Abstract
The overexpression of certain membrane-bound receptors is a hallmark of cancer progression and it has been suggested to affect the organization, activation, recycling and down-regulation of receptor-ligand complexes in human cancer cells. Thus, comparing receptor trafficking pathways in normal vs. cancer cells requires the ability to image cells expressing dramatically different receptor expression levels. Here, we have presented a significant technical advance to the analysis and processing of images collected using intensity based Förster resonance energy transfer (FRET) confocal microscopy. An automated Image J macro was developed to select region of interests (ROI) based on intensity and statistical-based thresholds within cellular images with reduced FRET signal. Furthermore, SSMD (strictly standardized mean differences), a statistical signal-to-noise ratio (SNR) evaluation parameter, was used to validate the quality of FRET analysis, in particular of ROI database selection. The Image J ROI selection macro together with SSMD as an evaluation parameter of SNR levels, were used to investigate the endocytic recycling of Tfn-TFR complexes at nanometer range resolution in human normal vs. breast cancer cells expressing significantly different levels of endogenous TFR. Here, the FRET-based assay demonstrates that Tfn-TFR complexes in normal epithelial vs. breast cancer cells show a significantly different E% behavior during their endocytic recycling pathway. Since E% is a relative measure of distance, we propose that these changes in E% levels represent conformational changes in Tfn-TFR complexes during endocytic pathway. Thus, our results indicate that Tfn-TFR complexes undergo different conformational changes in normal vs. cancer cells, indicating that the organization of Tfn-TFR complexes at the nanometer range is significantly altered during the endocytic recycling pathway in cancer cells. In summary, improvements in the automated selection of FRET ROI datasets allowed us to detect significant changes in E% with potential biological significance in human normal vs. cancer cells.
Keywords: Förster Resonance Energy Transfer (FRET), transferrin (Tfn), transferrin receptor (TFR), image processing, region of interest (ROI), strictly standardized mean differences (SSMD)
1. INTRODUCTION
The endocytic trafficking of membrane bound receptors has been shown to be defective in a wide range of human cancers [1–3]. Altered expression and/or activity levels of proteins involved in the regulation of membrane trafficking can affect the endocytosis, degradation and/or recycling of membrane bound receptors, leading to increased cell proliferation and motility as well as metastatic behavior [4,5]. Key endocytic proteins such as Rabaptin5, a Rab5 GAP involved in regulating endosome fusion [6] and Numb, a clathrin coat adaptor, have been found to be disrupted or significantly reduced in human cancers [7,8]. Moreover, the overexpression of certain membrane-bound receptors, which is a common occurrence during cancer progression [9–11], has been suggested to affect the organization, activation, recycling and down-regulation of receptor-ligand complexes in human cancer cells [2]. Receptor overexpression may provide an alternative manner to disrupt endocytic trafficking by saturating the limited availability of endocytic proteins involved in vesicle formation, sorting and fusion [1,12]. Therefore, it is crucial that we develop a better understanding of how the endocytic trafficking pathways of receptor-ligand complexes are regulated in normal cells and how these processes are disrupted in cancer [2]. However, comparing receptor trafficking pathways in normal vs. cancer cells requires the ability to image cells expressing dramatically different receptor expression levels, which can raise significantly technical issues when using standard fluorescence based imaging approaches.
Therefore, the development of novel quantitative imaging techniques is crucial to increase our understanding of the regulation of the intracellular endocytic trafficking of membrane receptors in both normal and cancer cells. Recently, Förster resonance energy transfer (FRET)-based imaging approaches have been developed to assay receptor dimerization/oligomerization as well as receptor-ligand interactions during endocytic trafficking pathways [13–16]. In particular, a quantitative FRET assay has been used to follow the endocytic/recycling trafficking of transferrin receptor (TFR) and its respective ligand, iron-bound transferrin (Tfn), at nanometer range resolution [17–22]. TFR is a well-known membrane-bound receptor which is responsible for cellular iron uptake upon the binding of iron-loaded Tfn [23]. Increased iron uptake is necessary to sustain the increased cell proliferation typical of cancer cells. Thus, elevated levels of TFR expression has been found in various malignancies (e.g., 74% breast carcinomas, 76% lung adenocarcinomas, and 93% lung squamous cell carcinomas) [24–31]. Thus, Tfn has been used as a target for molecular imaging techniques in breast tumors [32–35] and it has been widely used as carrier for anti-cancer drug/siRNA delivery systems [31,36,37]. We have capitalized on the homodimeric nature of the TFR [23,31] and employed FRET to quantitatively analyze the endocytic trafficking of Tfn-TFR complexes. As part of this assay, Tfn molecules conjugated with FRET donor or acceptor fluorophores are brought to nanometer-range proximity upon binding to the homodimeric TFR at the plasma membrane [17–22,38]. These Tfn-TFR complexes are internalized via clathrin-coated vesicles into endosomes, where the iron is released from Tfn upon endosomal acidification via ATPase proton pumps [23,39]. Then, the Tfn-TFR complexes are recycled back to the plasma membrane, where the apo-Tfn is released into the extracellular space [23,39]. FRET imaging was performed to test whether the TFR-Tfn receptor-ligand complexes undergo conformational changes throughout the endocytic recycling pathway as they are exposed to different endosomal luminal environments. Those conformational changes may lead to slight changes in the distance between Tfn molecules bound to TFR, which would result in variations in FRET levels since FRET is a highly sensitive proximity assay. Thus, changes in FRET levels may be due to the ability of TFR-Tfn complexes to undergo conformational changes that are mediated by alterations in the endocytic environment.
Intensity-based FRET using confocal microscopy is the most widely used imaging technique to perform nanometer scale measurements in live cells [15,17]. Although, the use of filter-based intensity FRET together with organic dye FRET pairs, such as Alexa Fluor 488 (AF488; donor) and 555 (acceptor; AF555) can be applied to most optical microscopy systems, confocal microscopy still provides the best approach to measure FRET signals in time-lapse and/or pulse-chase experiments using live-cell intensity-based FRET imaging. Confocal images are processed to remove background noise as well as spectral-bleedthrough (SBT) by the custom generated processed FRET (PFRET) algorithm Image J plugin software, as developed by Dr. Periasamy at the Keck Center for Cellular Imaging, University of Virginia [19,21,40–45] [17,19–21,40,42–46]. The PFRET algorithm removes SBT from uncorrected FRET images, taking into consideration intracellular variability of fluorophore expression levels. The PFRET algorithm allows for the calculation of apparent energy transfer efficiency (E%) as a percentage of the unquenched donor (D), providing an expression for the actual energy transferred from donor to acceptor molecules [19– 21,41–44]. However, apparent E% measurements do not allow for the separation between donors that participate in FRET events (FRET donors) and those that do not (non-FRET donors). To calculate precise energy transfer efficiencies (E) and distances between donor and acceptor fluorophores, one must determine an imaging factor (λ) that contains information about the donor and acceptor quantum efficiencies and the device detection efficiency [15,40,41,47,48]. Recently, the use of a standard donor-acceptor tandem construct with known energy transfer efficiency as measured by fluorescence lifetime has been used to determine the value of imaging factor λ and to normalize the relative FRET efficiencies [15,48]. However, generating such a construct is technically challenging, when using a system that involves the cellular uptake of fluorescently labeled ligand molecules. Furthermore, most biological experiments are by nature relative, since they require the comparison of a control vs. a test experiment in which cell samples undergo changes under specific conditions, such as time, temperature, pH, pharmacological treatments or genetic-based approaches. Given that the γ factor does not affect the relative comparison of E% measurements used in filter-based intensity FRET analysis, for simplicity, in our FRET analysis, we used γ = 1, as described previously [17,19–22,43–45]. Nevertheless, filter-based intensity FRET assays carried out with different imaging parameters will by definition have distinct γ factors. Importantly, previously, the TFR-Tfn model system has been characterized using both intensity- and lifetime-based imaging analysis in MDCK-PTR epithelial cells stably expressing TFR [17–22,49]. Similar E% values (15–20%) were detected demonstrating the strength of our intensity based FRET analysis [49].
There are two main issues that prevent the application of filter-based intensity FRET to an even wider variety of biological questions. The first issue is the application of filter-based intensity FRET to low signal FRET images such as those generated by the binding of AF488-Tfn and AF555-Tfn to TFR endogenously expressed in HME cells. The second is the ability to develop automated FRET image analysis that can reliably and consistently evaluate numerous images generated by time-lapse live cell imaging or high content high-throughput imaging. Previously, semi-automated quantitative FRET processing algorithms, such as the PFRET Image J plugin, FRET stoichiometry calculator[48], the FRET and colocalization analyzer[50], the PixFRET[51] and the RiFRET custom-based software[52], have been developed to implement SBT correction and to calculate precise E values. To analyze a large number of low FRET signal images processed by the PFRET algorithm, we developed a novel algorithm that implements standardized background correction procedures as well as the automated selection of defined x,y coordinates (non-grid) and non-overlapping ROIs based on different intensity and statistical based thresholding parameters[53]. Moreover, we have established a statistical signal-to-noise ratio (SNR) parameter, i.e. the strictly standardized mean difference (SSMD)[54–56], to validate changes to selected FRET ROI datasets as they affect the reliability and consistency of average E% values. Here, we have compared TFR-Tfn endocytic FRET in three well-known epithelial cell lines: human mammary epithelial (HME) cells, human breast cancer T47D cells and Madin-Darby canine kidney epithelial MDCK-PTR cells, a well-known epithelial cell model system[57]. These cell lines express TFR at various levels; whereas HME cells express low levels of TFR, T47D cells show significantly higher TFR expression levels[58] and MDCK-PTR cells, stably express human TFR at intermediate levels[59]. Our results suggest that the organization of Tfn-TFR complexes at nanometer range resolution is significantly altered during the endocytic recycling pathway in cancer cells. These results are particularly important since Tfn-TFR system is widely used to target anti-cancer drugs to tumors in vivo, making it crucial to analyze in detail the Tfn-TFR endocytic system in normal vs. cancer cells.
2. MATERIAL AND METHODS
2.1. Cells
MDCK–PTR cells are Madin-Darby canine kidney cells that stably express human TFR[19–22]. These cells are grown in Dulbecco’s modified eagle medium (DMEM)/10% fetal bovine serum (FBS)/Pen-Strep. HME cells (CC-2551) were purchased from Lonza and are grown in MEGM Complete media (CC-3051) with 10% fetal bovine serum, as recommended by the manufacturer. T47D (HTB-122) and BT549 (HTB133) cells were purchased from ATCC and cultured in RPMI-1640 media (30–2001, ATCC) as recommended by ATCC. The media formulation was supplemented with 50 units/mL penicillin and 50 µg/mL streptomycin from MP Biomedicals (091670249), and with 10% fetal bovine serum (30–2020 ATCC). All cells were grown in 5% CO2 at 37°C in a humidified incubation chamber, and were kept for less than 12 population doublings.
2.2. Tfn uptake
HME, T47D and MDCK-PTR cells were passaged onto MatTek glass bottom culture dishes and allowed to grow to ~75% confluency. For Tfn uptake experiments, cells were washed in phosphate buffered saline solution (PBS) and incubated in Tfn-free imaging media consisting of phenol red-free DMEM (LifeTechnologies, 31053), supplemented with 0.37 mg/mL sodium bicarbonate (Sigma, S7277), 5 mg/mL bovine serum albumin (Sigma, A9085), and 4.7 mg/mL HEPES (Sigma, H4034), at 37°C for 1 hour to clear internalized endogenous Tfn. For FRET confocal microscopy, cells were incubated with media containing donor only (AF488-Tfn), acceptor only (AF555-Tfn) or both at different acceptor:donor molar ratios (A:D) for different periods of time at 37°C: 60min for Figure 2–3 and 5 & 30min for the remaining figures. To increase A:D ratios, the concentration of acceptor AF555-Tfn was kept constant at 20µg/ml, while donor AF488-Tfn was added at 10µg/ml, 20µg/ml and 40µg/ml to generate A:D~2:1, A:D~1:1 and A:D~1:2, respectively. AF488-Tfn and AF555-Tfn were purchased from LifeTechnologies (~2 fluorophore molecules per Tfn molecule). Uptake times were terminated by washing cell chambers with ice-cold PBS followed by cell fixation using 4% formaldehyde in PBS for 15 minutes and mounted onto coverslips with 50% glycerol/1× PBS.
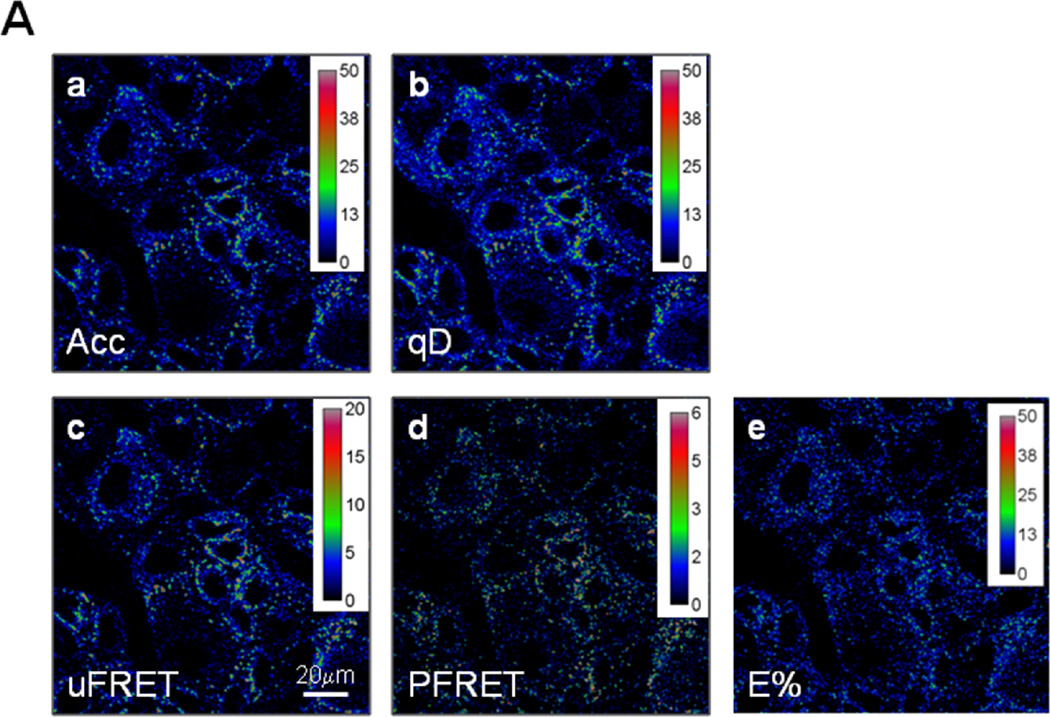
Figure 2. FRET analysis of TFR-Tfn complexes using ROI selection protocol.
Pseudo-color images show intracellular distribution of TFR-Tfn complexes upon internalization of AF555-Tfn and AF488-Tfn at D:A~0.5 for 60min at 37°C into HME cells. (A) Pseudocolor images of acceptor (Acc; panel a), quenched donor (qD; panel b), uncorrected FRET (uFRET; panel c), corrected FRET (PFRET; panel d) and E% levels (E%; panel e) show the typical endocytic pattern of irregular and punctate structures. PFRET and E% images confirm the existence of energy transfer between endogenous Tfn-AF488 (donor) and Tfn-AF555 (acceptor) in intracellular punctate endocytic structures in HME cells. Bar: 20 µm. (B) Different ROI datasets were applied to PFRET (panels a–d) and E% images (panels e–h). These ROI were selected using different values for intensity and statistical based ROI (10×10 pixels) selection parameters: ED=0 & %F=0 (panels a & e), ED=5 & %F=0 (panels b & f), ED=5 & %F=22 (panels c & g) and ED=5 & %F=30 (panels d & h). Bar: 20 µm.
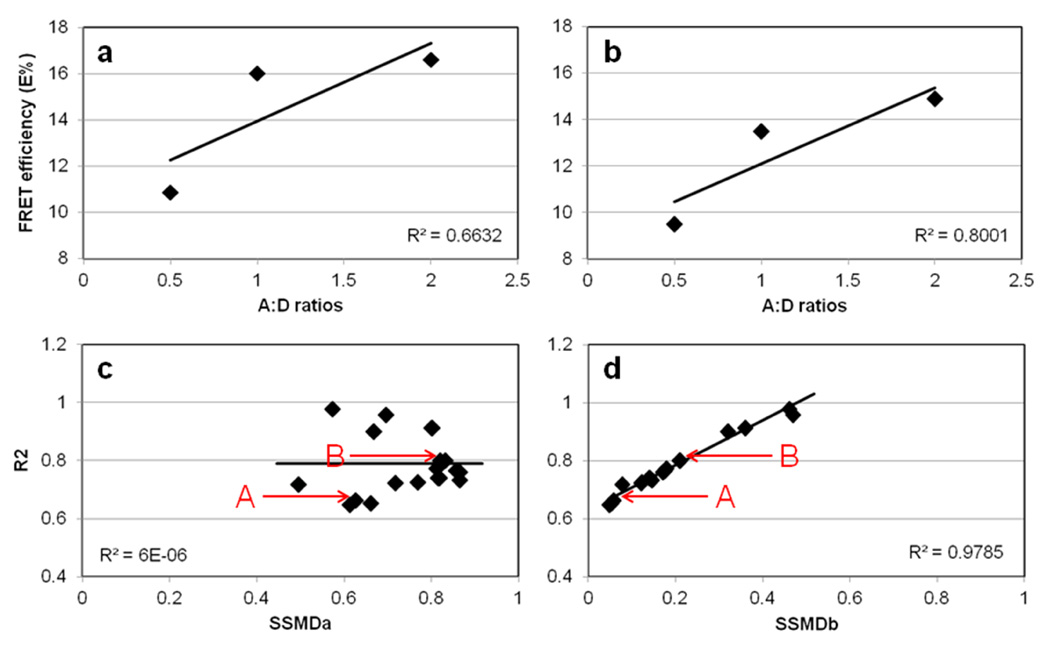
Figure 3. Validation of FRET ROI selection protocol using SSMD as a SNR related statistical parameter.
E% is calculated and plotted against A:D~1:2, A:D~1:1, and A:D~2:1 of TFR-Tfn complexes internalized into HMECs for 60min at 37°C. Despite lower FRET signal in HMECs, a trend for higher E% relative to increasing A:D ratios is clearly seen. The following ROI selection settings were used in panel a: ED=5 and %F=0 (Table 1, #11); and in panel b: ED=5 and %F=30 (Table 1, #15). The R2 of the E% vs. A:D relationship is increased when using %F=0 to %F=30, from R2=0.6632 to 0.8001, respectively. In panel c, the R2 values calculated for the E% vs. A:D relationship using different ROI selection settings (Table 1) were plotted against the SSMDa between A:D~2 vs. A:D~0.5. In panel d, the R2 values calculated for the E% vs. A:D relationship using different ROI selection settings (Table 1) were plotted against the SSMDb between A:D~2 vs. A:D~1. Whereas, R2 vs. SSMDa show an independent behavior, R2 vs. SSMDb shows a clear direct relationship with high R2=0.978. A and B (arrows) represent the values calculated using the settings used in panel a & b respectively (Table 1; #11 vs. #15). As shown in panel a & b and in Table 1, E% values show significant variance upon using different ROI datasets; SSMD analysis as shown in panel c & d and Table 1, is used to establish the best ROI dataset that shows the widest dynamic range of the E% signal. Thus, SSMD provides a FRET data quality assessment that combines signal range and variability measures in a concise summary number.
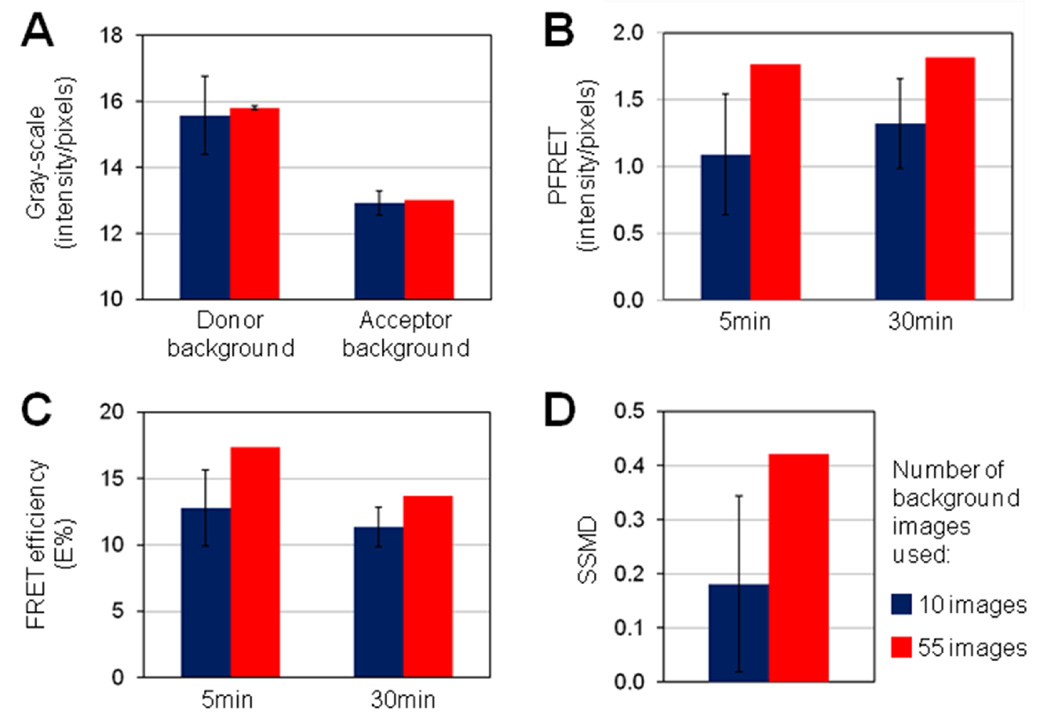
Figure 5. Role of background removal in FRET data variability and E% assay sensitivity.
10 (blue) or 50 (red) single-labeled images were used to determine instrument-related background values (panel a). Donor excitation/Donor emission donor channel images of single-labeled acceptor samples were used to determine donor background values to be removed from all donor channel images. Acceptor excitation/acceptor emission acceptor channel images of donor single-labeled samples were used to determine acceptor background values to be removed from all acceptor emission channel images, including acceptor and uFRET images. The following ROI settings were used for all 10 runs: ID=500, ED=5, STD=45 & %F=22. PFRET (panel b) and E% (panel c) averages collected at 5min vs. 30min internalization of TFR-Tfn complexes into HME cells are shown. In panel d, SSMD values using 5min vs 30min datapoints are shown. Increasing the number of images used to determine background values reduces the variability of E% values and increases the strength, reliability and sensitivity of E% assay. Error bars indicate standard deviation values from the average values of 10 ROI selection macro runs. Figure 4C images are one set of six set of images used to calculate the data presented in Figure 5.
2.3. Intensity based FRET microscopy
Images were acquired on a Zeiss LSM 510 META-NLO laser scanning microscope system using a Plan-Apochromat 63×/1.4 oil DIC objective and optimal acquisition settings to avoid saturation. In the AIM software (Zeiss USA, Thornwood, NY) the image display was configured to three channels, including donor excitation/donor emission (donor channel), acceptor excitation/acceptor emission (acceptor channel) and donor excitation/acceptor emission (FRET channel). Images were collected sequentially at 512×512 pixels, 8-bit depth, mean of 2 images and zoom 1×. PMT gain and black-level settings, laser power and pinhole were set at identical levels and remained unchanged for all image collections from double-labeled, single label acceptor and single label donor samples. Single-labeled images were used to correct SBT signal contamination in double-labeled images.
2.4. Precision FRET (PFRET) analysis
To calculate the energy transfer efficiency (E%), we used a quantitative analysis algorithm called precision FRET (PFRET) that removes the donor and acceptor SBT[40–45]. Three types of imaging conditions, i.e. donor channel (donor excitation wavelength/donor emission), acceptor channel (acceptor excitation wavelength/acceptor emission) and FRET channel (donor excitation wavelength/acceptor emission), are used to collect images at identical microscopy settings from three different types of cell samples, i.e. cells containing both AF488-Tfn and AF555-Tfn and cells containing only AF488-Tfn or AF555-Tfn; these single-labeled cells are used as reference specimens that will be used to remove SBT from the double-labeled cells. Images were collected from these three different types of cell samples using the donor, acceptor and FRET imaging channels. The following three images acquired from cells containing both AF488-Tfn & AF555-Tfn are named as follows: quenched donor (qD) image is collected using the donor channel; acceptor (Acc) image is collected using the acceptor channel; and uncorrected FRET (uFRET) image is collected using FRET channel. The nine images were first background-subtracted and then processed by the Image J PFRET (Precision FRET) software plugin to remove donor and acceptor SBT and generate PFRET and E% images. Both PFRET and E% images were generated using Image J PFRET software; all calculations are done with floating numbers and then the PFRET and E% values are converted to integers, to be saved as TIFF images (refs). E% images are calculated using a pixel-by-pixel approach that leads to high E% value variability, which is addressed by using the smooth Image J function to replace each pixel with the average of its 3×3 neighborhood pixels. Therefore, E% images should be used for visualization purposes only. A custom-written analysis program (Image J ROI selection macro) was applied to the PFRET images to select regions of interest (ROIs; 10×10 pixels) that show corrected FRET signal above threshold, excluding zero and saturated pixels[53]. Under our imaging conditions, there were less than 5% saturated pixels. The PFRET images were used to select the ROI dataset since they show the actual energy transferred from donor to acceptor fluorophores after background and spectral bleedthough removal. Due to their highly processed and corrected nature, PFRET images are ideal for ROI selection due to their high SNR. The selected ROIs were subsequently applied to all other images to extract the different averages of gray-scale fluorescence intensity values for the different parameters tested, including PFRET (actual energy transfer levels as per the PFRET SBT correction algorithm), uFRET, qD and Acc levels. These values were transferred to an Excel spreadsheet (Microsoft, Redmond, WA) for calculation of important parameters such as E%, unquenched donor (D), and calculated A:Dcalc ratios. All ROIs are included in the FRET analysis with the exception of those showing A:Dcalc values <0.1667 for Figure 3 and A:Dcalc <0.667 for all other figures. Energy transfer efficiency, E%, is calculated as a percentage of energy transfer in relation to the unquenched donor, i.e., D = qD + γ∙PFRET as described in the equation E% = 100 × (γ∙PFRET/D) or E% = 100 × [1 – (qD/D)]. γ value is a function of the quantum yield of the fluorophores and of the detection setup. Since all our imaging conditions remain constant γ value does not affect the interpretation of the relative E% data when E% is calculated assuming γ =1. Each FRET dataset includes at least 5–6 images for double-labeled samples and up to 50 single-labeled images.
3. RESULTS AND DISCUSSION
3.1. Automated selection of ROIs for intensity based FRET analysis
Advanced imaging analysis requires the processing of images according to mathematical algorithms developed towards the measurement of parameters that provide information about the relationship between the objects identified in the image that cannot be obtained from direct visualization of the original images. For example, E% is a parameter that provides information on whether two labeled molecules are in nanometer range proximity. To calculate E% values, images need to be processed pixel-by-pixel using specific SBT correction algorithms resulting in a completely processed image, i.e. corrected FRET image (PFRET image, Figure 2d), which shows the energy transfer levels in the image. In standard FRET data analysis, visual inspection is employed to select appropriate ROIs that show higher levels of energy transfer in PFRET corrected images and to identify their x,y pixel coordinates. Alternatively, pixel by pixel analysis can be employed by using thresholding-based approaches. Both approaches have limitations. Although pixel by pixel analysis is appropriate to visualize E% changes with high spatial resolution, it may lead to high E% value variability due to the low SNR of intensity-based FRET images. In contrast, manual ROI selection allows for calculating E% averages thus reducing pixel-to-pixel variability but it is inherently biased and time-consuming when applied to multiple image analysis. Therefore, we have developed an automated image analysis method as an Image J macro that selects ROIs showing FRET signal above certain thresholds within cellular images[53].
Here we have established an Image J based background removal, SBT correction and ROI selection FRET analysis protocol that includes seven steps as described in Figure 1. A total number of nine images are collected from cells containing AF488-Tfn (donor single-labeled), AF555-Tfn (acceptor single-labeled) or AF488-Tfn and AF555-Tfn (double-labeled) cell samples employing three imaging channels (donor, acceptor and FRET), using a Zeiss LSM 510 confocal microscope, as described previously [17,19–22,43–45]. Then, images are opened in Image J and saved in tif format in several different folders (donor, acceptor and FRET folders), depending on the imaging channels used to collect those images (Figure 1). For background subtraction, twenty ROIs (20×20 pixels) are randomly selected over 512×512 pixel single-labeled images. The mean values of all ROIs are then averaged to calculate the background value to be removed from the single- and double-labeled images, as described below (Figure 1a, A1). The acceptor channel images collected from donor single-labeled cells are used to calculate acceptor-related background values and subsequently remove them from acceptor emission images including acceptor and FRET channel images from single and double labeled cells (Figure 1a, A2-A3). The donor channel images collected from acceptor single labeled cells are used to calculate donor-related background values and subsequently remove them from all donor channel images from single and double labeled cells (Figure 1a, A4-A5). A large number of images should be used for background determination. The Image J ROI selection macro provides several options on how to select the images used for background correction: computer-based random selection, manual selection and use of all available images. Then, background-subtracted donor, acceptor and FRET channel images are processed by the PFRET software Image J plugin, which removes donor and acceptor SBT pixel-by-pixel on the basis of matched gray-scale fluorescence intensity levels between the double-label specimen and the single-label reference specimens to generate the corrected FRET images (PFRET images) that represent the actual energy-transfer levels (PFRET levels).
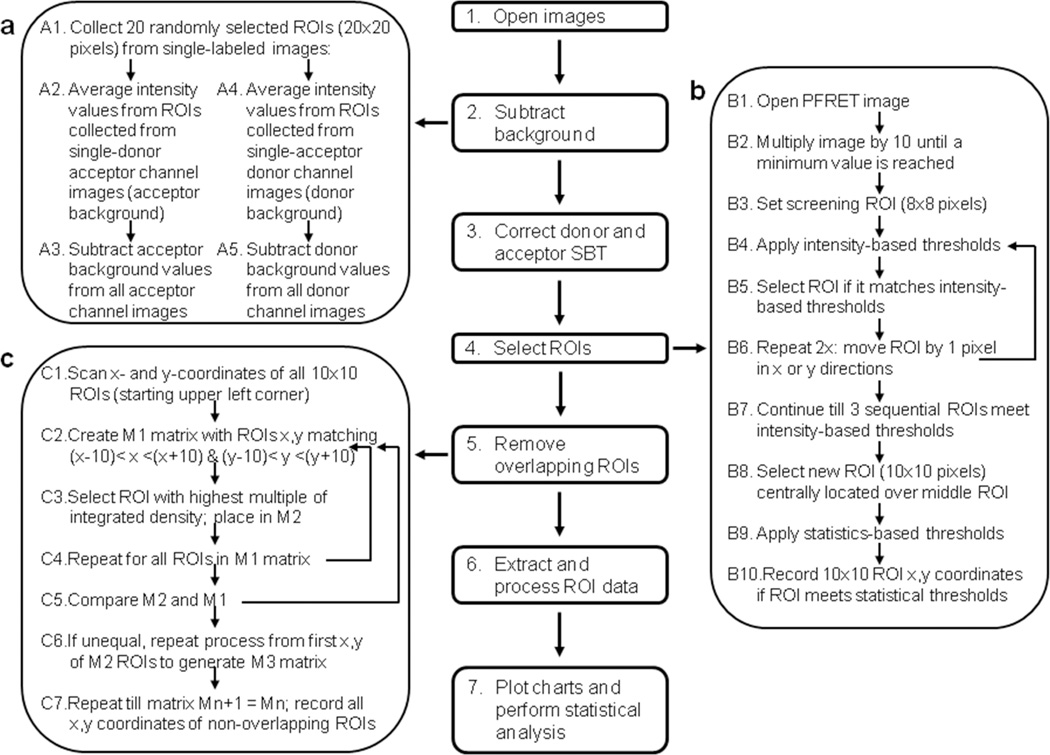
Figure 1. Schematic representation of background substraction, SBT correction and ROI selection FRET analysis protocol.
Seven steps are required for the FRET analysis protocol: 1) Zeiss format lsm image opening, 2) background removal, 3) SBT correction (PFRET software), 4) ROI selection, 5) overlapping ROI removal, 6) Extract and processing of FRET related ROI data, and 7) plot charts and perform statistical analysis. Three of those steps in the Image J ROI selection macro are described in detail, including background removal (panel a), ROI selection (panel b) and removal of overlapping ROIs (panel c).
To select FRET ROIs from PFRET images, we have developed a custom generated Image J ROI selection macro that comprises image manipulation to increase FRET signal contrast, definition of intensity and statistically based thresholds to select above-threshold ROIs, removal of overlapping ROIs and recording of the x,y coordinates of the selected ROIs. PFRET images are manipulated to enhance FRET signal contrast and allow for easier identification of above threshold ROIs even in images with low FRET signal. PFRET images, which show a very narrow range of pixel values, i.e. 0–20 values, after SBT correction, are multiplied by 10 until a minimum value (e.g. 5 or 10) or a pre-set maximum number of multiplications (e.g. 40) are reached (Figure 1b). Throughout this manuscript we have used a multiplication factor equal to 40 and a minimum pixel value of 10, unless otherwise noted. Then, a screening ROI (8×8 pixels) is set at the upper left corner of the manipulated PFRET image. Several gray-scale intensity-based parameters are measured for the initial 8×8 box, including the mean pixel integrated density (ID), i.e. product of number of pixels per mean gray value, and mean pixel intensity difference (edge detection, ED), which comprises the difference between the mean pixel intensity of 8×8 box and that of a smaller 6×6 pixel box, centered in the original 8×8 box. Pre-set ID=500 to 1500 and ED=0 to 5 values are used to select the ROIs for further analysis. This step is repeated twice following the move of the previously selected ROI by 1 pixel in the×or y directions. If three sequential 8×8 ROIs reach the minimum thresholds, then a 10×10 pixel (2.79µm×2.79µm) ROI is selected, centered on the middle 8×8 ROI. The possibility of selecting a smaller ROI (8×8 pixels) is included in the FRET ROI selection macro. The selected ROIs are then analyzed for statistically based parameters, including the standard deviation (STD) of the gray-scale values used to generate the mean pixel integrated intensity and the percentage of non-zero pixels (Area Fraction; %F). The x,y coordinates of the ROIs that meet the pre-set thresholds for these parameters are recorded and saved in a text file (Figure 1b). Since the screening ROI moves on the×as well as the y direction, overlapping ROIs are generated upon meeting threshold selection, leading to the possibility of bias due to the increasing number of ROIs in locations showing high intensity signal. To remove overlapping ROIs, ROI matrixes (R1) for each x,y coordinate comprising +/− 10×or y pixels are generated. Then the ROI with the highest multiple of ID parameter is selected from that matrix R1 (Figure 1c) and placed in second matrix (R2). This step is repeated for all overlapping ROIs within matrix R1 and R1 and R2 are compared; if unequal a new matrix (R3) is generated and process is repeated. If R1=R2, all overlapping ROIs have been removed. The possibility of allowing 1 or 2 pixel overlap between ROIs is also allowed in the ROI macro; alternatively no overlap entails that no ROIs overlap with each other. Upon final selection of non-overlapping ROIs matching the minimum thresholds for the intensity as well as the statistically based parameters, their x,y coordinates are applied to the original non-manipulated background-subtracted donor, acceptor, FRET channel images as well as to the PFRET images to extract ROI FRET datasets, including, qD, uFRET and Acc, PFRET, D, A:Dcalc ratios, and E% that can be used to plot charts and perform statistical analysis (Figure 1).
3.2. Validation of automated selection of FRET ROIs using E% vs. A:D ratio relationship
Previously, the TFR-Tfn model system has been characterized using intensity- and lifetime- based imaging analysis in MDCK-PTR epithelial cells stably expressing TFR [49]. The TFR-Tfn FRET based assay showcases the capabilities of FRET analysis to dissect the trafficking of membrane-bound receptor-ligand complexes [17], and demonstrate their clustered organization due to their homodimeric nature and potential for the formation of higher order clusters [19–21,44]. Here, we test whether detectable FRET occurs between AF488-Tfn and AF555-Tfn upon binding to endogenous TFR and subsequent co-internalization into HME cells for 60min. PFRET images confirm the existence of energy transfer between endogenous Tfn-AF488 (donor) to Tfn-AF555 (acceptor) in intracellular punctate endocytic structures (Figure 2A). qD (panel a), Acc (panel b), uFRET (panel c), PFRET (panel d) and E% (panel e) images provide important information about the morphology pattern of these parameters; as expected, the typical endocytic pattern of irregular and punctate structures is clearly detected across all images (Figure 2A).
A major challenge to intensity-based FRET analysis is the ability to select quality FRET ROI datasets from images showing low FRET signal with very low SNR, as exemplified by those showing the internalization of human AF488-Tfn and AF555-Tfn into HME cells upon their binding to endogenous human TFR (Figure 2A & 4A). HME cells internalized with AF555-Tfn and AF488-Tfn at increasing A:D ratios ~0.5, ~1 & ~2, for 60min at 37°C, were imaged and processed for FRET analysis using the Image J background removal, SBT correction and ROI selection protocol as described in Figure 1. ROIs were selected from PFRET images using different Image J ROI selection parameters, such as ED=0 vs. 5, and %F=0 vs. 30. In Figure 2B, the FRET ROI dataset coordinates were applied to PFRET (panels a–d) and to E% images (panels e–h); increasing ED and/or %F values lead to a markedly reduced number of selected ROIs and an increasingly improved visual match to the presence of detectable PFRET and E% levels (compare Figure 2B, panels a–b (PFRET images) and e–f (E% images) vs. panels c–d (PFRET images) and g–h (E% images).
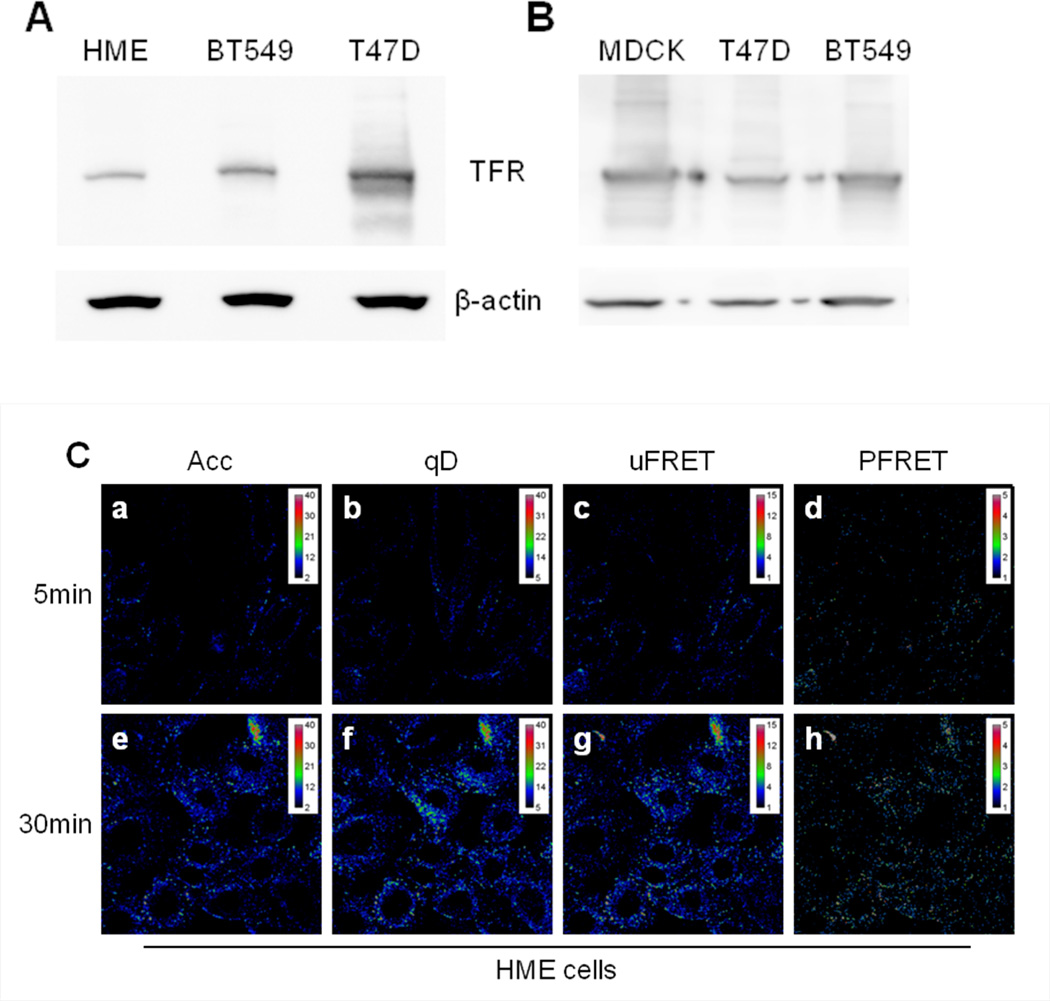
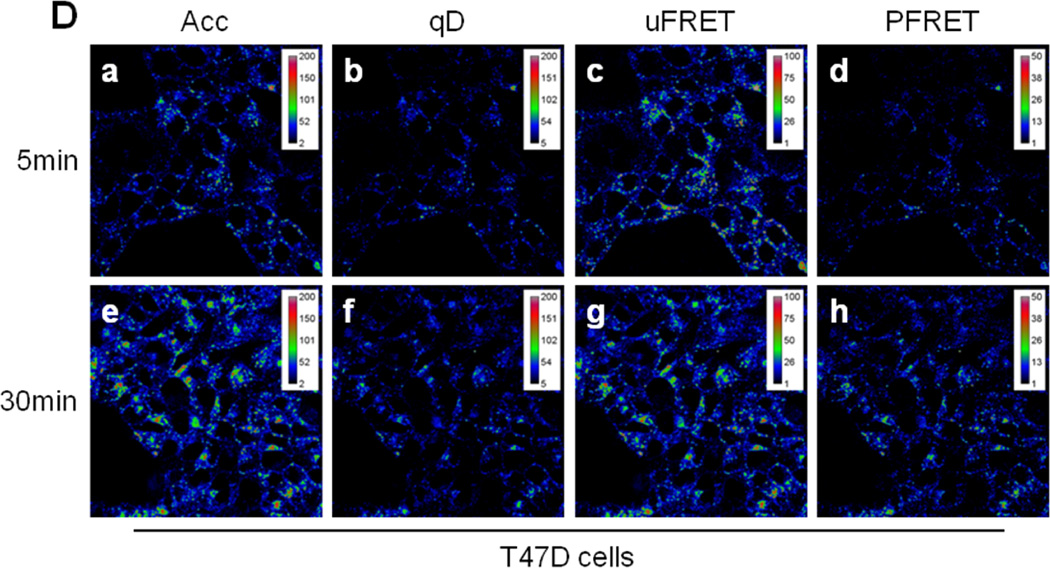
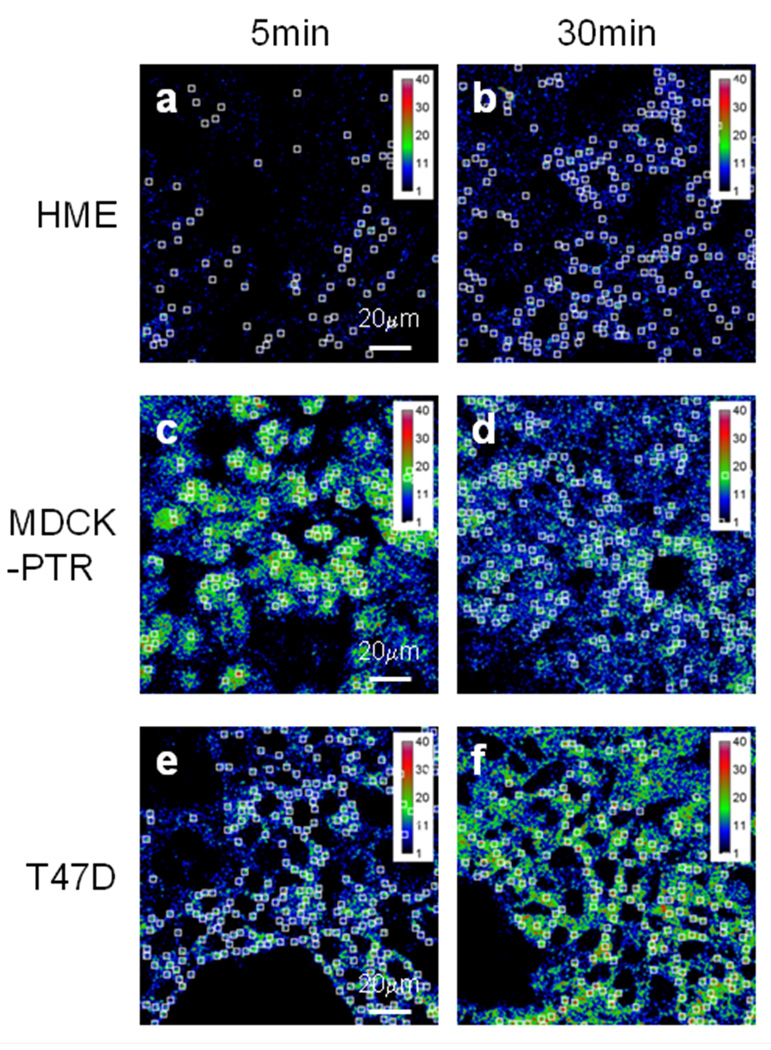
Figure 4. FRET analysis of TFR-Tfn complexes during endocytic recycling pathway in normal vs. cancer cells.
(A) Western blot of human TFR in HME, BT-549, and T47D cells. Blots indicate higher expression of human TFR in human breast cancer cell lines BT549 and T47D compared to normal HME cells. (B) Western blot of human TFR in MDCK-PTR, BT-549, and T47D cells. Blots indicate similar expression levels of human TFR in human breast cancer cell lines BT549 and T47D compared to MDCK-PTR stably expressing human TFR. (C-D) Pseudo-color images show intracellular distribution of TFR-Tfn complexes in HME (Panel C) and T47D (Panel D) cells after 5min (panels a–d) and 30min (panels e–h) internalization at 37°C of AF488-Tfn and AF555-Tfn at A:D~2. Pseudocolor images of acceptor (Acc; panel a & e), quenched donor (qD; panel b & f), uncorrected FRET (uFRET; panel c & g) and corrected FRET (PFRET; panel d & h) show the typical endocytic pattern of irregular and punctate structures. A peripheral distribution is detected at 5min, while at 30min, a peri-nuclear accumulation is observed. PFRET images confirm the existence of energy transfer between endogenous Tfn-AF488 (donor) and Tfn-AF555 (acceptor) in intracellular punctate endocytic structures in HME and T47D cells, in spite of the dramatically different levels of human TFR expressed by these two cell types. Bar: 20 µm. Pseudo-color ranges were generated using ImageJ Rainbow RGB look-up table.
Next, our main goal was to validate the ROI selection procedure by testing the ability of different ROI selection macro settings (intensity and/or statistical based settings) to influence a measure of assay performance that is calculated based on data extracted from the selected ROI coordinate datasets. Several approaches have been used to measure assay performance in high-throughput drug screening [55,56,60]. Both Z’ factor and SSMD have been demonstrated to provide an adequate value metric of the quality and strength of high-throughput drug screening assays [54,61]. Recently, the Z’ parameter was applied to evaluate the quantitative performance of FRET-based fluorescence lifetime imaging (FLIM) assays [62]. Here, we have used SSMD to validate the Image J ROI selection macro. SSMD consists of the ratio between the difference of the means and the standard deviation of the difference between two populations, such as positive and negative controls, such as treated vs. untreated samples or early vs. later time points. For independent populations, SSMD= (μ1-μ2)/√(σ1 2+ σ2 2) in which μ represents mean values and σ represents standard deviation [54,56,60,61]. Moreover, this equation defines E% SNR, since it measures the size of signal change (i.e. dynamic range of the assay E% signal) in relation to the variability of the signal difference, providing a data quality assessment that combines signal range and variability measures in a concise summary number [60]. Another issue concerning the ability to evaluate the quality of FRET ROI datasets relates to lack of well characterized E%-mediated linear relationships that can be used to determine the best FRET ROI dataset. Therefore, to experimentally demonstrate that SSMD can be used as a quality assessment measure of each selected FRET ROI dataset in cells showing low signal FRET, we determined SSMD for E% vs. A:D ratios, a well-established positively dependent linear relationship that indicates clustered and specific protein-protein interactions[63–66], such as those underlying the FRET detected between donor-Tfn and acceptor-Tfn bound to homodimeric TFR throughout the binding, internalization and recycling of Tfn-TFR complexes[17–22]. The R2 values of the E% vs. A:D from images subjected to Image J ROI selection macro at ED=5 & %F=0 vs ED=5 & %F=30 were determined; these relationhips are shown in Figure 3a–b. The R2 is supposed to increase from 0 to 1 value as the E% is positively dependent on increasing A:D ratios[63–66]. Then, SSMD values were determined for the E% values at A:D~2 vs. 0.5 (SSMDa) and E% values at A:D~2 vs. 1 (SSMDb) (Figure 3c–d); the E% values and standard deviation values were generated from data extracted from images subjected to a wide variety of E, %F and STD Image J ROI selection macro values and then R2, SSMDa and SSMDb were calculated as shown in Table 1. As shown in Figure 3d, a tight linear relationship (R2=0.9875) is detected between increasing SSMDb (E% at A:D~2 vs. E% at A:D~1) values and R2 values. In contrast, at larger dynamic ranges such as the ones detected between E% at A:D~2 and E% at A:D~0.5, such a correlation between SSMDa (E% at A:D~2 vs. E% at A:D~0.5) is not detected (Figure 3c), suggesting that the importance of the different FRET ROI selection settings is reduced when SSMD> 0.5. However, when SSMD <0.5, it is important to select the adequate Image J ROI selection settings, such as ED=5 and %F>22. Since increasing %F values above 30 leads to a marked reduction in the number of ROIs we kept the Image J ROI selection settings at ED=5 and %F=22 for the rest of the experiments in this manuscript, unless specifically noted.
Table 1.
Data point values for Figure 3
| Tests | ED | %F | STD | ROI count |
SSMDa E%A:D~2 vs. E%A:D~0.5 |
SSMDb E%A:D~2 vs. E%A:D~1 |
R2 E% vs. A:D |
|---|---|---|---|---|---|---|---|
| #1 | 0 | 0 | 0 | 6676 | 0.4953 | 0.0779 | 0.7172 |
| #2 | 0 | 0 | 35 | 6032 | 0.6127 | 0.0476 | 0.6483 |
| #3 | 0 | 22 | 0 | 4236 | 0.7184 | 0.1209 | 0.7228 |
| #4 | 0 | 22 | 35 | 4065 | 0.7692 | 0.1215 | 0.7244 |
| #5 | 0 | 30 | 0 | 1557 | 0.8569 | 0.1746 | 0.7652 |
| #6 | 0 | 30 | 35 | 1538 | 0.8649 | 0.1697 | 0.7593 |
| #7 | 0 | 35 | 0 | 648 | 0.8025 | 0.3592 | 0.9116 |
| #8 | 0 | 35 | 35 | 647 | 0.7999 | 0.3592 | 0.9122 |
| #9 | 0 | 30 | 45 | 1440 | 0.8661 | 0.1452 | 0.7322 |
| #10 | 0 | 38 | 0 | 358 | 0.6960 | 0.4685 | 0.9579 |
| #11* | 5 | 0 | 0 | 5483 | 0.6267 | 0.0571 | 0.6632 |
| #12 | 5 | 0 | 35 | 5012 | 0.6611 | 0.0527 | 0.6528 |
| #13 | 5 | 22 | 0 | 3220 | 0.8162 | 0.1401 | 0.7406 |
| #14 | 5 | 22 | 35 | 3159 | 0.8188 | 0.1389 | 0.7389 |
| #15* | 5 | 30 | 0 | 1179 | 0.8324 | 0.2099 | 0.8001 |
| #16 | 5 | 30 | 35 | 1176 | 0.8212 | 0.2077 | 0.7995 |
| #17 | 5 | 35 | 0 | 500 | 0.6685 | 0.3189 | 0.9010 |
| #18 | 5 | 35 | 35 | 500 | 0.6685 | 0.3189 | 0.9010 |
| #19 | 5 | 30 | 45 | 1125 | 0.8130 | 0.1794 | 0.7714 |
| #20 | 5 | 38 | 0 | 280 | 0.5730 | 0.4601 | 0.9774 |
Notes:
#11 corresponds to data point A in Figure 3c–d
#15 corresponds to data point B in Figure 3c–d
3.3. FRET analysis of TFR-Tfn complexes during endocytic recycling pathway in normal vs. cancer cells
To develop a better understanding of how the endocytic trafficking pathways of receptor-ligand complexes are regulated in normal cells and how these processes are disrupted in cancer we have compared Tfn-TFR trafficking pathways in normal vs. cancer cells, which requires the ability to image cells expressing dramatically different receptor expression levels. Here, we have compared TFR protein expression levels in three well-known epithelial cell lines: normal human mammary epithelial (HME) cells, human breast cancer T47D cells and Madin-Darby canine kidney epithelial MDCK-PTR cells, a well-known epithelial cell model system [57]. TFR protein levels are higher expressed in T47D cancer cells and in MDCK-PTR cells in comparison to the HME cells (Figure 4A–B). To test whether a higher TFR expression level correlated with increased Tfn internalization in cancer cells, a functional analysis of TFR expression was performed using a Tfn uptake fluorescence imaging-based assay. Using a time course experiment (5 vs. 30min), we concluded that T47D cells show a significantly increased total amount of Tfn internalization compared to normal HMECs at the indicated time points (Figure 4C–D, panels Acc, a & e).
One interesting characteristic of Tfn-TFR complexes is that they remain associated throughout the endocytic recycling pathway. Tfn is released at the extracellular space when the receptor-ligand complexes get exposed to the media neutral pH [23,39]. Therefore, energy transfer between donor and acceptor-labeled Tfn-TFR complexes occurs from their internalization until their delivery to the plasma membrane via the endocytic recycling pathway. Since E% is a relative expression of distance, slight changes in E% levels may indicate alterations in the conformation of Tfn-TFR complexes due to changes in endocytic environment, such as the pH acidification, the iron release from the Tfn molecule and the putative association/dissociation of regulatory proteins with the Tfn-TFR complexes. To test whether we can detect changes in E% levels during the Tfn-TFR complexes endocytic recycling pathway, HME cells were internalized with AF488-Tfn and AF555-Tfn at A:D~2 at 37°C for 5min vs. 30min. After fixation, which was shown not to affect E% levels between Tfn-TFR complexes as well as other receptor-ligand complexes [19–21], cells were imaged and processed for FRET analysis using the Image J background removal, SBT correction and ROI selection protocol as described in Figure 1. In Figure 4C–D, 5min (panels a–d) and 30min (panels e–f) post-internalization endocytic staining patterns are shown for AF488-Tfn (qD, panels a & e), AF555-Tfn (Acc, panels b & f), uFRET (panels c & g) and PFRET (panels d & h) in HME (Figure 4C) vs. T47D cells (Figure 4D). As expected both in HME and T47D cells, there is less overall intensity at 5min in all images than at 30min since we are using a steady state internalization approach, leading to intracellular accumulation with longer internalization periods. As a consequence of higher Acc and qD levels at 30min, PFRET levels also show increased levels. However, since E% is calculated as a percentage of energy transfer in relation to the unquenched donor, higher PFRET absolute levels may not represent higher E% levels, even at similar A:D ratios. Nevertheless, it is clear that HME cells show significantly lower Tfn internalization and thus PFRET levels than T47D cells.
3.4. Role of background removal in intensity-based FRET analysis of low PFRET signal images
The first step of the Image J ROI selection macro consists in the background removal from both donor and channel images. To do that we have normally used up to 10 single labeled images to calculate the donor and acceptor background values as an average of 20 randomly selected ROIs (20×20 pixels), as described above. However, when a reliability test of 10 runs was performed, donor and acceptor background values calculated from 10 randomly selected images out of 50 total images show a significantly higher level of variability then those that are similarly calculated from all of the 50 images (Figure 5a). The non-overlapping ROI selection method using the following selection parameters: ID=500, ED=5, STD=45 & %F=22, was applied to the PFRET images generated from HME cells as shown in Figure 4C and then FRET data was extracted, as described above. A:Dcalc levels are unchanged upon background removal with values calculated from 10 vs. 50 images (unpublished results). In contrast, uFRET (unpublished results) and PFRET (Figure 5b) levels show higher standard deviation values upon background removal using values calculated from 10 images than 50 images, suggesting that high variability of donor and acceptor background values leads to lower SNR levels in uFRET and PFRET values both at 5 and 30min internalization. E% values also show higher variability when calculated from images subjected to background removal using 10 vs, 50 images to calculate donor and acceptor background values (Figure 5c). Interestingly, when 50 images were used and the standard deviation of background values reduced, E% values show a significant decrease from 5 to 30min internalization, suggesting that reduced variability of donor and acceptor background values leads to higher FRET SNR ratio and thus increased sensitivity of E% assays (Figure 5c). To test this hypothesis, SSMD was calculated between E% values at 5 vs. 30min internalization from assays in which 10 vs 50 images were used to calculated donor and acceptor background values. As shown in Figure 5d, SSMD shows lower values as well as higher standard deviation values when using 10 vs. 50 images to calculate background values. Importantly, using at least 50 images to calculate background values leads to higher assay sensitivity as shown by increased SSMD values as well as by the ability to detect statistically significant differences between E% values generated at 5 or at 30min internalization of Tfn-TFR complexes (p-value10>0.05 –n.s.-; p-value50< 0.001 –sig-).
3.5. Comparison of different automated methods to select ROIs from low PFRET signal images
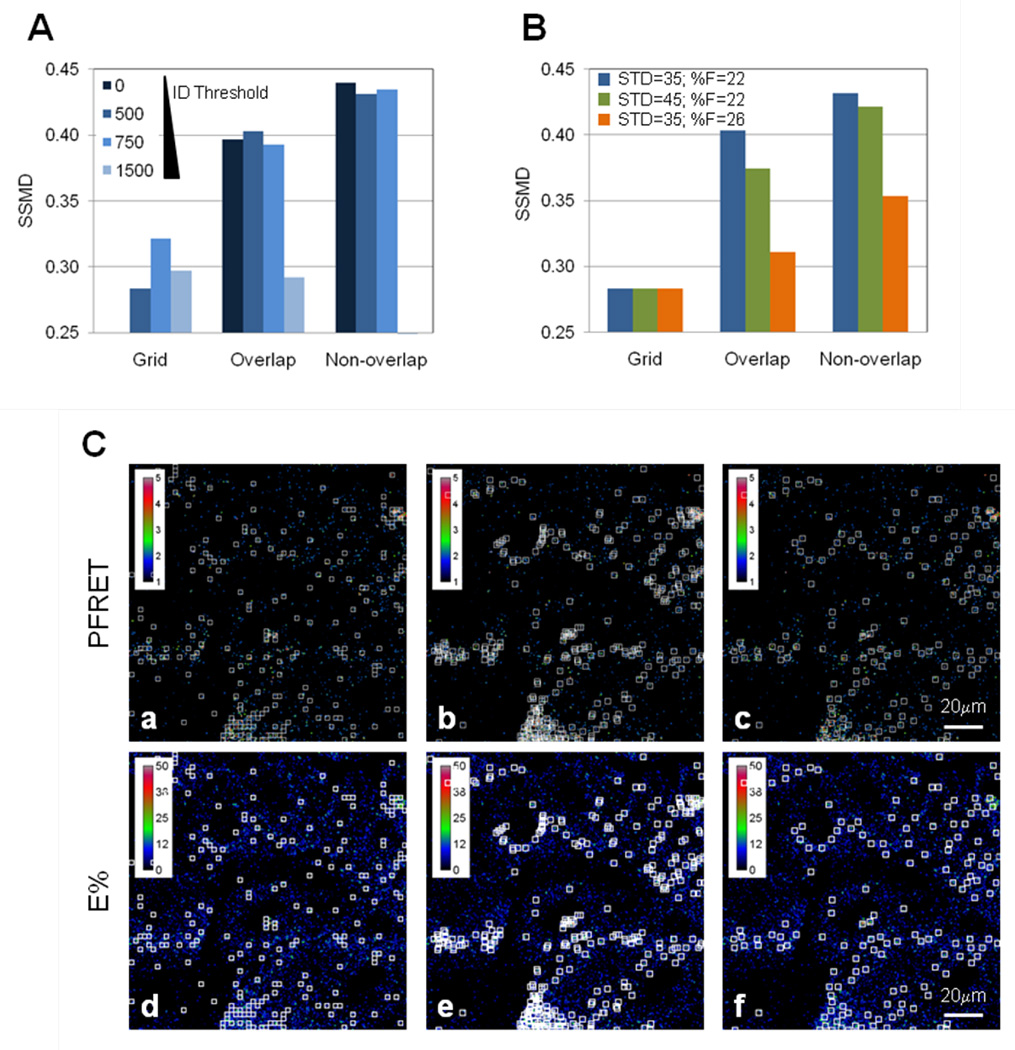
In the Image J ROI selection macro, ROIs can be chosen that are non-overlapping (Figure 1), overlapping or on a pre-determined grid. These ROI selection options were run on the PFRET images collected from the 5min and 30min pre-internalization of Tfn-TFR complexes using the following selection minimum thresholds: ED=5 and ID=0, 500, 750, 1500, STD=35; %F=22. SSMD values were then calculated based on the E% and standard deviation values, as described above. As shown in Figure 6A, higher SSMD values are detected using the non-overlapping option of the Image J ROI selection macro. In contrast, the ROI grid option shows the lowest SSMD level indicating its lower reliability and sensitivity to detect minor E% changes. Increasing ID improves SSMD values in the grid selection method but not on the non-overlap method suggesting that other parameters such as the edge detection (E) or %F are predominant in their effect on the quality of the non-overlapping ROI datasets. However, high ID threshold values, e.g. 1500, reduce drastically the number of ROIs selected leading to reduced SSMD values in all three ROI selection methods used. As expected, removing overlapping ROIs improves SNR ratio as shown by the higher SSMD values generated using the non-overlapping ROI selection method in comparison with that of overlapping ROIs. Next, the followed minimum thresholds were applied: ED=5 and ID=500 values together with different STD and %F values, such as STD=35 & %F=22, STD=45 & %F=22 and STD=35 & %F=26, to the Image J ROI selection macro using the PFRET images collected from the 5min and 30min pre-internalization of Tfn-TFR complexes. As shown in Figure 6B, ROIs selected by the grid method show very low SSMD values compared with those generated using non-overlapping methods. The following ROI selection parameters: ED=5, ID=500, STD=35 & %F=22 lead to the best SSMD values (>0.4) for the 5min vs. 30min FRET assay of Tfn-TFR complexes, suggesting that increasing STD and %F above those values does not lead to improved SSMD values, most probably because it leads to dramatic reduction of ROI numbers. In Figure 6C, the selected ROIs are shown on PFRET (panels a–c) and E% (panels d–f) for the following ROI selection parameters: ED=5, ID=500, STD=35 & %F=22, using the grid method (panels c & d), overlapping approach (panel b & e) and non-overlapping method (panel c & f). The ability to select ROIs on the×and y axis allows for a more flexible ROI selection that matches the correct PFRET pixel distribution more accurately both visually and quantitatively using the SSMD SNR evaluation parameter. Furthermore, the ability to remove non-overlapping ROIs by selecting the best ROI (highest level of gray-scale ID, as described above) improves SNR ratio as shown by higher SSMD values and reduces potential bias. In summary, these results show that non-overlapping non-grid ROI selection methods are superior in their ability to collect quality ROI datasets and subsequently E% datasets in comparison with overlapping and grid based ROI selection methods.
Figure 6. Different ROI selection methods affect SSMD levels.
(A) Different ROI selection methods such as using a pre-set grid, overlapping ROIs and non-overlapping ROIs, were used under different ROI selection settings such as ED=5, increasing ID values from 0 to 1500 and STD=0 and %F=0. Nonoverlapping ROI datasets show the highest SSMD levels (>0.4). (B) Increasing statistical based thresholds for ROI selection above 35 for STD and 22 for %F does not improve SSMD >0.4 suggesting that STD=35 together with %F=22 are the adequate ROI selection settings for this cell-based FRET system and imaging conditions. (C) ROIs selected by grid method are shown in PFRET (panel a) and E% (panel d) images. Overlapping ROIs are shown in PFRET (panel b) and E% (panel e) images. Non-overlapping ROIs are shown in PFRET (panel c) and E% (panel f) images. The selected ROIs are shown for 30min internalization FRET and E% images. The ROI settings used are: ID=500, ED=5, STD= 35 and %F= 22. Bar: 20 µm.
3.6. Role of ROI selection parameters in SSMD SNR analysis
The Image J ROI selection macro includes several ways to manipulate the PFRET image with the main goal of increasing the PFRET image contrast, i.e. the dynamic range of pixel intensity. Then, ROIs are selected on the manipulated PFRET images based on intensity and statistical-based thresholds. Image manipulation includes multiplication of pixel values by 10 until either a minimum value (e.g. 5 or 10) or a pre-set maximum number of multiplications (e.g. 40) are reached. It is important to stress that the final FRET data is extracted from the background-subtracted and non-manipulated donor, acceptor, uFRET and PFRET images using the selected ROI x,y coordinates. If the maximum number of multiplications is set at 10 or higher with the other ROI selection parameters as follows: ID=500, ED=5, STD=35 & %F=22, the SSMD values remain constant above or below 0.4 when the pre-set minimum value equals 10 or 5, respectively (results not shown). These results suggest that a multiplication factor above 10 should increase the dynamic range of pixel intensity enough that minimum values above 10 are reached. Such an increase in pixel intensity appears sufficient to allow an accurate ROI selection (SSMD >0.4) and therefore throughout this manuscript we have used a multiplication factor equal to 40 and a minimum pixel value of 10, unless otherwise noted. However, setting an adequate number for the multiplication factor and minimum value depends on the background level and SNR of the original FRET images. It is therefore appropriate, when using a different FRET cell based system and/or different imaging conditions, to test different multiplication factors and minimum pixel values to determine which value would lead to the best ROI dataset as shown by SSMD values > 0.4.
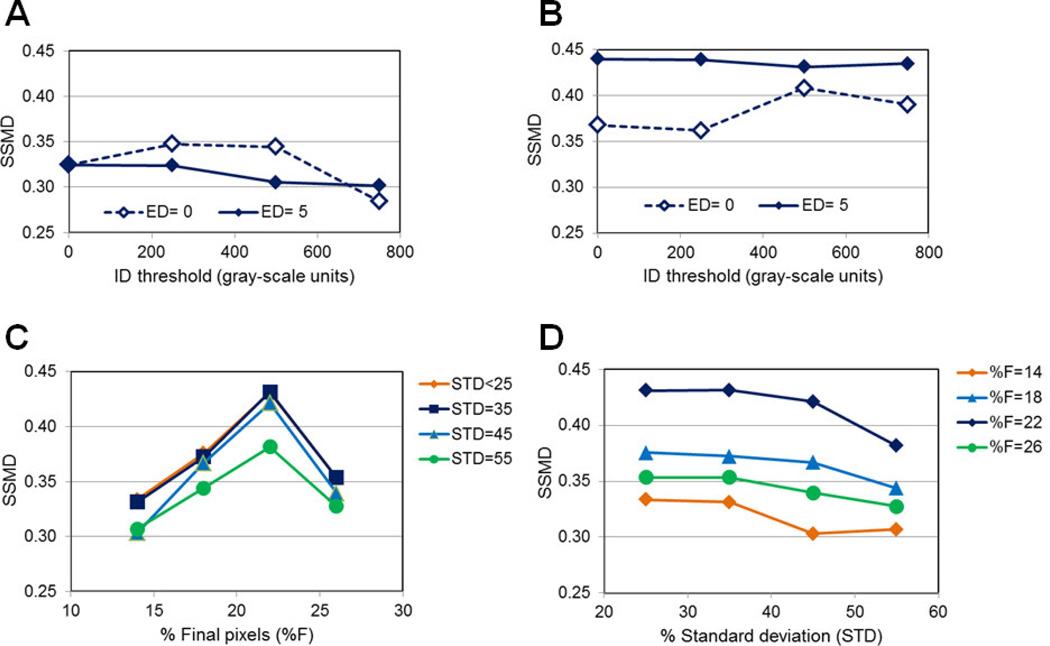
In Figure 7A–B, the role of ID on the quality of ROI selection is tested. In the absence of statistical-based parameters, choosing ID between 200–600 plus ED=5 lead to the best SSMD values ~0.35 (Figure 7A). However, in the presence of STD=35 and %F=22 plus ED=5, the SSMD values show a significant increase, independently of ID values; even with ED=0, SSMD values are above 0.35 when using STD=35 and %F=22 (Figure 7B). These results suggest that statistical-based thresholds are predominant over the intensity based thresholds, resulting in ROI datasets that generate good quality SSMD values (> 0.4). The ED intensity-based parameter addresses edge detection, which is particularly important when evaluating images containing endocytic punctate structures that show significant differences between the periphery and the center of ROIs. However, in other types of images such as those including plasma membrane staining it is advised to keep ED=0 to allow for the detection of ROIs with similar values at the periphery and center.
Figure 7. Role of intensity- and statistically-based parameters in ROI selection.
(A) Increasing ID levels were used to test the role of ID in ROI selection when ED=0 or 5 and STD=0 and %F=0 via SSMD analysis. (B) Increasing ID levels were used to test the role of ID in ROI selection when ED=0 or 5 and STD=22 and %F=35 via SSMD analysis. Statistically based parameters have a predominant role in selecting good quality ROI datasets as shown by SSMD>0.4 values. (C) Increasing %F values at different STD values were used to test the role of %F in ROI selection when ID=500 & ED=5 via SSMD analysis. (D) Increasing STD values at different %F values were used to test the role of STD in ROI selection when ID=500 & ED=5 via SSMD analysis. %F=22 & STD=35 are shown as the adequate values for good quality ROI selection, as shown by SSMD>0.4 values.
In Figure 7C–D, we test different combinations of STD and %F values to determine which statistical-based threshold shows primacy in the selection of good quality ROI datasets that lead to SSMD>0.4 values between 5 and 30min TFR-Tfn internalization at 37°C. Considering Figure 7C–D, only %F=22 with STD=25–45 show SSMD >0.4 values indicating that for these particular biological system and imaging conditions %F=22 is the adequate parameter to select good quality ROI datasets. We have applied this Image J ROI selection macro to other biological systems using intensity based FRET confocal microscopy and in our experience the following ROI selection settings of ID=500, ED=5, %F=22 and STD=35 lead in the majority of cases to the selection of adequate ROI datasets. We recommend using a control vs. treatment condition that would allow for the use of SSMD based approaches to test the quality of ROI datasets. Testing a few different parameters values around the ones indicated above should allow for a rather quick determination of the ideal ROI selection parameters for the specific images analyzed, which would result in SSMD > 0.4–0.5 values.
3.7. FRET levels during TFR-Tfn endocytic trafficking in normal vs. cancer cells
Previously, we have used MDCK-PTR cells stably overexpressing human TFR as a model system to study the FRET behavior of TFR-Tfn complexes[19–21] and other receptors such as pIgA-R[43–45] and LDL-R (unpublished data). FRET analysis using intensity-based confocal microscopy as well as fluorescence lifetime microscopy (FLIM), indicate that TFR-Tfn complexes show a clustered behavior during their endocytic trafficking in MDCK cells[49]. In Figure 8, we compared the internalization of AF488-Tfn and AF555-Tfn into MDCK-PTR, HME and T47D cells at an A:D~2 for 5min (Panels a, c & e) vs 30min (Panels b, d & f) at 37°C. After fixation, cells were imaged and processed for FRET analysis using the Image J background removal, SBT correction and ROI selection protocol, as described above for HME cells (Figure 2B & 6C). E% images show the ROI dataset selected for HME (Panels a–b), MDCK-PTR (Panels c–d) and T47D (Panels e–f) using the Image J ROI selection macro with the following ROI selection parameters: ID=500, ED=5, STD=35 and %F=22. In all cells, the ROI dataset matches well visually with E% levels across the images (Figure 8).
Figure 8. FRET ROI selection during endoctic trafficking of TFR-Tfn complexes in HMEC, MDCK-PTR and T47D cells.
Non-overlapping ROIs were applied to E% images of 5min (panels a, c & e) vs. 30min (panels b, d & f) TFR-Tfn endocytic trafficking in HME (panels a–b), MDCK-PTR (panels c–d) and T47D (panels e–f) cells. The ROI settings used are: ID=500, ED=5, STD= 35 and %F= 22. Bar: 20 µm.
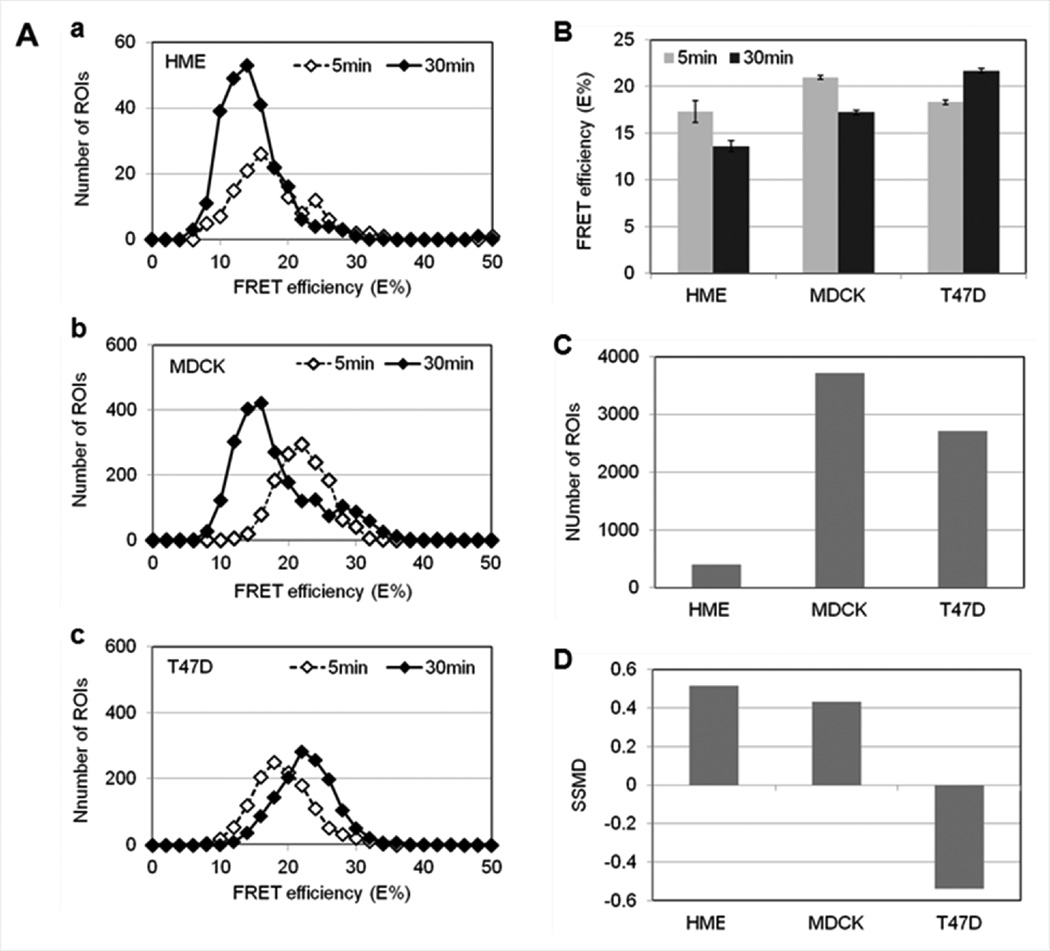
To address E% differences in a quantitative manner, selected ROI coordinates were applied to HME, MDCK-PTR and T47D FRET images and average E% values extracted as described above in Figure 5C. As shown in Figure 9, in both HME and MDCK-PTR cells, FRET analysis shows a significant decrease of E% levels between 5min and 30min of endocytic trafficking of TFR-Tfn complexes. Histogram analysis shows the E% distribution over the ROI dataset, demonstrating the difference between E% levels at 5min vs. 30min both in HME (Panel a) and MDCK-PTR (Panel b) cells. However, in T47D breast cancer cells (Panel c), a reverse behavior is detected with an increase of E% levels between 5min and 30min of endocytic trafficking of TFR-Tfn complexes. These results are clearly observed when ROI averages are calculated in Figure 9B; both E% decrease from 5min to 30min in MDCK-PTR and HME cells as well as E% increase from 5min to 30min in T47D cells are statistically significant when using t-test analysis (p>0.001). Although the number of ROIs collected from 6 images is dramatically lower in HME cells than in MDCK-PTR and T47D cells (Figure 9C), SSMD values remain above 0.4 in MDCK-PTR and HME cells, whereas SSMD values show negative values ~0.4–0.5 in T47D cells (Figure 9D). The substantial decrease in ROIs collected in HME images is due to the reduced level of TFR expression in HME cells when compared to that in MDCK-PTR and T47D cells (Figure 4A–B) as well as the lower level of Tfn internalization by HME vs. T47D cells (Figure 4C–D). In summary, despite differences in TFR expression levels, HME cells behave similarly to MDCK-PTR cells but not to T47D cancer cells. These results suggest a marked difference in endocytic trafficking of TFR-Tfn complexes in normal vs. cancer cells.
Figure 9. Changes in E% levels detected during endoctic trafficking of TFR-Tfn complexes in HMEC, MDCK-PTR and T47D cells.
(A) Histogram analysis of E% distribution at 5min (white diamonds) vs. 30min (black diamonds) internalization of TFR-Tfn complexes into HME (Panel a), MDCK-PTR (Panel b) and T47D (Panel c) cells. (B) E% averages at 5min (gray) vs. 30min (black) internalization of TFR-Tfn complexes into HME, MDCK-PTR and T47D cells. Error bars indicate 95% confidence interval. (C) Number of ROIs selected for HME, MDCK-PTR and T47D cells. (D) SSMD analysis. Although the number of ROIs decreases dramatically in HME cells due to their reduced level of TFR expression, SSMD values remain >0.4 for both MDCK and HME datasets. In contrast, T47D cells show negative SSMD values indicating a reversed E% behavior during TFR-Tfn endocytic trafficking.
4. CONCLUSION
Here, we have presented a significant technical advance to the image analysis and processing of images collected using intensity based FRET confocal microscopy. Intensity-based approaches remain as the FRET imaging analysis most widely used by cell and molecular biologists, since the majority of microscopy instruments, e.g. wide-field, confocal, spinning disk, two-photon and TIRF microscopes, can be used to collect FRET images using this technique. Although, intensity based FRET does not require special microscopy accessory instruments, such as FLIM, or complex reference FRET proteins, such as FRET stoichiometry analysis, it does necessitate a significant post-collection image processing protocol, which has been provided in available and accessible software programs. However, these software programs have focused primarily in providing SBT correction algorithms. Here we have developed an automated ROI selection Image J macro, which allows for the selection of ROI datasets, based on intensity and statistical based thresholds, within cellular images with reduced FRET signal.
Another important technical advance provided here is the demonstration of a statistical SNR parameter, e.g. SSMD, to validate the quality and reliability of FRET analysis, in particular of ROI selection. SSMD indicates the effect size of a difference between a control and a treatment situation. In FRET imaging analysis, we propose the use of SSMD as a relative measurement indicating the SNR between images collected from control cells and those from cells subjected to different protocol conditions such as incubation time or drug treatments. SSMD may have many other applications in the evaluation of improvements to already developed imaging processing approaches allowing for a clear validation of whether such an improvement actually leads to an increased SNR of the biological experiment tested.
Finally, the Image J ROI selection macro together with SSMD as an evaluation parameter of SNR levels, allowed us to demonstrate that TFR-Tfn complexes show significantly different E% behavior during their endocytic trafficking in normal vs. cancer cells. Since E% is a relative measure of distance, we propose that these changes in E% levels represent conformational changes in TFR-Tfn complexes during endocytic pathway. TFR-Tfn complexes are exposed to dramatically different environments during their intracellular transport. Changes in pH, iron binding and interaction with accessory proteins all may lead to complex but significant conformational changes in the TFR-Tfn complexes, which remain tightly associated throughout intracellular transport. Importantly similar changes in E% levels have been clearly demonstrated in two different cell lines, showing endogenous (HMEC) or exogenous (MDCK) expression of human TFR, respectively. In contrast, T47D cancer cells showing high TFR expression levels, undergo a reverse behavior with increased E% levels during TFR-Tfn endocytic trafficking. Furthermore, improvements in automated selection of ROI datasets allowed us to detect minor but significant changes in E% with potential biological significance in human normal v. cancer cells. In summary, technical upgrades in ROI selection in intensity based FRET analysis together with the use of SSMD, a SNR-related parameter, to validate the quality of those ROI datasets, allowed us to demonstrate that FRET signal decreases during the endocytic trafficking of TFR-Tfn receptor-ligand complexes in cells expressing endogenous or exogenous human TFR. We propose that slight but significant E% changes may indicate the occurrence of conformational changes in TFR-Tfn complexes during their endocytic recycling pathway.
In summary, we have established the importance of background noise removal as well as of appropriate FRET ROI selection as implemented by a novel statistically validated custom generated image analysis algorithm for increased reliability and reproducibility of intensity-based FRET. This ROI selection algorithm collects strongly reliable FRET data even from low signal FRET HME cells expressing reduced levels of endogenous TFR. Furthermore, the use of this ROI selection algorithm allows for the detection of minor but significant E% changes that may indicate conformational changes undergone by TFR-Tfn complexes via the endocytic pathway, providing important insights into the regulation of the endocytic trafficking of TFR-Tfn complexes in normal vs. cancer cells that express dramatically different levels of TFR. These results are particularly important since Tfn-TFR system is widely used to target anti-cancer drugs to tumors in vivo, making it crucial to analyze in detail the Tfn-TFR endocytic system in normal vs. cancer cells.
Highlights.
ImageJ macro uses intensity and statistical thresholds to select ROIs in FRET images.
SSMD is used as a relative measurement of signal to noise ratio between FRET images.
FRET analysis shows TFR-Tfn conformational changes during their endocytic pathway.
TFR-Tfn FRET efficiency levels show differences between normal and cancer cells.
TFR-Tfn conformational changes are altered during endocytic transport in cancer cells.
ACKNOWLEDGEMENTS
We would like to thank Dr. Joseph Mazurkiewicz, Dr. Alejandro Adam and Anupam Das (Albany Medical College) for their helpful comments and assistance. We would also like to thank Dr. Ammasi Periasamy, Dr. Yuansheng Sun, Ye Chen and Horst Wallrabe from the Keck Center for Cellular Imaging at the University of Virginia for their support and helpful comments. We would like to thank the numerous summer undergraduate research assistants that worked in this project throughout the years; this work would not have possible without the help of Christine Kim, Ashley Cronin, Becky Cardin, Nadia Lomakina, Stephen Okaine, Michael Sullivan, Amanda Tower, Alyssa Trigger at Albany Medical College and Andrew Reilly at the College of St. Rose. 6.
ABBREVIATIONS
- Acc
Acceptor
- A:D
Internalized acceptor:donor ratios
- A:Dcalc
Calculated A:D ratios
- AF488
Alexa Fluor 488
- AF555
Alexa Fluor 555
- AF488-Tfn
Alexa Fluor 488-Transferrin
- AF555-Tfn
Alexa Fluor 555-Transferrin
- D
Donor
- E%
Energy transfer efficiency
- ED
Edge detection
- FRET
Förster Resonance Energy Transfer
- HME
Human mammary epithelial cells
- ID
Integrated density
- qD
Quenched donor
- MDCK-PTR
Madin-Darby canine kidney epithelial cells stably expressing human TFR
- %F
Area fraction
- PBS
Phosphate buffered saline solution
- PFRET
Precision FRET
- uFRET
Unquenched FRET
- ROI
Region of interest
- SBT
Spectral bleed-through
- SNR
Signal to noise ratio
- SSMD
Strictly standardized mean differences
- STD
Standard deviation
- Tfn
Transferrin
- TFR
Transferrin receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nature reviews. Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 2.Abella JV, Park M. Breakdown of endocytosis in the oncogenic activation of receptor tyrosine kinases. American journal of physiology. Endocrinology and metabolism. 2009;296:E973–E984. doi: 10.1152/ajpendo.90857.2008. [DOI] [PubMed] [Google Scholar]
- 3.Bache KG, Slagsvold T, Stenmark H. Defective downregulation of receptor tyrosine kinases in cancer. The EMBO journal. 2004;23:2707–2712. doi: 10.1038/sj.emboj.7600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitra S, Cheng KW, Mills GB. Rab25 in cancer: a brief update. Biochemical Society transactions. 2012;40:1404–1408. doi: 10.1042/BST20120249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agola JO, Jim Pa, Ward HH, Basuray S, Wandinger-Ness A. Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clinical genetics. 2011;80:305–318. doi: 10.1111/j.1399-0004.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magnusson MK, Meade KE, Brown KE, Arthur DC, Krueger LA, Barrett AJ, Dunbar CE. Rabaptin-5 is a novel fusion partner to platelet-derived growth factor beta receptor in chronic myelomonocytic leukemia. Blood. 2001;98:2518–2525. doi: 10.1182/blood.v98.8.2518. [DOI] [PubMed] [Google Scholar]
- 7.Pece S, Confalonieri S, R Romano P, Di Fiore PP. NUMB-ing down cancer by more than just a NOTCH. Biochimica et biophysica acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. The Journal of cell biology. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Experimental cell research. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandal MV, Madshus IH. Epidermal growth factor receptor and cancer: control of oncogenic signalling by endocytosis. Journal of cellular and molecular medicine. 12:1527–1534. doi: 10.1111/j.1582-4934.2008.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefebvre J, Ancot F, Leroy C, Muharram G, Lemière A, Tulasne D. Met degradation: more than one stone to shoot a receptor down. FASEB journal. 2012;26:1387–1399. doi: 10.1096/fj.11-197723. [DOI] [PubMed] [Google Scholar]
- 12.French AR, Sudlow GP, Wiley HS, Lauffenburger DA. Postendocytic trafficking of epidermal growth factor-receptor complexes is mediated through saturable and specific endosomal interactions. The Journal of biological chemistry. 1994;269:15749–15755. [PubMed] [Google Scholar]
- 13.Liu P, Ahmed S, Wohland T. The F-techniques: advances in receptor protein studies. Trends in endocrinology and metabolism. 2008;19:181–190. doi: 10.1016/j.tem.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Ciruela F, Vilardaga J-P, Fernández-Dueñas V. Lighting up multiprotein complexes: lessons from GPCR oligomerization. Trends in biotechnology. 2010;28:407–415. doi: 10.1016/j.tibtech.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeug A, Woehler A, Neher E, Ponimaskin EG. Quantitative Intensity-Based FRET Approaches—A Comparative Snapshot. Biophysical Journal. 2012;103:1821–1827. doi: 10.1016/j.bpj.2012.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Wallrabe H, Seo S, Periasamy A. FRET microscopy in 2010: the legacy of Theodor Förster on the 100th anniversary of his birth. Chemphyschem. 2011;12:462–474. doi: 10.1002/cphc.201000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barroso M, Sun Y, Wallrabe H, Periasamy A. Nanometer-scale measurements using FRET and FLIM microscopy. In: Gilmore A, editor. Luminescence: the Instrumental Key to the Future of Nanotechnology. Pan Stanford: 2013. In Press. [Google Scholar]
- 18.Barroso MM. Quantum dots in cell biology. The journal of histochemistry and cytochemistry. 2011;59:237–251. doi: 10.1369/0022155411398487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallrabe H, Chen YE, Periasamy A, Barroso M. Issues in confocal microscopy for quantitative FRET analysis. Microscopy research and technique. 2006;69:196–206. doi: 10.1002/jemt.20281. [DOI] [PubMed] [Google Scholar]
- 20.Wallrabe H, Bonamy G, Periasamy A, Barroso M. Receptor complexes cotransported via polarized endocytic pathways form clusters with distinct organizations. Molecular biology of the cell. 2007;18:2226–2243. doi: 10.1091/mbc.E06-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Periasamy A, Wallrabe H, Chen Y, Barroso M. Quantitation of Protein – Protein Interactions: Confocal FRET Microscopy. Methods in Cell Biology. 2008;89:569–598. doi: 10.1016/S0091-679X(08)00622-5. [DOI] [PubMed] [Google Scholar]
- 22.McGrath N, Barroso M. Quantum dots as fluorescence resonance energy transfer donors in cells. Journal of biomedical optics. 2008;13:031210. doi: 10.1117/1.2939417. [DOI] [PubMed] [Google Scholar]
- 23.Luck AN, Mason AB. Transferrin-mediated cellular iron delivery. Current topics in membranes. 2012;69:3–35. doi: 10.1016/B978-0-12-394390-3.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatter KC, Brown G, Trowbridge IS, Woolston RE, Mason DY. Transferrin receptors in human tissues: their distribution and possible clinical relevance. Journal of clinical pathology. 1983;36:539–545. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue T, Cavanaugh PG, Steck Pa, Brünner N, Nicolson GL. Differences in transferrin response and numbers of transferrin receptors in rat and human mammary carcinoma lines of different metastatic potentials. Journal of cellular physiology. 1993;156:212–217. doi: 10.1002/jcp.1041560128. [DOI] [PubMed] [Google Scholar]
- 26.Prost AC, Ménégaux F, Langlois P, Vidal JM, Koulibaly M, Jost JL, Duron JJ, Chigot JP, Vayre P, Aurengo A, Legrand JC, Rosselin G, Gespach C. Differential transferrin receptor density in human colorectal cancer: A potential probe for diagnosis and therapy. International journal of oncology. 1998;13:871–875. doi: 10.3892/ijo.13.4.871. [DOI] [PubMed] [Google Scholar]
- 27.Shindelman JE, Ortmeyer AE, Sussman HH. Demonstration of the transferrin receptor in human breast cancer tissue. Potential marker for identifying dividing cells. International journal of cancer. 1981;27:329–334. doi: 10.1002/ijc.2910270311. [DOI] [PubMed] [Google Scholar]
- 28.Jian J, Yang Q, Huang X. SRC regulates TYR20 phosphorylation of transferrin receptor-1 and potentiates breast cancer cell survival. The Journal of biological chemistry. 2011;286:35708–35715. doi: 10.1074/jbc.M111.271585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryschich E, Huszty G, Knaebel HP, Hartel M, Büchler MW, Schmidt J. Transferrin receptor is a marker of malignant phenotype in human pancreatic cancer and in neuroendocrine carcinoma of the pancreas. European journal of cancer. 2004;40:1418–1422. doi: 10.1016/j.ejca.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 30.Daniels TR, Delgado T, Rodriguez Ja, Helguera G, Penichet ML. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clinical immunology. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Daniels TR, Bernabeu E, Rodríguez Ja, Patel S, Kozman M, Chiappetta Da, Holler E, Ljubimova JY, Helguera G, Penichet ML. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochimica et biophysica acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Högemann-Savellano D, Bos E, Blondet C, Sato F, Abe T, Josephson L, Weissleder R, Gaudet J, Sgroi D, Peters PJ, Basilion JP. The transferrin receptor: a potential molecular imaging marker for human cancer. Neoplasia. 2003;5:495–506. doi: 10.1016/s1476-5586(03)80034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochimica et biophysica acta. 2012;1820:264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan L, Hao Y, Wang S, Korotcov A, Zhang R, Wang T, Califano J, Gu X, Sridhar R, Bhujwalla ZM, Wang PC. Visualizing Head and Neck Tumors In Vivo Using Near- Infrared Fluorescent Transferrin Conjugate. Molecular imaging. 2008;7:42–49. [PubMed] [Google Scholar]
- 35.Holland JP, Evans MJ, Rice SL, Wongvipat J, Sawyers CL, Lewis JS. Annotating MYC status with (89)Zr-transferrin imaging. Nature medicine. 2012;18:1586–1591. doi: 10.1038/nm.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi CHJ, Alabi Ca, Webster P, Davis ME. Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dufès C, Robaian M Al, Somani S. Transferrin and the transferrin receptor for the targeted delivery of therapeutic agents to the brain and cancer cells. Therapeutic delivery. 2013;4:629–640. doi: 10.4155/tde.13.21. [DOI] [PubMed] [Google Scholar]
- 38.Venugopal V, Chen J, Barroso M, Intes X. Quantitative tomographic imaging of intermolecular FRET in small animals. Biomedical optics express. 2012;3:3161–3175. doi: 10.1364/BOE.3.003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maxfield FR, McGraw TE. Endocytic recycling. Nature reviews. Molecular cell biology. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Periasamy A. Additional correction for energy transfer efficiency calculation in filter-based Forster resonance energy transfer microscopy for more accurate results. Journal of biomedical optics. 2010;15:020513. doi: 10.1117/1.3407655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Elangovan M, Periasamy A. FRET Data Analysis: The Algorithm. In. In: Periasamy A, Day R, editors. Molecular Imaging: FRET Microscopy and Spectroscopy. New York: Oxford University Press; 2005. pp. 126–145. [Google Scholar]
- 42.Chen Y, Periasamy A. Intensity range based quantitative FRET data analysis to localize protein molecules in live cell nuclei. J Fluoresc. 2006;16:95–104. doi: 10.1007/s10895-005-0024-1. [DOI] [PubMed] [Google Scholar]
- 43.Elangovan M, Wallrabe H, Chen Y, Day RN, Barroso M, Periasamy A. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29:58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- 44.Wallrabe H, Elangovan M, Burchard A, Periasamy A, Barroso M. Confocal FRET microscopy to measure clustering of ligand-receptor complexes in endocytic membranes. Biophysical journal. 2003;85:559–571. doi: 10.1016/S0006-3495(03)74500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallrabe H, Stanley M, Periasamy A, Barroso M. One- and two-photon fluorescence resonance energy transfer microscopy to establish a clustered distribution of receptor-ligand complexes in endocytic membranes. Journal of biomedical optics. 2003;8:339–346. doi: 10.1117/1.1584444. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Mauldin JP, Day RN, Periasamy A. Characterization of spectral FRET imaging microscopy for monitoring nuclear protein interactions. Journal of microscopy. 2007;228:139–152. doi: 10.1111/j.1365-2818.2007.01838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh DR, Raicu V. Comparison between whole distribution- and average-based approaches to the determination of fluorescence resonance energy transfer efficiency in ensembles of proteins in living cells. Biophysical journal. 2010;98:2127–2135. doi: 10.1016/j.bpj.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoppe A, Christensen K, Swanson Ja. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophysical journal. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallrabe H, Periasamy A, Kim C, Talati R, Barroso M. Confocal FRET and FLIM Microscopy to Characterize the Distribution of Membrane Receptors. Proceedings of SPIE. 2006;vol. 6089:2424. [Google Scholar]
- 50.Hachet-haas M, Converset N, Marchal O, Matthes H, Gioria S, Galzi J, Lecat S. FRET and Colocalization Analyzer — A Method to Validate Measurements of Sensitized Emission FRET Acquired by Confocal Microscopy and Available as an ImageJ Plug-in. Microscopy Research and Technique. 2006;956:941–956. doi: 10.1002/jemt.20376. [DOI] [PubMed] [Google Scholar]
- 51.Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microscopy research and technique. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
- 52.Roszik J, Lisboa D, Szöllosi J, Vereb G. Evaluation of intensity-based ratiometric FRET in image cytometry--approaches and a software solution. Cytometry. Part A. 2009;75:761–767. doi: 10.1002/cyto.a.20747. [DOI] [PubMed] [Google Scholar]
- 53.Talati R, Barroso M. Methods And Apparatuses For Analyzing Digital Images To Automatically Select Regions Of Interest Thereof. U.S. Patent Application US 8,126,2, 11/769,036. 2007
- 54.Zhang XD. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. Journal of biomolecular screening. 2011;16:775–785. doi: 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
- 55.Coma I, Herranz J, Martin J. Statistics and decision making in high-throughput screening. Methods in Molecular Biology. 2009;565:69–106. doi: 10.1007/978-1-60327-258-2_4. [DOI] [PubMed] [Google Scholar]
- 56.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE. Statistical methods for analysis of high- throughput RNA interference screens. Nature Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barroso M, Sztul ES. Basolateral to apical transcytosis in polarized cells is indirect and involves BFA and trimeric G protein sensitive passage through the apical endosome. The Journal of cell biology. 1994;124:83–100. doi: 10.1083/jcb.124.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang DC, Jiang XP, Elliott RL, Head JF. Inhibition of growth of human breast carcinoma cells by an antisense oligonucleotide targeted to the transferrin receptor gene. Anticancer research. 2001;21:1777–1787. [PubMed] [Google Scholar]
- 59.Abe K, Zhao L, Periasamy A, Intes X, Barroso M. Non-Invasive In Vivo Imaging of Breast Cancer Cell Internalization of Transferrin by Near Infrared FRET. PlosOne. 2013 doi: 10.1371/journal.pone.0080269. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gubler H. Assay Data Quality assessment. Methods in Molecular Biology. 2009;552:79–95. doi: 10.1007/978-1-60327-317-6_6. [DOI] [PubMed] [Google Scholar]
- 61.Zhang XD, Lacson R, Yang R, Marine SD, McCampbell A, Toolan DM, Hare TR, Kajdas J, Berger JP, Holder DJ, Heyse JF, Ferrer M. The use of SSMD-based false discovery and false nondiscovery rates in genome-scale RNAi screens. Journal of biomolecular screening. 2010;15:1123–1131. doi: 10.1177/1087057110381919. [DOI] [PubMed] [Google Scholar]
- 62.Alibhai D, Kelly DJ, Warren S, Kumar S, Margineau A, Serwa Ra, Thinon E, Alexandrov Y, Murray EJ, Stuhmeier F, Tate EW, Neil Maa, Dunsby C, French PMW. Automated fluorescence lifetime imaging plate reader and its application to Förster resonant energy transfer readout of Gag protein aggregation. Journal of biophotonics. 2012;11:1–11. doi: 10.1002/jbio.201200185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renz M, Daniels BR, Vámosi G, Arias IM, Lippincott-Schwartz J. Plasticity of the asialoglycoprotein receptor deciphered by ensemble FRET imaging and single-molecule counting PALM imaging. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2989–E2997. doi: 10.1073/pnas.1211753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kenworthy a K, Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. The Journal of cell biology. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kenworthy a K, Petranova N, Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Molecular biology of the cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pentcheva T, Spiliotis ET, Edidin M. Cutting edge: Tapasin is retained in the endoplasmic reticulum by dynamic clustering and exclusion from endoplasmic reticulum exit sites. Journal of immunology. 2002;168:1538–1541. doi: 10.4049/jimmunol.168.4.1538. [DOI] [PubMed] [Google Scholar]