative transcriptionally altered proteins in retarded and normal embryos is depicted in Figure 3. The more abundant class (39.1%) contains proteins involved in membrane structure and function. A further dissection of this major class showed that a large number of the proteins are glycoproteins, which are also the second most abundant class (34.8%) (Figure 3). The major functions summarized (signaling, molecule secretion, transport of macromolecules and ions, cell-cell junctions, cell adhesion, synapses) are associated with both basic and specialized membrane functions. The group of developmental (7.0%) and homeobox (2.7%) proteins are involved in the regulation and coordination of embryonic growth. All the features and functional classes described in Figure 3 were further interrogated and clarified from the analysis of microarray data through the Gene Ontology Tool and the results are shown in Table 2. The most abundant classes related to developmental processes and in particular cell and tissue differentiation, including cellular components and extracellular matrix. Over 80 de-regulated transcripts in retarded embryos were related to nervous system development. This included neuron cell differentiation and other functional features such as ion transport, cell-cell signaling and response to external stimuli (hormones and growth factors). The latter mechanisms are also more generally associated with the maintenance of cellular homeostasis, which would seem to have been compromised in the retarded embryos (Table 2). Pivotal aspects of basal metabolism such as oxidation-reduction reactions, biosynthetic processes, and amino acid and lipid metabolism, also differed between normal and retarded embryos (Table 2). Transcription factors and regulatory proteins also seemed to differ between normal and retarded embryos (Table 2).
Figure 3. Bioinformatic data analysis.
The results of the SP/PIR TOOL analysis of the data are summarized and the class and percentage of transcripts are reported.
Table 2. Gene Ontology analysis for molecular functions, subclasses, and percentage, of the differentially expressed genes (DEGs), for normal and retarded buffalo embryos on Day 27 of development.
| Molecular function | Subclasses | DEGs (%) |
| Developmental process | Ion/cation transport; Neuron differentiation; Cell adhesion; Oxidation reduction | 25.0 |
| Homeostatic process | Neurological processes; Ion homeostasis; Regulation of hormone levels; Cell-cell signalling | 16.3 |
| Cell differentiation | Cellular process; Cellular component biogenesis; Biological regulation | 15.0 |
| Nervous system development | Cell differentiation; Neurogenesis | 12.7 |
| Morphogenesis | Multicellular organismal development; Cellular component morphogenesis | 11.9 |
| Response to external stimuli | Response to stress; Response to chemical stimuli; Response to wounding; Inflammatory response; Blood coagulation | 10.1 |
| Organic acid metabolic process | Biosynthetic process; Amino acid metabolic process; Lipid metabolic process | 7.5 |
Ingenuity Pathway Analysis (IPA)
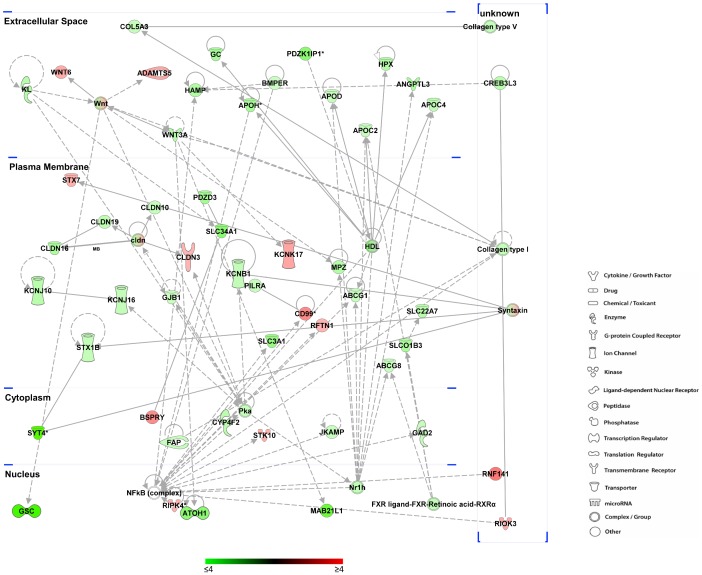
The Ingenuity Pathway Analysis provided further detail on the functional pathways expressed between normal and retarded embryos, visualizing the molecular networking among different pathways and cellular compartments. The network of altered transcripts involved in lipid metabolism (Figure 4) is a clear example of the large range of functions altered in retarded embryos. The transcripts (57) described in this network are associated with all cellular compartments and a large number of the proteins are localized in the plasma membrane. These proteins are involved in a wide range of pathways and functions including ion, vitamin and protein transport (KCNB1, GC, SLC34A1), tight junctions (CLDN10), transcriptional regulators (GSC,), enzymes (GAD2) and apoliproteins (APOD, APOH).
Figure 4. Lipid metabolism network.
The scheme shows the altered transcripts involved in lipid metabolism pathways (according to IPA algorithm). The putative cellular and extracellular localization of each transcript is also shown. The colour of each transcript indicates the expression status: the colour scale from green to red indicates the degree of down- and up-regulation respectively. The legend of shapes correspondence is also reported.
Fifteen of the differentially expressed transcripts in retarded embryos (12 down-regulated and 3 up-regulated) are associated with the complement/coagulation cascade (Table 3). These included members of the serine protease family (SERPINE1, SERPIND1 and SERPINA1). SERPINE1 interacts with PLAT to regulate the activity of plasminogen (PLG) that in turn regulates the complement cascade (C5, C8A) (Table 3). The transcripts of SERPINE1 and C5 are two of the ten transcripts whose altered expression was confirmed by qRT-PCR in both normal and retarded embryos (Figure 2, Table 1).
Table 3. Coagulation/complement cascade genes.
| Gene Symbol | Description | Direction of change |
| CD55 | CD55 molecule, decay accelerating factor for complement | +2.04 |
| CPB2 | carboxypeptidase B2 (plasma) | −3.25 |
| F2 | coagulation factor II (thrombin) | −1.88 |
| F9 | coagulation factor IX | −3.9 |
| F13B | coagulation factor XIII, B polypeptide | −4.86 |
| C5 | complement component 5 | −2.24 |
| C8A | complement component 8, alpha polypeptide | −1.70 |
| C8B | complement component 8, beta polypeptide | −1.72 |
| FGB | fibrinogen beta chain | −1.59 |
| MBL2 | mannose-binding lectin (protein C) 2, soluble | −1.54 |
| PLG | plasminogen | −1.89 |
| PLAU | plasminogen activator, urokinase | +1.76 |
| SERPINA 5 | SERPINE peptidase inhibitor, clade A (antitrypsin) 5 | −1.75 |
| SERPIND1 | SERPIN peptidase inhibitor, clade D (heparin cofactor) 1 | −1.97 |
| SERPINE1 | SERPIN peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1) member 1 | +2.16 |
Gene identifier, description, and direction of change, of the differentially expressed genes involved in the coagulation/complement cascade for retarded compared with normal buffalo embryos on Day 27 of development are reported.
Discussion
Buffalo embryos that undergo late embryonic mortality have retarded growth on Day 25 of development compared with embryos that continue to develop [13], [14]. A comparison was therefore made of the transcriptome profiles for normal and retarded embryos on Day 27. At this stage of development embryos are preparing to attach to the uterine endometrium which represents a critical phase in the establishment of a pregnancy. Analysis of the transcriptome revealed that retarded embryos undergo extensive changes in the expression of genes involved in a wide range of biological mechanisms associated with developmental processes and cell differentiation. The large number of abnormally expressed protein associated to membrane structure and functions, correlates with the abundance and heterogeneity of this class of proteins [27].
The altered transcripts are involved in signalling, ion transport and molecule secretion, cell junction and adhesion, extra cellular matrix components. All these functions allow the response to internal and external stimuli, contributing to the maintenance of the homeostatic equilibrium and enabling cell and tissue differentiation [28]. Further these molecular features constitute the prerequisite for an efficient embryonic-maternal communication that is pivotal for the establishment and achievement of a successful pregnancy [29]. Annexin A2 (ANXA2), a calcium-dependent phospholipid-binding protein is a component of the extra cellular matrix and like the ANXA3 transcript, were up-regulated in this study. ANXA2 is a pleiotropic gene playing a role in regulation of cellular growing and adhesion and in the transduction of cell signals. It is involved in the maintenance of placentation [30] and is important for embryo adhesion to the endometrium [31]. Cell adhesion is required for embryo morphogenesis [28]. ANXA2 and ANXA1 proteins were decreased in caruncles that were adjacent to the same embryos that were retarded in the present study [13].
Developmental processes were the predominantly altered group in retarded embryos and included nervous system differentiation and functioning. In mammals, neurulation is the next developmental step after gastrulation [32]. The altered transcripts cover a wide range of functions ranging from axonal morphogenesis and synaptic conformation and activity (e.g. neurexin1, neuroglin), growth factor (e.g. neurotrophin) and transcription regulators (e.g. homeobox LHX8).
Several other components of the homeobox gene class that were down-regulated in retarded embryos are functionally associated with developmental processes. This is perhaps not a surprising finding considering the role of these proteins as regulative molecules in cell, tissue and organ differentiation. One of these genes, Goosecoid, is the first gene expressed in the organizer region of all vertebrates [32]. The homeobox gene POU5F1 (previously known as OCT4), which has a pivotal role in the precocious steps of embryogenesis, and is a marker of cell pluripotency [33], was up-regulated in retarded embryos. These genes regulate the expression of a large number of other transcripts involved in stemness and differentiation, so they need a very fine spatiotemporal tuning. Noteworthy, POU5F1, in coordination with other two pivotal factors (SOX2 and NANOG), represses the expression of genes involved in lineage commitment including also GSC.
Cellular metabolism was another group of altered processes that emerged from the in silico analysis. A number of altered transcripts are involved in redox reactions. This class of chemical processes is associated with basic cell biochemical pathways and mechanisms such as energetic metabolism and hormone production. Lipid and steroid metabolism is linked to biological oxidation and hormone secretion and function [34]. The lipid metabolism pathways outlined in Figure 4 highlights the close interplay that exists between the different structural and functional pathways that showed altered expression. The altered transcripts (57) cover a wide range of functions including the homeobox Goosecoid (GSC) described above. Similarly, amino acid metabolism, including tryptophan and alanine, encompasses important cellular biochemical patterns and influences protein synthesis. Amino acid biochemistry, similarly to other processes of cellular metabolism, constitutes a crucial aspect of in vivo and in vitro embryo development [35].
Complement cascade and coagulation in placenta tissues are known to be involved in pregnancy failure. In humans and mice, altered activity in coagulation and complement cascade, in both maternal and embryonic counterparts, is associated with higher rates of pregnancy failure [36]. Studies in humans (hatching blastocysts, [37]) and mice (foetuses, [38]) showed specific roles for the complement and coagulation cascade in spontaneous loss of pregnancies. In retarded embryos, 15 transcripts were altered, and most (12/15) showed decreased expression. Two members of the SERPIN family (SERPIND1 and SERPINA5) were down regulated and one (SERPINE1) was up-regulated in retarded embryos. In the previous proteome study, SERPINE1 was up-regulated in the chorionamnion of buffalo embryos retarded on Day 25 and SERPINA3 (a1-antichymotrypsin) was up-regulated in caruncles adjacent to these embryos [13].
The extensive dysregulation observed in retarded embryos compared with normal embryos suggested that important regulative patterns were altered in retarded embryos. A group of dysregulated transcripts, involved in several processes, code for transcription factors including homeobox (e.g. goosecoid, down regulated), helix loop helix proteins, (e.g. BHLHE41 (previously known as BHLHB3, up regulated), and zinc finger proteins (e.g. FEZF1, down regulated). Other down-regulated genes (e.g Neuronatin) are known to be imprinted in several mammalian species [39]. Few non-coding RNAs (ncRNA, e.g. MIR2315) were dysregulated; however, it is likely that there was an underestimation of this component because of the lack of full annotation of the transcripts on the bovine array and the heterologous hybridization utilized. Epigenetic mechanisms have been widely established as a pivotal level of transcriptome regulation [40], [41]. Also, there is substantial evidence for a role of genomic imprinting and ncRNA in embryonic differentiation and development [42], [43].
Notwithstanding the limitations of a heterologous system, it does provide an important initial understanding of the transcriptome for species that lack an appropriately annotated genome [23] and the results offer a first insight at large scale level of the molecular events. The heterologous platform chosen for this experiment is enriched for transcripts involved in the bovine reproductive tissues. In our studies, buffaloes that had retarded embryos on Day 25 did not show an increase in circulating concentrations of P4 from Day 10 to Day 20 after mating [3], [13]. Progesterone influences the structure and function of the uterine endometrium, which impacts on the developing embryo [44]–[46]. Accordingly, a lack of progesterone can be presumed to be the underlying cause of retarded growth in buffalo embryos and the differences in transcriptome (present study) and proteome [13] profiles between retarded and normal buffalo embryos. Further evidence of the relationship between P4 and late embryonic mortality in buffaloes is that mortality can be reduced by treatment with GnRH agonist, hCG or P4 on Day 25 after mating [47].
In summary, the present study has demonstrated notable differences in the transcriptome of buffalo embryos that had normal and retarded growth on Day 27 after mating. A major class of altered transcripts were glycoproteins variously involved in membrane structure and function including cell adhesion, ion transport, cell-cell junction and communication, signalling, and secretion. The main biological processes associated with altered gene expression were developmental processes including nervous system differentiation, cellular and chemical homeostasis, ion transport, and general metabolism. We may conclude that changes in the transcriptome of retarded embryos impact on the ability of these embryos to attach to the uterine endometrium and establish a pregnancy.
Funding Statement
This work was supported by a grant from Ministry of University and Research P.R.I.N./MIUR 2008 code 2008X2PNYX_005 (MS), the Italian Association for Cancer Research (Grant IG-13176), the University of Salerno (Fondi FARB 2011–2013), EU COST Action ‘SEQAHEAD: Next Generation Sequencing Data Analysis Network’ (BM1006). GN was supported by a ‘Mario e Valeria Rindi’ fellowship of the Italian Foundation for Cancer Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Campanile G, Neglia G (2007) Embryonic mortality in buffalo cows. Ital J Anim Sci 6: 119–129. [Google Scholar]
- 2. Campanile G, Baruselli PS, Neglia G, Vecchio D, Gasparrini B, et al. (2010) Ovarian function in the buffalo and implications for embryo development and assisted reproduction. Anim Reprod Sci 121: 1–11. [DOI] [PubMed] [Google Scholar]
- 3. Russo M, Vecchio D, Neglia G, Pacelli C, Prandi A, et al. (2010) Corpus luteum Function and Pregnancy Outcome in Buffaloes during the Transition Period from Breeding to Non-Breeding Season. Reprod Domest Anim 45: 988–991. [DOI] [PubMed] [Google Scholar]
- 4. Di Francesco S, Neglia G, Vecchio D, Rossi P, Russo M, et al. (2012) Influence of season on corpus luteum structure and function and AI outcome in the Italian Mediterranean buffalo (Bubalus bubalis). Theriogenology 78: 1839–1845. [DOI] [PubMed] [Google Scholar]
- 5. Campanile G, Neglia G, Gasparrini B, Galiero G, Prandi A, et al. (2005) Embryonic mortality in buffaloes synchronized and mated by AI during the seasonal decline in reproductive function. Theriogenology 63: 2334–2340. [DOI] [PubMed] [Google Scholar]
- 6. Spencer TE, Sandra O, Wolf E (2008) Genes involved in conceptus-endometrial interactions in ruminants: insights from reductionism and thoughts on holistic approaches. Reproduction 135: 165–179. [DOI] [PubMed] [Google Scholar]
- 7. Carter F, Rings F, Mamo S, Holker M, Kuzmany A, et al. (2010) Effect of elevated circulating progesterone concentration on bovine blastocyst development and global transcriptome following endoscopic transfer of in vitro produced embryos to the bovine oviduct. Biol Reprod 83: 707–719. [DOI] [PubMed] [Google Scholar]
- 8. Lonergan P (2011) Influence of progesterone on oocyte quality and embryo development in cows. Theriogenology 76: 1594–1601. [DOI] [PubMed] [Google Scholar]
- 9. Vecchio D, Neglia G, Gasparrini B, Russo M, Pacelli C, et al. (2012) Corpus luteum development and function and relationship to pregnancy during the breeding season in the Mediterranean buffalo. Theriogenology 77: 1811–1815. [DOI] [PubMed] [Google Scholar]
- 10. Neglia G, Natale A, Esposito G, Salzillo F, Adinolfi L, et al. (2008) Effect of prostaglandin F-2 alpha at the time of AI on progesterone levels and pregnancy rate in synchronized Italian Mediterranean buffaloes. Theriogenology 69: 953–960. [DOI] [PubMed] [Google Scholar]
- 11. Mann GE, Lamming GE (1999) The influence of progesterone during early pregnancy in cattle. Reprod Domest Anim 34: 269–274. [Google Scholar]
- 12. Santos JEP, Thatcher WW, Chebel RC, Cerri RLA, Galvao KN (2004) The effect of embryonic death rates in cattle on the efficacy of estrus synchronization programs. Anim Reprod Sci 82–83: 513–535. [DOI] [PubMed] [Google Scholar]
- 13. Balestrieri ML, Gasparrini B, Neglia G, Vecchio D, Strazzullo M, et al. (2013) Proteomic profiles of the embryonic chorioamnion and uterine caruncles in buffaloes (Bubalus bubalis) with normal and retarded embryonic development. Biol Reprod 88: 119. [DOI] [PubMed] [Google Scholar]
- 14. Neglia G, Vecchio D, Russo M, Di Palo R, Pacelli C, et al. (2012) Efficacy of PGF(2 alpha) on Pre-ovulatory Follicle and Corpus Luteum Blood Flow. Reprod Domest Anim 47: 26–31. [DOI] [PubMed] [Google Scholar]
- 15. Clemente M, Lopez-Vidriero I, O′Gaora P, Mehta JP, Forde N, et al. (2011) Transcriptome Changes at the Initiation of Elongation in the Bovine Conceptus. Biol Reprod 85: 285–295. [DOI] [PubMed] [Google Scholar]
- 16. Mamo S, Rizos D, Lonergan P (2012) Transcriptomic changes in the bovine conceptus between the blastocyst stage and initiation of implantation. Anim Reprod Sci 134: 56–63. [DOI] [PubMed] [Google Scholar]
- 17. Mamo S, Mehta JP, McGettigan P, Fair T, Spencer TE, et al. (2011) RNA Sequencing Reveals Novel Gene Clusters in Bovine Conceptuses Associated with Maternal Recognition of Pregnancy and Implantation. Biol Reprod 85: 1143–1151. [DOI] [PubMed] [Google Scholar]
- 18. Forde N, Lonergan P (2012) Transcriptomic Analysis of the Bovine Endometrium: What is Required to Establish Uterine Receptivity to Implantation in Cattle? J Reprod Develop 58: 189–195. [DOI] [PubMed] [Google Scholar]
- 19. Mamo S, Mehta JP, Forde N, McGettigan P, Lonergan P (2012) Conceptus-Endometrium Crosstalk During Maternal Recognition of Pregnancy in Cattle. Biol Reprod 87: 6. [DOI] [PubMed] [Google Scholar]
- 20. Ushizawa K, Herath CB, Kaneyama K, Shiojima S, Hirasawa A, et al. (2004) cDNA microarray analysis of bovine embryo gene expression profiles during the pre-implantation period. Reprod Biol Endocrin 2: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strazzullo M, Rossetti C, Fusco G, Campanile C, Vecchio D, et al. (2010) Genomic characterization and chromosomal mapping of 5 river buffalo skeletal muscle differentiation master genes. Cytogenet Genome Res 128: 221–227. [DOI] [PubMed] [Google Scholar]
- 22. Nazar RN, Chen P, Dean D, Robb J (2010) DNA Chip Analysis in Diverse Organisms with Unsequenced Genomes. Mol Biotechnol 44: 8–13. [DOI] [PubMed] [Google Scholar]
- 23. Davey MW, Graham NS, Vanholme B, Swennen R, May ST, et al. (2009) Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genomics 10: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robert C, Nieminen J, Dufort I, Gagne D, Grant JR, et al. (2011) Combining Resources to Obtain a Comprehensive Survey of the Bovine Embryo Transcriptome Through Deep Sequencing and Microarrays. Mol Reprod Dev 78: 651–664. [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 26. Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 27. Almen MS, Nordstrom KJ, Fredriksson R, Schioth HB (2009) Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol 7: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barone V, Heisenberg CP (2012) Cell adhesion in embryo morphogenesis. Curr Opin Cell Biol24: 148–153. [DOI] [PubMed] [Google Scholar]
- 29. Ostrup E, Hyttel P, Ostrup O (2011) Embryo-maternal communication: signalling before and during placentation in cattle and pig. Reprod Fert Develop 23: 964–975. [DOI] [PubMed] [Google Scholar]
- 30. Talbot NC, Powell AM, Caperna TJ, Garrett WM (2010) Proteomic analysis of the major cellular proteins of bovine trophectoderm cell lines derived from IVP, parthenogenetic and nuclear transfer embryos: Reduced expression of annexins I and II in nuclear transfer-derived cell lines. Anim Reprod Sci 120: 187–202. [DOI] [PubMed] [Google Scholar]
- 31. Garrido-Gomez T, Dominguez F, Quinonero A, Estella C, Vilella F, et al. (2012) Annexin A2 is critical for embryo adhesiveness to the human endometrium by RhoA activation through F-actin regulation. FASEB J 26: 3715–3727. [DOI] [PubMed] [Google Scholar]
- 32. Oestrup O, Hall V, Petkov SG, Wolf XA, Hyldig S, et al. (2009) From zygote to implantation: morphological and molecular dynamics during embryo development in the pig. Reprod Domest Anim 44 Suppl 339–49. [DOI] [PubMed] [Google Scholar]
- 33. Maruotti J, Munoz M, Degrelle SA, Gomez E, Louet C, et al. (2012) Efficient derivation of bovine embryonic stem cells needs more than active core pluripotency factors. Mol Reprod Dev 79: 461–477. [DOI] [PubMed] [Google Scholar]
- 34. Takahashi M (2012) Oxidative stress and redox regulation on in vitro development of mammalian embryos. J Reprod Develop 58: 1–9. [DOI] [PubMed] [Google Scholar]
- 35. Menezo Y, Lichtblau I, Elder K (2013) New insights into human pre-implantation metabolism in vivo and in vitro. J Assist Reprod Gen 30: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tincani A, Cavazzana I, Ziglioli T, Lojacono A, De Angelis V, et al. (2010) Complement activation and pregnancy failure. Clin Rev Allerg Immu 39: 153–159. [DOI] [PubMed] [Google Scholar]
- 37. Parks JC, McCallie BR, Janesch AM (2011) Schoolcraft WB, Katz-Jaffe MG (2011) Blastocyst gene expression correlates with implantation potential. Fertil Steril 95: 1367–1372. [DOI] [PubMed] [Google Scholar]
- 38. Girardi G, Yarilin D, Thurman JM, Holers VM, Salmon JE (2006) Complement activation induces dysregulation of angiogenic factors and causes fetal rejection and growth restriction. J Exp Med 203: 2165–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gu T, Su X, Zhou Q, Li X, Yu M, et al. (2012) Molecular characterization of the Neuronatin gene in the porcine placenta. PloS one 7: e43325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scarano MI, Strazzullo M, Matarazzo MR, D′Esposito M (2005) DNA methylation 40 years later: Its role in human health and disease. J Cell Physiol 204: 21–35. [DOI] [PubMed] [Google Scholar]
- 41. Leeb M, Wutz A (2012) Establishment of epigenetic patterns in development. Chromosoma 121: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barlow DP (2011) Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet 45: 379–403. [DOI] [PubMed] [Google Scholar]
- 43. Yang Z, Dong D, Zhang Z, Crabbe MJ, Wang L, et al. (2012) Preferential regulation of stably expressed genes in the human genome suggests a widespread expression buffering role of microRNAs. BMC Genomics 13 Suppl 7S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clemente M, de la Fuente J, Fair T, Al Naib A, Gutierrez-Adan A, et al. (2009) Progesterone and conceptus elongation in cattle: a direct effect on the embryo or an indirect effect via the endometrium? Reproduction 138: 507–517. [DOI] [PubMed] [Google Scholar]
- 45. Beltman ME, Roche JF, Lonergan P, Forde N, Crowe MA (2009) Evaluation of models to induce low progesterone during the early luteal phase in cattle. Theriogenology 72: 986–992. [DOI] [PubMed] [Google Scholar]
- 46. Forde N, Beltman ME, Duffy GB, Duffy P, Mehta JP, et al. (2011) Changes in the Endometrial Transcriptome During the Bovine Estrous Cycle: Effect of Low Circulating Progesterone and Consequences for Conceptus Elongation. BBiol Reprod 84: 266–278. [DOI] [PubMed] [Google Scholar]
- 47. Vecchio D, Neglia G, Di Palo R, Prandi A, Gasparrini B, et al. (2010) Is a Delayed Treatment with GnRH, hCG or Progesterone Beneficial for Reducing Embryonic Mortality in Buffaloes? Reprod Domest Anim 45: 614–618. [DOI] [PubMed] [Google Scholar]