Reducing lignin content in stems and roots of alfalfa results in an increased nodule number phenotype.
Abstract
Reduction of lignin levels in the forage legume alfalfa (Medicago sativa) by down-regulation of the monolignol biosynthetic enzyme hydroxycinnamoyl coenzyme A:shikimate hydroxycinnamoyl transferase (HCT) results in strongly increased digestibility and processing ability of lignocellulose. However, these modifications are often also associated with dwarfing and other changes in plant growth. Given the importance of nitrogen fixation for legume growth, we evaluated the impact of constitutively targeted lignin modification on the belowground organs (roots and nodules) of alfalfa plants. HCT down-regulated alfalfa plants exhibit a striking reduction in root growth accompanied by an unexpected increase in nodule numbers when grown in the greenhouse or in the field. This phenotype is associated with increased levels of gibberellins and certain flavonoid compounds in roots. Although HCT down-regulation reduced biomass yields in both the greenhouse and field experiments, the impact on the allocation of nitrogen to shoots or roots was minimal. It is unlikely, therefore, that the altered growth phenotype of reduced-lignin alfalfa is a direct result of changes in nodulation or nitrogen fixation efficiency. Furthermore, HCT down-regulation has no measurable effect on carbon allocation to roots in either greenhouse or 3-year field trials.
The plant cell wall represents a major determinant of plant structure and is of fundamental importance in plant growth and development, resistance to pathogen attack, quality of plant-based foods and fibers, and processing of biomass for fuels. The plant cell wall is composed of cellulose, hemicellulose, pectin, and lignin. High levels of lignification in secondary cell walls limit the accessibility of degrading enzymes to cellulose; therefore, lignin down-regulation via genetic modification has been used to improve forage quality (Guo et al., 2001) and enhance saccharification efficiency for bioethanol production (Chen and Dixon, 2007; Fu et al., 2011). However, depending on which gene of the lignin pathway is targeted, improvements in bioprocessing efficiency may be compromised due to altered plant growth and development. For example, some transgenic reduced-lignin tobacco (Nicotiana tabacum) lines show biomass reduction, while in other cases, there is no visible phenotype (Chabannes et al., 2001). Transgenic alfalfa (Medicago sativa) lines down-regulated in hydroxycinnamoyl coenzyme A:shikimate hydroxycinnamoyl transferase (HCT) exhibit the greatest improvements in saccharification efficiency, as well as the greatest reduction in yield, among a series of lines independently down-regulated at different steps in the monolignol pathways (Chen and Dixon, 2007; Shadle et al., 2007). Growth inhibition of HCT down-regulated alfalfa plants is associated with hormonal imbalances, altered GA perception, and a massive induction of defense gene transcripts (Gallego-Giraldo et al., 2011b). However, blocking the accumulation of the defense signal salicylic acid (SA) in these plants results in a substantial recovery of growth (Gallego-Giraldo et al., 2011a).
Lignin-deficient mutants have been widely characterized, although most broad phenotypic studies have focused on shoot morphology. In fact, not much is known about the expression and regulation of lignin biosynthetic genes in the roots of wild-type and lignin-modified plants. One study, however, looked at the compact root architecture1 (cra1) mutant of the model legume Medicago truncatula, which is characterized by short thick roots, likely resulting from the deregulation of root cell elongation. This mutant has less lignin but increased root flavonoid content (Laffont et al., 2010). Considering the significance of belowground carbon allocation and soil residence time of roots to the long-term carbon sequestration potential of plants, the importance of belowground carbohydrate reserves for the regrowth of perennial grasses, grazed forage legumes, and trees, and the importance of root architecture and chemistry to drought resistance and interactions with soil microorganisms, understanding the effects of modified aboveground/belowground biomass on belowground responses represents a gap in our current knowledge.
Legumes such as alfalfa form symbiotic associations with nitrogen-fixing soil bacteria called rhizobia. This symbiosis results in the formation of root nodules, where reduced nitrogen from rhizobia is exchanged with reduced carbon from the host photosynthate. Root nodules enable legumes to grow in nitrogen-poor soils; therefore, they play a critical role in sustainable agricultural production worldwide (Peoples et al., 2009; Ferguson et al., 2010). The legume-rhizobia association begins with signal exchange between the symbiotic partners. This is initiated by flavonoid and isoflavonoid compounds exuded through the plant roots when compatible rhizobia are sensed in the rhizosphere (Peters et al., 1986; Peters and Long, 1988; Long, 2001). The rhizobial symbionts recognize specific flavonoid signals and respond by producing lipochitooligosaccharide signals (nodulation [Nod] factors; Long, 2001). Recognition of specific Nod factors by the host plant initiates a symbiotic signal transduction pathway in the roots, leading to infection, nodule development, and symbiotic nitrogen fixation. This complex interaction has been reviewed previously in detail (Oldroyd and Downie, 2008; Ferguson et al., 2010; Oldroyd et al., 2011).
Because lignin modification is known to impact flavonoid biosynthesis and hormone signaling (Chen et al., 2003; Gallego-Giraldo et al., 2011a, 2011b), it is important to know whether the reduced growth phenotype of low-lignin alfalfa plants may result, if only in part, from altered establishment and/or functioning of the rhizobial symbiosis. Several legume mutants affected in different steps of nodule initiation, development, and autoregulation exhibit root growth defects (Wopereis et al., 2000; Veereshlingam et al., 2004; Bright et al., 2005; Kuppusamy et al., 2009; Popp and Ott, 2011), although nodulation was not altered in the short roots of the cra1 lignin mutant (Laffont et al., 2010). Some legume mutants can also exhibit hypernodulation phenotypes and normal root growth (Penmetsa and Cook, 1997; Yokota et al., 2009; Schnabel et al., 2011). These apparently contradictory results mean that it is not clear how root growth and nodulation may be impacted by the modification of lignin biosynthesis.
We here describe how lignin modification impacts root growth, nodulation, and carbon/nitrogen allocation in alfalfa. Enhanced nodulation of HCT antisense alfalfa is associated with increased constitutive accumulation of flavonoids. This altered root phenotype, however, does not significantly affect nitrogen content or carbon sequestration in greenhouse- or field-grown plants.
RESULTS
Root Characteristics of Reduced-Lignin Alfalfa
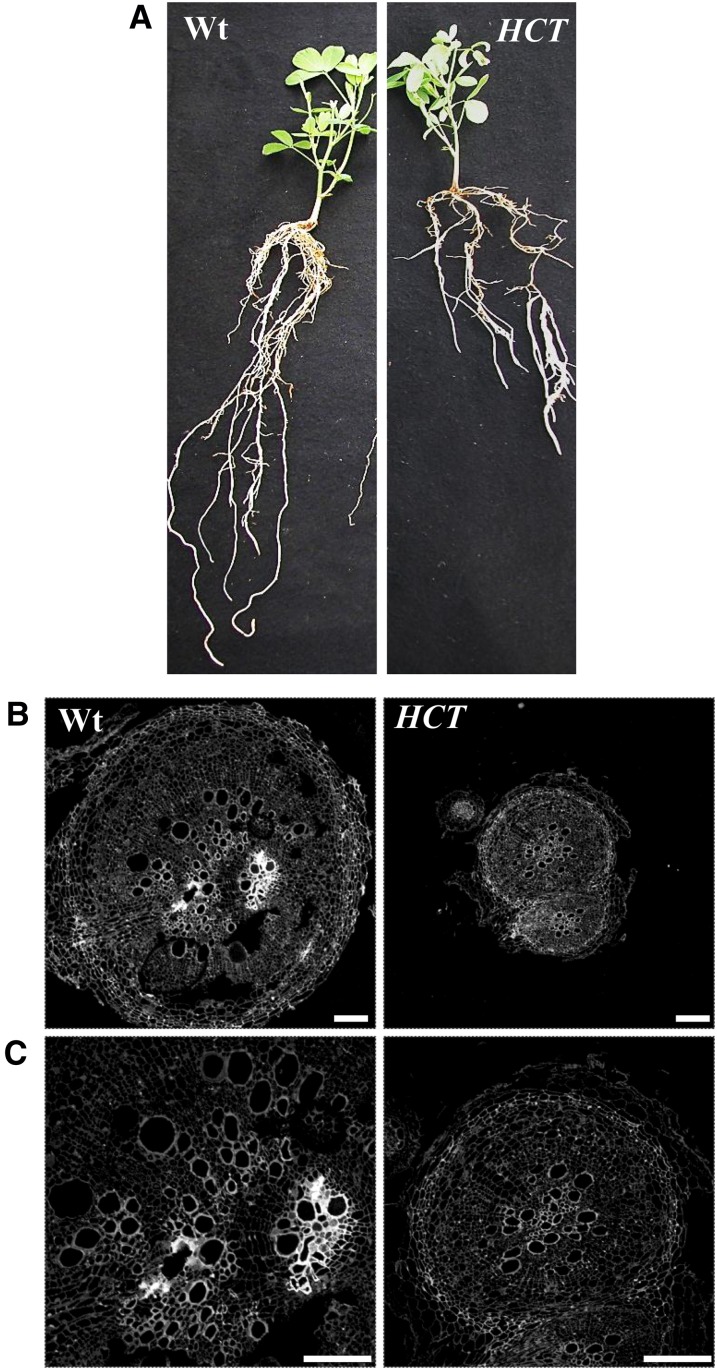
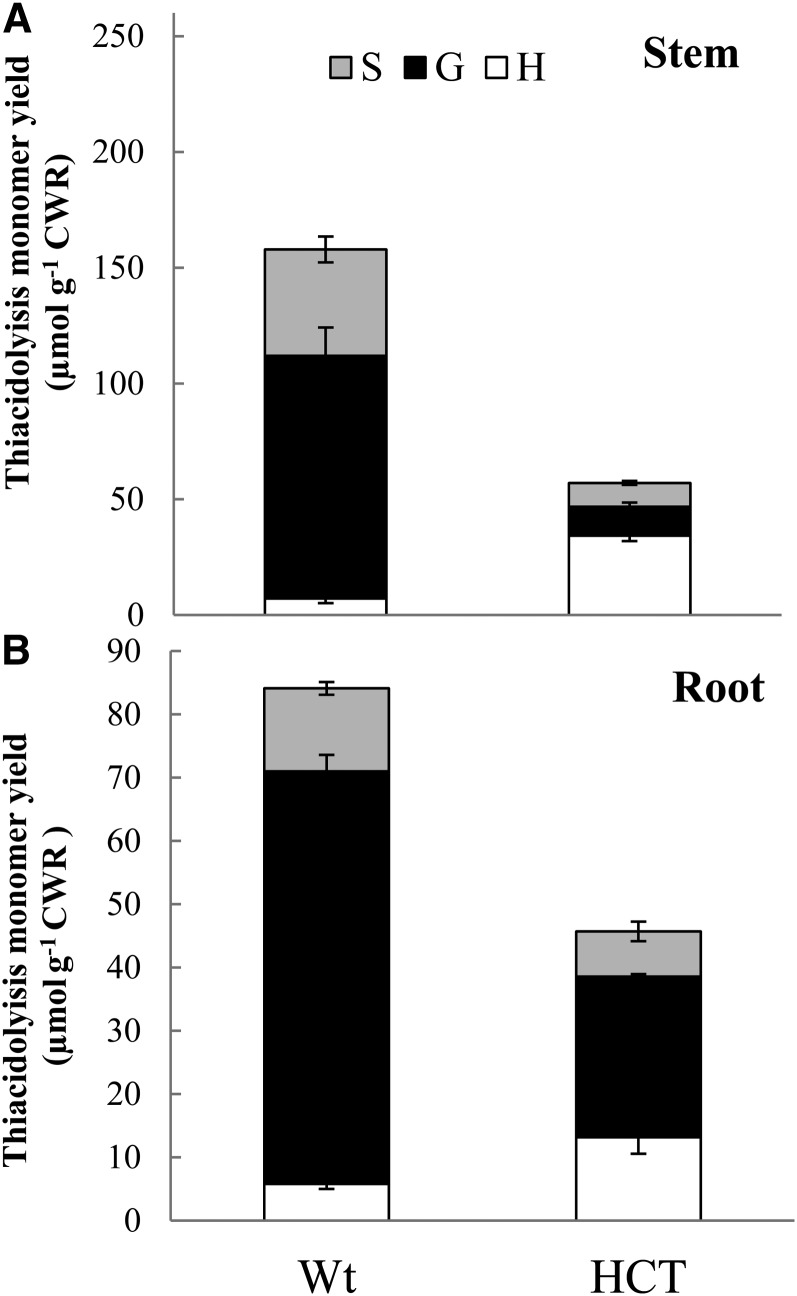
Previous studies have shown that HCT down-regulated alfalfa plants exhibit a dwarf phenotype (Shadle et al., 2007; Gallego-Giraldo et al., 2011a, 2011b); however, these studies did not evaluate the effects of lignin down-regulation on root growth and development. Therefore, we performed a phenotypic characterization of roots from HCT down-regulated alfalfa lines on plants with a confirmed reduction of HCT transcript levels in roots assessed by quantitative real-time (qRT)-PCR (Supplemental Fig. S1). HCT antisense alfalfa plants showed reduced root length and root diameter compared with wild-type roots, although the number of lateral roots was not significantly different (Table I; Fig. 1A). Root cell anatomy in cross sections of HCT antisense lines was similar to that of the wild type, but lignin autofluorescence was reduced (Fig. 1, B and C). This apparent reduction in lignin levels was confirmed by thioacidolysis, which revealed decreased levels of both guaiacyl and syringyl lignin monomer units, with an increase in the p-hydroxyphenyl monomer units (Fig. 2B) in the roots, characteristic of the lignin composition previously reported in stems of HCT down-regulated plants (Shadle et al., 2007) and also found here (Fig. 2A). However, the extent of reduction of total lignin content in the roots as a result of HCT down-regulation was significantly less than in the stems (by 65% in stem, 40% in root; Fig. 2).
Table I. Root phenotypes of HCT antisense and wild-type alfalfa plants.
Root length was measured in 5-week-old plants. Numbers of nodules per root were determined after 3 weeks of inoculation with S. meliloti. Five independent roots per plant were randomly chosen from the total root system to count the number of nodules formed. Results are means of nine plants by line ± se. Values with different letters are significantly different (P < 0.05).
| Line | Root Length | Root Diameter | No. of Lateral Roots per Root | No. of Nodules per Root |
|---|---|---|---|---|
| mm | ||||
| Wild type | 148 ± 25 a | 0.2 ± 0.05 a | 15 ± 2 a | 4 ± 1.2 a |
| HCT | 95 ± 25 b | 0.1 ± 0.08 b | 13 ± 2 a | 14 ± 2.4 b |
Figure 1.
Root growth and microscopic analysis of lignin deposition of wild-type (Wt) and HCT down-regulated alfalfa. A, Overall root phenotypes of 5-week-old plants. B, Cross sections (10-μm thickness) made in the same root material presented in A, at the upper part of the root close to the stem intersection. Total lignin was determined by UV autofluorescence. All settings for these micrographs were identical. C, Closeups of roots showing a decrease in UV autofluorescence in the HCT down-regulated line. Bars in B and C = 200 μm. [See online article for color version of this figure.]
Figure 2.
Lignin content and composition in roots of wild-type (Wt) and HCT-down-regulated alfalfa, determined by thioacidolysis. Analysis was performed in 2-month-old roots. Results are means ± sd of three biological replicates. Each biological replicate consisted of at least three complete root systems. CWR, Cell wall residue; G, guaiacyl unit; H, p-hydroxyphenyl unit; S, syringyl unit.
Impact of Lignin Modification on Nodulation Efficiency
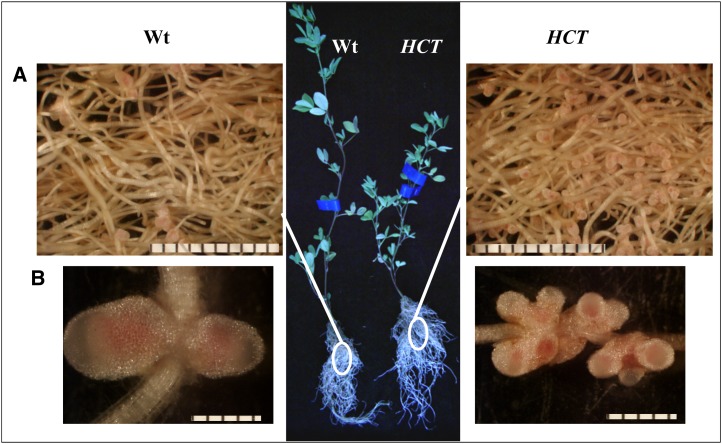
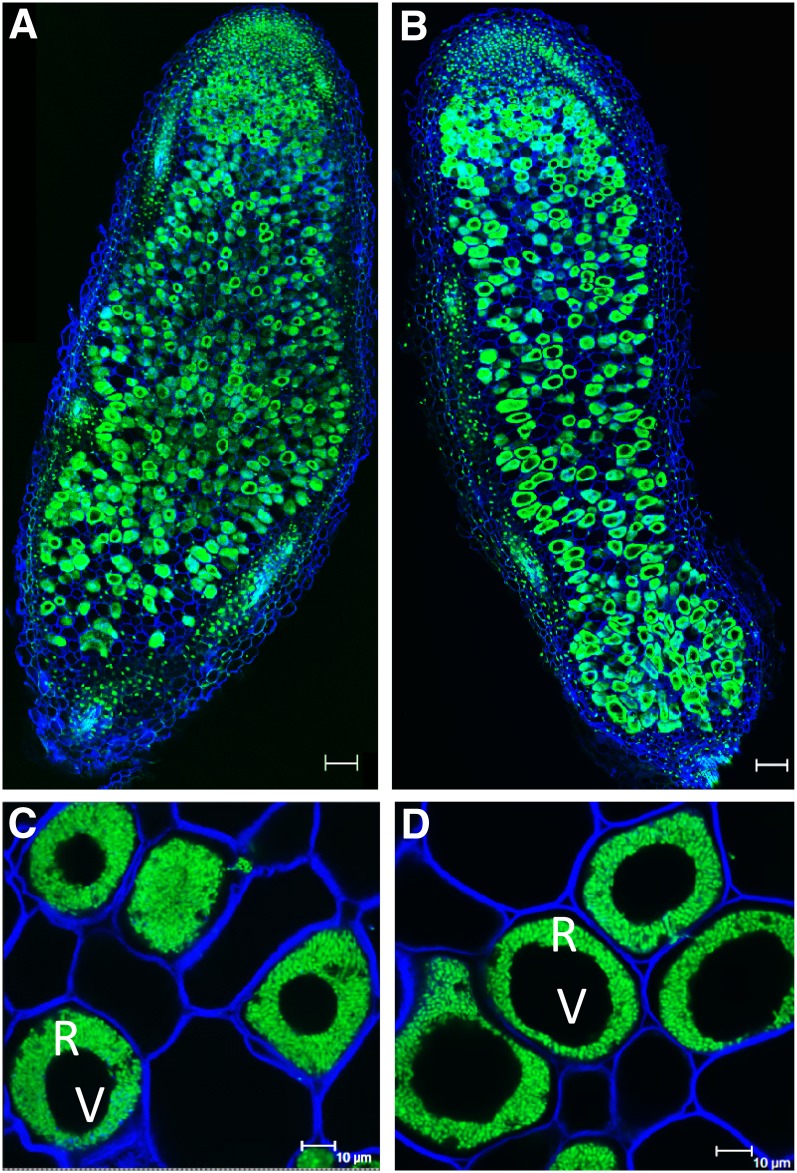
Nodulation assays were performed by inoculating roots of 5-week-old plants with Sinorhizobium meliloti Sm1021 under greenhouse conditions. Although mature alfalfa plants have a long taproot, the root system is highly branched, with fibrous roots, bearing most of the nodules, proliferating in the upper part of the soil profile. At 18 d post inoculation (dpi), roots from HCT down-regulated lines showed an increased nodule number phenotype (Fig. 3). The number of nodules per root was 3.5 times higher in the HCT antisense plants than in the wild type, although the size of the nodules on the reduced-lignin lines was smaller (Table I; Fig. 3B). Both HCT antisense and wild-type nodules displayed normal phenotypic features, being elongated and accumulating the pink pigment leghemoglobin (Fig. 3). Accumulation of leghemoglobin is suggestive of functional nitrogen fixation (Ott et al., 2005). The reduction of atmospheric nitrogen is catalyzed by the oxygen-sensitive bacterial nitrogenase complex. Leghemoglobin acts as a buffer for free oxygen, providing just enough oxygen for rhizobial respiration while maintaining low enough oxygen levels for efficient nitrogenase activity (Appleby, 1984; Ott et al., 2005). The presence of rhizobia inside nodules was analyzed using confocal microscopy (Fig. 4). Low-magnification imaging (Fig. 4, A and B) showed fully infected nodules in both wild-type and HCT antisense lines, with uninfected cells alternating with infected cells, which displayed a typical doughnut shape, with a large central vacuole and the cytoplasmic ring fully packed with rhizobia. At higher magnification, individual bacteroids (the endoreduplicated, elongated, nitrogen-fixing form of rhizobia) could be observed (Fig. 4, C and D). In both wild-type and HCT antisense lines, the bacteroids exhibited a normal, elongated appearance. Furthermore, nodulation led to similar increases in transcript levels of two N2 fixation marker genes, plant leghemoglobin1 and bacterial nitrogenase (Supplemental Fig. S2), suggesting that nodules on HCT antisense plants are functional in their capacity for nitrogen fixation. This was confirmed by acetylene reduction assay of three independent lines; nodules from HCT down-regulated lines reduced acetylene (a proxy for nitrogenase activity) at the same rate as nodules from wild-type lines (Supplemental Fig. S3).
Figure 3.
Overall plant growth and nodulation of roots from wild-type (Wt) and HCT down-regulated alfalfa plants after 18 d of S. meliloti inoculation. A, Closeup of roots showing nodules. Bars = 10 mm. B, Closeup of groups of nodules on control and HCT down-regulated plants. Bars = 4 mm.
Figure 4.
S. meliloti-infected root nodules of wild-type (A and C) and HCT1 (B and D) plants. Nodules were harvested at 18 dpi, fixed, sliced into 50-µm-thick sections, and stained with the Syto13 DNA dye and with calcofluor to reveal cell walls. Visualization was by confocal microscopy. Green fluorescence corresponds to rhizobia, and blue fluorescence corresponds to plant cell walls. Bars in A and B = 100 µm; C and D = 10 μm. R, Cytoplasmic ring packed with rhizobia; V, central vacuole.
Phytohormone Levels in Roots of Reduced-Lignin Alfalfa
The dwarf/branched phenotype of HCT antisense alfalfa plants is associated with changes in the levels of several phytohormones in the aboveground sections of the plant (Gallego-Giraldo et al., 2011b), and phytohormones are involved in the control of nodulation (Oldroyd and Downie, 2008). To determine if hormone levels in roots are similarly affected and may correlate with the reduced root growth and enhanced nodulation phenotypes described above, the levels of a range of phytohormones in 5-week-old noninoculated roots were determined. Levels of two GAs, GA1 and GA4, were higher in the roots of HCT antisense lines than in the wild type (Table II), while cytokinin and auxin levels were not significantly different between roots of HCT antisense and wild-type lines. Auxin transport was also measured in roots, both basipetally and acropetally, and was essentially the same in HCT antisense and wild-type lines (Supplemental Fig. S4).
Table II. Phytohormone levels (ng g−1 fresh weight) in roots of HCT antisense and wild-type alfalfa.
Results are means of three biological replicates ± se. Values with different letters are significantly different between wild-type and HCT down-regulated lines (P < 0.05).
| Hormone | Wild Type | HCT |
|---|---|---|
| Abscisic acid | 1.7 ± 0.1 a | 3.3 ± 0.2 b |
| Jasmonic acid | 24.2 ± 3.7 a | 40.6 ± 2.1 b |
| Jasmonic acid conjugated to Ile | 1.1 ± 0.1 a | 1.5 ± 0.03 b |
| SA | 41.0 ± 7.1 a | 18.0 ± 0.5 b |
| GA4 | 0.03 ± 0.002 a | 1.0 ± 0.5 b |
| GA1 | 0.2 ± 0.02 a | 0.5 ± 0.03 b |
| Indoleacetic acid | 13.4 ± 0.5 a | 11.5 ± 1.1 a,b |
| Trans-zeatin | 0.07 ± 0.01 a | 0.09 ± 0.01 a |
| Isopentenyl adenine | 0.04 ± 0.001 a | 0.04 ± 0.04 a |
| Dihydrozeatin | 0.02 ± 0.03 a | 0.03 ± 0.01 a |
Stems of HCT antisense alfalfa exhibited elevated levels of SA, which in turn accounts for the high expression of pathogenesis-related (PR) proteins and, at least in part, for the reduced growth of the aerial portions of the plant (Gallego-Giraldo et al., 2011a, 2011b). In this study, however, SA levels were lower in the roots of HCT antisense lines compared with the wild type; in contrast, levels of two other plant stress hormones, abscisic and jasmonic acids, increased in roots of HCT antisense lines compared with the wild type (Table II).
Flavonoid Levels in Roots of Low-Lignin Alfalfa
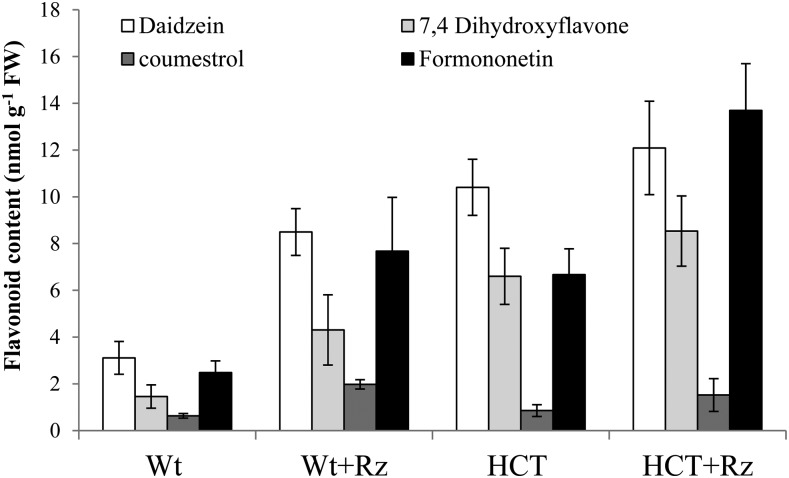
We have recently shown an increase in flavonoid metabolites as a result of HCT down-regulation in aboveground organs of alfalfa (Gallego-Giraldo et al., 2011b), representing either a spillover from the lignin pathway or a secondary stress response. It is well known that flavonoids play a crucial role in inducing nodulation in Medicago species plants (Zhang et al., 2009); therefore, the increased nodule numbers of HCT antisense plants might reflect increased flavonoid levels. To remove the possible confounding effect due to the presence of multiple glycosides of phenolic compounds in plants, samples were enzymatically hydrolyzed prior to analysis of the resulting aglycones. HPLC analysis revealed a significant increase in the levels of two isoflavones, namely daidzein and formononetin, and one flavone, 7,4′-dihydroxyflavone, in HCT antisense roots compared with the wild type in the absence of inoculation (Fig. 5). Levels of the isoflavonoid-derived coumestrol remained largely unchanged. Although the flavones chrysoeriol and luteolin are the most potent S. meliloti nod gene-inducing flavonoids secreted by alfalfa (Hartwig et al., 1990), 7,4′-dihydroxyflavone and formononetin glycosides are also released from alfalfa roots and can activate S. meliloti nod genes (Hartwig et al., 1990; León-Barrios et al., 1993).
Figure 5.
Soluble phenolic aglycones in roots of wild-type (Wt) and HCT down-regulated alfalfa, 2 d before and 18 d after inoculation with S. meliloti (+Rz). Results are means ± sd of three biological replicates. Each biological replicate consisted of at least three complete root systems. FW, Fresh weight.
Transcriptome Changes in the Roots of HCT Down-Regulated Plants
Based on previous microarray analysis, the dwarf phenotype of HCT down-regulated alfalfa stems is associated with a massive up-regulation of PR genes (Gallego-Giraldo et al., 2011b). To establish whether the changes in root growth found in HCT down-regulated lines also correlate with plant defense gene overexpression, we performed a genome-wide transcript profiling in roots of HCT down-regulated plants using the Affymetrix Medicago Gene Chip. As a result, 143 differentially expressed genes (compared with the wild type) were identified (ratio values under 0.5 are down-regulated and those above 2.0 are up-regulated), of which only three were down-regulated (Supplemental Table S1; namely, the HCT target gene, one putative auxin transporter, and one unknown protein). Analysis of the functional categories of genes up-regulated in HCT down-regulated roots revealed an enrichment in transcription factors (33 genes, 23%) followed by primary metabolism (25 genes, 18%), flavonoid biosynthesis [13 genes, 9.5%, including several leucoanthocyanidin dioxygenase genes and (iso)flavonoid O-methyltransferases], receptor kinase signaling (10 genes, 7%), and hormone- and transport-related genes (nine genes, 6%; Supplemental Table S2). Consistent with the reduced levels of SA, PR genes were not overexpressed in the roots.
Effects of Nodulation on Lignin Deposition
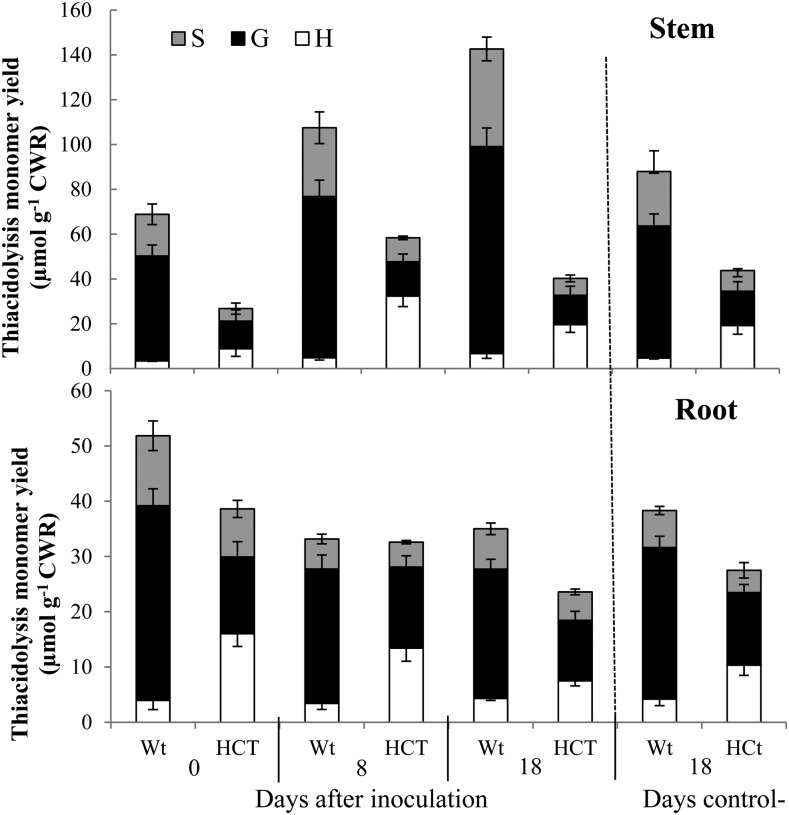
After establishing that lignin modification impacts nodulation efficiency, we next examined whether bacterial symbiosis impacts lignin deposition in alfalfa roots or stems. Lignin content and composition were determined by thioacidolysis in plants prior to and until 18 dpi. In stems of wild-type plants, there was a clear increase in lignin content as a percentage of total cell wall material with time after inoculation, and the values in stems of inoculated plants were greater than in stems of uninoculated plants with regular fertilization at 18 dpi (Fig. 6). In contrast, inoculation did not increase the lignification in stems of HCT antisense plants at 18 dpi. Opposite to the pattern in the stems, the lignin content as a percentage of total cell wall material decreased almost 60% in roots of wild-type plants by 18 dpi compared with the uninoculated plants. This does not appear to be the result of inoculation with S. meliloti, since the levels of lignin were similar in inoculated and uninoculated roots at 18 dpi. A similar pattern was observed in the HCT antisense lines (Fig. 6). Although aboveground biomass was significantly enhanced by nodulation in both wild-type and HCT antisense lines (Table III), root biomass exhibited only a small increase in both wild-type and HCT antisense lines following nodulation, and the root biomass at 18 dpi was not significantly different from that of uninoculated control roots (Table III).
Figure 6.
Effects of nodulation on lignin content and composition in stem and in root of wild-type (Wt) and HCT down-regulated alfalfa. S. meliloti inoculation was performed in 6-week-old plants. The negative control consisted of plants growing in parallel without bacterial inoculation. Results are means ± sd of three biological replicates. Each biological replicate consisted of at least three complete root systems. CWR, Cell wall residue; G, guaiacyl unit; H, hydroxyphenyl unit; S, syringyl unit.
Table III. Effects of nodulation on biomass, nitrogen content, and carbon content of wild-type and HCT down-regulated alfalfa stems and roots grown in the greenhouse.
Results are means ± sd of five biological replicates in the case of biomass and three biological replicates in the case of nitrogen and carbon. Values with different letters are significantly different (P < 0.05).
| Tissue | dpi | Line | Biomass | Nitrogen Content | Carbon Content |
|---|---|---|---|---|---|
| g dry weight per plant | mg per g dry weight | ||||
| Stem | 0 | Wild type | 0.38 ± 0.04 a | 15.1 ± 1.3 a | 164.4 ± 7.3 a |
| HCT | 0.25 ± 0.06 b | 17.4 ± 1.2 a | 168.1 ± 8.9 a | ||
| 8 | Wild type | 0.41 ± 0.07 a | 14.6 ± 2.1 a | 284.1 ± 22.0 b | |
| HCT | 0.22 ± 0.03 b | 17.5 ± 0.6 a | 259.1 ± 5.3 b | ||
| 18 | Wild type | 1.20 ± 0.09 c | 25.8 ± 0.6 b | 319.0 ± 17.4 c | |
| HCT | 0.80 ± 0.06 d | 24.6 ± 0.8 b | 353.1 ± 22.9 c | ||
| Control | Wild type | 0.53 ± 0.1 a | 15.3 ± 1.3 a | 189.7 ± 28.9 a | |
| HCT | 0.32 ± 0.1 a,b | 15.7 ± 1.9 a | 175.3 ± 21.3 a | ||
| Root | 0 | Wild type | 0.18 ± 0.01 a | 32.0 ± 4.1 a | 264.1 ± 50.2 a |
| HCT | 0.12 ± 0.02 b | 29.8 ± 4.9 a | 265.1 ± 51.2 a | ||
| 8 | Wild type | 0.34 ± 0.04 c | 35.1 ± 1.7 a | 382.1 ± 31.2 b | |
| HCT | 0.18 ± 0.01 a | 29.1 ± 2.1 a | 335.2 ± 29.8 a,b | ||
| 18 | Wild type | 0.25 ± 0.01 d | 22.0 ± 0.7 b | 523.4 ± 66 c | |
| HCT | 0.17 ± 0.01 a | 21.8 ± 0.7 b | 580.1 ± 86.6 c | ||
| Control | Wild type | 0.28 ± 0.05 c,d | 19.6 ± 4.8 b | 224.4 ± 80.4 a | |
| HCT | 0.20 ± 0.02 a,d | 20.6 ± 2.4 b | 233.7 ± 30.1 a | ||
Carbon and Nitrogen Allocation of Greenhouse-Grown Plants
Nitrogen and carbon contents were measured in both stems and roots during nodulation to determine whether the differences in lignin deposition and nodulation described above impacted the overall carbon/nitrogen allocation. Total nitrogen content increased after 18 dpi in both HCT antisense and wild-type stems (Table III). In contrast, there was an opposite trend in roots, where the N2 content decreased about 1.4-fold in both genotypes between 8 and 18 dpi (Table III). Overall, this suggests that reallocation of nitrogen from root to stem as a result of nodulation is not negatively impacted by the reduction of lignin levels via HCT down-regulation.
Carbon content increased following nodulation in both stems and roots of HCT antisense and wild-type lines (Table III). Unlike nitrogen, total carbon content as a percentage of dry weight was similar in both the wild-type and HCT down-regulated genotypes. The fact that total carbon levels in roots appear to be unaffected by reduction in lignin content suggests that lignin reduction may have little impact on overall carbon sequestration and that carbon is channeled into other compounds following the down-regulation of lignin.
Nodulation of Reduced-Lignin Plants Grown in the Field
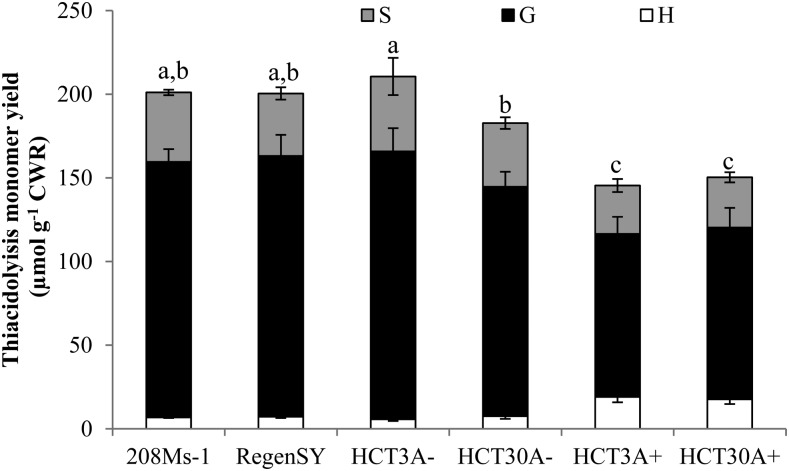
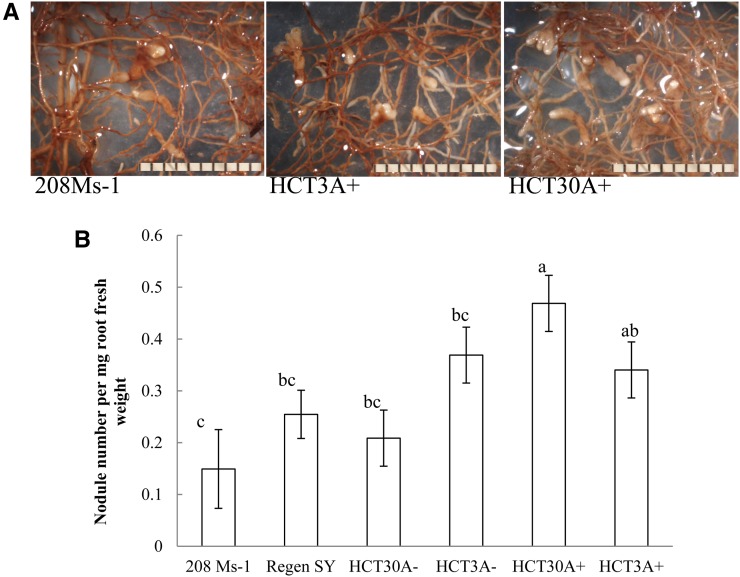
The HCT antisense transgene (events 3a and 30a) was introgressed into a commercial genetic background (208Ms-1) to generate F1 progeny and determine whether the hypernodulation phenotype of HCT down-regulated alfalfa was consistent when plants were grown under field conditions. This was necessary because the transformation-competent line cv Regen SY4D, which is the genetic background for the HCT antisense construct, has poor agronomic performance. The segregation of the transgene in the full-sib progeny with or without the RNA interference construct (labeled as HCT3A+ and HCT3A−, respectively) was used to evaluate row plots side by side (down-regulated plants versus non-down-regulated plants). After 3 years of field growth at one location, roots were harvested from the non-down-regulated events HCT3A− and HCT30A− and the corresponding down-regulated HCT30A+ and HCT3A+ lines, the cv Regen SY4D control (genetic background), and the female parent 208Ms-1. The reduction in the levels of HCT transcripts from HCT down-regulated events grown in the field was confirmed (Supplemental Fig. S5A), and this level of HCT transcript down-regulation (between 40% and 50%) was found to impact root lignin content in field-grown plants (Fig. 7). The reduction in lignin content in the roots of HCT30A+ and HCT3A+ plants grown in the field was associated with an increase in p-hydroxyphenyl monomers characteristic of HCT down-regulation (Fig. 7). The numbers of nodules on the roots of field-grown plants that had been inoculated with rhizobia at the seedling stage were also evaluated. The number of nodules expressed on a root dry weight basis was two times greater in the HCT30A+ lines than in the 208Ms-1 parental line and the corresponding HCT30A− full-sib progeny (Fig. 8). The HCT3A+ line also produced more nodules than the maternal parent 208Ms-1 but did not differ when compared with the non-HCT down-regulated full-sibs (HCT3A−; Fig. 8B).
Figure 7.
Lignin content and composition in roots of alfalfa plants grown in the field for 3 years. Lignin was measured in the parental line 208Ms-1, non-down-regulated HCT−, and down-regulated HCT+ events. The cv Regen SY4D was used as a negative control. Results are means ± sd of four biological replicates in the case of parental and negative controls and eight biological replicates for the HCT events. Values with different letters are significantly different (P < 0.05). CWR, Cell wall residue; G, guaiacyl unit; H, p-hydroxyphenyl unit; S, syringyl unit.
Figure 8.
Root nodulation of alfalfa plants grown in the field for 3 years. A, Closeups of roots showing nodules in the parental 208Ms-1 line and two independent HCT down-regulated lines (HCT3A+ and HCT30A+). Bars = 10 mm. B, Nodule number in the parental line 208Ms-1, non-down-regulated HCT−, and down-regulated HCT+ events. The cv Regen SY4D was used as a negative control. Results are means ± sd of four biological replicates in the case of parental and negative controls and eight biological replicates for the HCT events. Values with different letters are significantly different (P < 0.05). [See online article for color version of this figure.]
Biomass and Nitrogen and Carbon Allocation in Field-Grown Plants
Biomass yield of both the aboveground biomass and roots of HCT down-regulated events grown in the field was reduced (Table IV). The nitrogen content in the roots was greater in the HCT30A+ and HCT3A+ lines than in their corresponding null segregant controls (Table IV). In contrast, carbon content remained essentially unchanged for all events, other than a slight increase in the stems of HCT3A+ plants (Table IV). Overall, the phenotypic results obtained from field-grown plants were consistent with those previously identified in the greenhouse.
Table IV. Biomass, nitrogen content, and carbon content in stems and roots of alfalfa plants grown in the field for 3 years.
Values are from the parental line 208Ms-1, non-down-regulated null-segregant HCT−, and down-regulated HCT+ events. The cv Regen SY4D was used as a negative control. Nitrogen and carbon content values in stems are means of three values from different harvests. Results are means ± sd of four biological replicates in the case of parental and null segregant controls and eight biological replicates for the HCT events. Values with different letters are significantly different (P < 0.05).
| Tissue | Line | Biomass | Nitrogen Content | Carbon Content |
|---|---|---|---|---|
| g dry weight per plant over 3 years | mg per g dry weight | |||
| Stem | 208Ms-1 | 424 ± 43 a | 12.6 ± 0.2 a | 135.3 ± 3.6 a |
| Regen SY4D | 317 ± 36 b,c | 13.2 ± 0.2 b | 133.3 ± 4.2 a | |
| HCT30A− | 340 ± 30 b,c | 13.2 ± 0.2 b | 134.7 ± 4.2 a | |
| HCT3A− | 298 ± 27 b,c | 13.1 ± 0.2 b | 134.4 ± 4.4 a | |
| HCT30A+ | 254 ± 37 c | 13.7 ± 0.2 b,c | 136 ± 4.3 a | |
| HCT3A+ | 280 ± 29 b,c | 13.7 ± 0.3 b,c | 137 ± 5.4 a,b | |
| Root | 208Ms-1 | 122 ± 9 a | 5.16 ± 0.4 a | 166.4 ± 2.3 a |
| Regen SY4D | 119 ± 14 a,b | 5.65 ± 0.7 a | 158.5 ± 7 a | |
| HCT30A− | 114 ± 13 a,b | 6.03 ± 0.8 a | 165.2 ± 6 a | |
| HCT3A− | 86 ± 9 b,c | 5.70 ± 0.4 a | 165.4 ± 5.6 a | |
| HCT30A+ | 76 ± 6 c | 8.11 ± 1 b | 163.8 ± 8.5 a | |
| HCT3A+ | 97 ± 13 a,b,c | 7.29 ± 0.6 b | 162.7 ± 10 a | |
DISCUSSION
Nodulation in Roots of Reduced-Lignin Alfalfa Plants
The roots of HCT down-regulated reduced-lignin plants had increased numbers of nodules per root biomass in both greenhouse and field experiments. The increased nodule number in the HCT down-regulated lines was not associated with an overall improvement in nitrogen fixation. In the M. truncatula sickle mutant, a hypernodulation phenotype is associated with auxin transport inhibition (Prayitno et al., 2006), whereas local auxin transport is normal in roots of HCT down-regulated plants, resembling the hypernodulation mutant super numeric nodules with short root length and local normal auxin transport (van Noorden et al., 2006)
The analysis of noninoculated reduced lignin plants suggests that increased flavone production and or GA accumulation may underlie the increased nodule number phenotype of the HCT down-regulated roots, although further investigation of root metabolic and transcriptional changes in response to rhizobial inoculation is needed. The increased flavonoid production correlates with the up-regulation in the HCT down-regulated lines of eight genes related to the flavone and isoflavone biosynthetic pathways, suggesting that flavonoid production in these lines represents an active, secondary response to lignin reduction rather than simply metabolic spillover. The levels of 7,4′-dihydroxyflavone are around four times higher in HCT down-regulated roots than in wild-type roots and even higher compared with nodulated roots of wild-type plants. This molecule attracts bacteria to the root and activates rhizobial nod gene expression, leading to the production and secretion of Nod factors (Zhang et al., 2009). However, it is possible that the increased levels of the two isoflavones, daidzein and formononetin, also play a role in the increased nodule number phenotype. Daidzein stimulates the DNA-binding activity of NodD1 to nod gene promoters of S. meliloti (Peck et al., 2006). Moreover, nodulation in isoflavone-deficient soybean (Glycine max) roots can be restored by using genistein-hypersensitive rhizobial cells, independent of auxin transport restoration, suggesting that induction of nod signal biosynthesis by isoflavones is a critical component of the nodulation signaling mechanism (Subramanian et al., 2006).
A significant body of evidence also points to elevated GA levels, as recorded in the HCT down-regulated roots, as a potential cause of hypernodulation. For example, mutations in pea (Pisum sativum) that reduce GA levels strongly inhibit nodule development, and exogenous application of GA rescues the mutant phenotype (Ferguson et al., 2005). Moreover, GA response genes and GA biosynthetic pathway genes are up-regulated after rhizobial infection in Sesbania rostrata and soybean (Lievens et al., 2005; Hayashi et al., 2012), and constitutive GA signaling mutants can also exhibit reduced nodule formation (Ferguson et al., 2011). Interestingly, GA4 is the main accumulated bioactive GA in HCT down-regulated alfalfa roots, compared with GA1 or GA3 in pea and Lotus japonicus (Ferguson et al., 2005, 2011; Maekawa et al., 2009), although knowledge of the role of this bioactive form in nodulation is limited.
Root versus Stem Growth Regulation in Reduced-Lignin Plants
HCT antisense reduced-lignin plants are characterized by growth reductions in both the stem and root. Significant differences in phytohormone levels and flavonoid accumulation exist between the two tissues; for instance, GAs are overproduced in HCT down-regulated roots (this work) but not in stems (Gallego-Giraldo et al., 2011b). This overproduction of GAs in roots might explain the shortened root phenotype. GA accumulation is positively associated with cell elongation over a certain range (Maekawa et al., 2009), but high GA accumulation (more than 15 times the level of GA4 in HCT down-regulated roots compared with the wild type in this study) can cause a reduction of root cell elongation (Fagoaga et al., 2007; Maekawa et al., 2009). In stems of HCT down-regulated plants, the situation is clearly different: GA levels are slightly reduced compared with the wild type, but the defense phytohormones SA, jasmonic acid, and abscisic acid overaccumulate (Gallego-Giraldo et al., 2011b). Furthermore, previous (Gallego-Giraldo et al., 2011b) and present transcriptome data (Supplemental Table S1) from both tissues support the fact that very different genetic reprogramming is taking place in the two tissue types: in the stem, there is a very strong overexpression of PR genes (Gallego-Giraldo et al., 2011b) not found in the roots; rather, a subset of GA biosynthesis pathway genes is induced in these tissues.
Although there is an increase in flavonoid levels in both tissues, this increase is characterized by the formation of different flavonoid aglycones. In the stem, apigenin glucoside is the main flavone affected, while apigenin is not detected at all in the roots, and a different flavone, 7,4′-dihydroxyflavone, along with the isoflavones daidzein and formononetin, accumulate instead. Although all of these compounds originate from the same biosynthetic pathway, different genes/enzymes may participate in their production in the two tissue types. For example, microarray analysis showed the up-regulation of different members of the chalcone synthase and isoflavone synthase gene families in stems of the HCT down-regulated plants compared with the ones up-regulated in roots (Gallego-Giraldo et al., 2011b). M. truncatula roots treated with exogenous formononetin are affected in root growth (Laffont et al., 2010). Therefore, the reduction in root length in HCT down-regulated plants is, similar to the increased nodule number phenotype, possibly the consequence of the increases in both GA and isoflavone levels.
Carbon/Nitrogen Allocation during Nodulation in Low-Lignin Plants
Nitrogen and carbon levels increased during nodulation in the stems of both wild-type and HCT down-regulated alfalfa grown under greenhouse conditions. In parallel, aerial biomass yield increased in comparison with the uninoculated plants. In the roots, carbon content increased during nodulation, while nitrogen levels remained stable. The observation that the increased number of nodules found in HCT down-regulated roots does not correlate with increased nitrogen fixation is consistent with reports of Medicago species hypernodulating mutants (Jeudy et al., 2010). Interestingly, relative lignin content decreased by 32% in control and by 41% in HCT down-regulated roots following nodulation. This reduction could reflect the diversion of carbon flux from lignin to flavonoids during rhizobia interaction. Remarkably, there were no differences between the control and HCT down-regulated plants in terms of overall nitrogen and carbon content per unit of biomass in the greenhouse experiments.
The findings from this study regarding the aboveground and belowground biomass yields, nodulation, and nitrogen and carbon partitioning were reproduced in the field experiments. HCT down-regulated lines clearly exhibited the enhanced nodule number phenotype when grown under field conditions, possibly associated with increased flavonoid production (measured indirectly as increased expression of chalcone synthase and isoflavone synthase), and yet the plants showed a reduction in overall biomass yield. Interestingly, there were no reductions in overall nitrogen or carbon content per unit of biomass in HCT down-regulated plants compared with controls; in fact, nitrogen content of the HCT+ roots was somewhat elevated. More notable, however, is the fact that lignin reduction did not result in reduced belowground carbon sequestration in field-grown plants, other than that arising from the reduced root biomass. It remains to be determined whether the carbon allocated to the roots of low-lignin plants has a reduced residence time in the soil as a consequence of the reduced recalcitrance of the cell wall material.
MATERIALS AND METHODS
Plant Growth Conditions and Nodulation Assay
Alfalfa (Medicago sativa ‘Regen SY4D’) plants were grown at 22°C under long-day conditions in the greenhouse. All experiments were performed with clonally propagated F1 transgenic plants as described previously (Reddy et al., 2005; Chen et al., 2006; Shadle et al., 2007). The lines analyzed were antisense HCT (30a-HCT1, 3a-HCT2, and 29a-HCT3), an empty vector control line (CK48-Wt1), and two wild-type control lines (Ctl1-Wt2 and Ctrl49-Wt3). The wild-type (Wt1–Wt3) and HCT (HCT1–HCT3) antisense cuttings were transferred to small pots containing a sterile substrate of turface and vermiculite (2:1) and were subjected to nitrogen starvation for 6 d with a low-nutrient solution (Broughton and Dilworth, 1971). Five weeks after transplanting, each plant was inoculated with a 50-mL suspension (optical density at 600 nm = 0.03) of Sinorhizobium meliloti wild-type strain Sm1021, as described previously (Ardourel et al., 1994; Pislariu et al., 2012). No further fertilization was applied to inoculated plants prior to harvesting at 8 and 18 dpi, although plants were watered regularly with deionized water. Control plants were watered with nutrient solution in parallel with inoculated plants. After 18 dpi, nodulated root segments were harvested, fixed in 2.5% (v/v) glutaraldehyde in 0.1 m PIPES, pH 7.2, for 1 h, and extensively washed with 0.1 m PIPES. Nodules were embedded in 5% (w/v) low-melting-point agarose and sliced into 50-µm-thick sections using a Vibratome 1000 Plus. Syto13 staining was performed as described (Haynes et al., 2004). After rinsing with 80 mm PIPES, staining with calcofluor white stain (Sigma-Aldrich) was performed according to the manufacturer’s instructions. Confocal images were acquired using a Leica TCS SP2 AOBS confocal laser scanning microscope.
Visualization of Lignin
For visualization of total lignin autofluorescence, similar portions of alfalfa roots were fixed with 3% (v/v) glutaraldehyde in phosphate-buffered saline buffer (pH 7.4), dehydrated in a graded ethanol series, and embedded in LR White resin (London Resin). The resin was then polymerized at 55°C for 3 d. Serial 0.5-μm sections were cut with a diamond knife on a Leica EM UC7 ultramicrotome (Leica Microsystems) or an RMC MT-X ultramicrotome (Boeckeler Instruments). Semithin sections were immersed in 50% (v/v) glycerol and observed with a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems) illuminated with a 405-nm blue diode laser, and emission was detected at 460 nm. In order to obtain meaningful comparisons, laser intensity, pinhole, and photomultiplier gain settings were kept constant between samples.
Acetylene Reduction Assay
The acetylene reduction assay to assess nitrogen fixation ability was performed on plants at 18 dpi. The entire root system of each plant was placed in a sealed test tube, and the assay was conducted as described previously (Oke and Long, 1999). When the assay was completed, the samples were dried and acetylene reduction was calculated on a root dry weight basis.
Profiling of Soluble Phenolic Compounds
Fresh, clean root tissue was ground in liquid N2, and 0.5 to 1.0 g was extracted two times in 10 volumes of 80% (v/v) methanol at 4°C. Extracts were combined and dried under a stream of N2. Dried residues were resuspended by vortexing in 1 mL of 100% methanol. A total of 500 μL of the resuspended material was subjected to enzymatic hydrolysis as described previously (Gallego-Giraldo et al., 2011b). Of these, 40 μL was injected onto an ODS2 reverse-phase column (5-μm particle size, 4.6 × 250 mm) and eluted in 1% (v/v) phosphoric acid with an increasing gradient of acetonitrile (0–5 min, 5%; 5–10 min, 5%–10%; 10–25 min, 10%–17%; 25–30 min, 17%–23%; 30–65 min, 23%–50%; 65–69 min, 50%–100%) at a flow rate of 1 mL min−1 for 70 min. Compound identities were determined by comparing UV spectra and the retention times with those of authentic standards. Additionally, the identities of the four main peaks present in the HLPC trace were confirmed by analyzing the same extract using HPLC/mass spectrometry on an HP 1100 liquid chromatograph coupled with a Bruker Esquire ion-trap mass spectrometer equipped with an electrospray source. Separation was achieved using a Waters Spherisorb 2 (i.e. ODS2) reverse-phase column (5-µm particle, 250 × 4.6 mm) under the following conditions: solvent A, 1% acetic acid; solvent B, acetonitrile; gradient, 5% to 50% B in 60 min at 0.8 mL min−1. Ion charge control was set at 30,000 with a maximum acquired time of 100 ms. Mass spectra were recorded over a range of 50 to 2,200 mass-to-charge ratio. Compounds were identified by comparing the UV and mass spectra. Flavonoid content was quantified by scoring the peak areas at 280 nm using calibration curves generated with authentic standards of daidzein, formononetin, 7,4′-dihydroxyflavone, and coumestrol.
Measurement of Phytohormone Levels and Auxin Transport
Phytohormone levels were determined as described previously (Gallego-Giraldo et al., 2011b) from 1 g of fresh, clean root material. Basipetal and acropetal auxin transport was measured using a modification of previously published protocols (Shin et al., 2005; Laffont et al., 2010). Six-week-old HCT (HCT1–HCT3) and wild-type (Wt1–Wt2) plants were grown on turface and vermiculite (2:1) in the greenhouse. After harvesting and root cleanup, the plants were laid on plates containing Fåhraeus medium (Catoira et al., 2000). Root tips were removed from the main root, and then agar blocks of 2 mm3 supplemented with 136 nCi [3H]indoleacetic acid (i.e. a specific activity of 20 Ci mmol−1; 1 Ci/mL; American Radiolabeled Chemicals) in the presence or absence of 10 mm NPA (an auxin transport inhibitor) were placed immediately next to the root tips, for measurement of transport in the basipetal direction, and next to the root basipetal (cut) end, for measurement of transport in the acropetal direction. The donor block was separated from the medium by a strip of Parafilm (to prevent diffusion of the [3H]indoleacetic acid through the agar). The plants were fixed to the medium plate using staples, and plates were placed vertically in a box and covered with aluminum foil. They were incubated at 20°C, and the agar blocks were removed after 1.5, 2, 4, and 6 h of incubation. Root regions closest to the donor blocks of approximately 0.5 mm in length were excised and discarded. The remaining root segments of approximately 5 cm in length were excised and placed into scintillation vials each containing 5 mL of scintillation fluid (Ready-HP solution; Beckman). After overnight extraction at 4°C, the radioactivity was measured using a Beckman Coulter LS6500 scintillation counter. This experiment was performed two times in the case of basipetal transport and one time in the case of acropetal transport. Each biological replicate consisted of five roots from one independent line of HCT down-regulated and wild-type plants.
Measurement of Carbon and Nitrogen Contents
The complete root systems and stems of plants harvested at different time intervals (0, 8, and 18 d) after rhizobia inoculation (three plants per harvest) were frozen and ground in liquid nitrogen. About 30 mg of the dry ground root material was submitted to the Analytical Services Laboratory under the plant analysis program at Pennsylvania State University for carbon and nitrogen determination using the combustion method on the Elementar VarioMax (Elementar Analysensysteme).
Measurement of Lignin Content and Composition
Lignin was determined by the thioacidolysis method (Lapierre et al., 1985, 1995) using approximately 20 mg of cell wall residue samples reacted with 3 mL of 0.2 m BF3 etherate in an 8.75:1 dioxane:ethanethiol mixture. Lignin-derived monomers were identified and quantified by gas chromatography-mass spectrometry as their trimethylsilyl derivatives on an Agilent 6890 gas chromatograph equipped with a DB-5MS column (60 m × 0.25 mm × 0.25 μm) attached to a 5973 mass spectrometer operated in electron-impact mode (70 eV) with a scan range of 60 to 650 mass-to-charge ratio.
qRT-PCR and Microarray Analyses
Total RNA was purified from noninoculated root tissue using TRIzol reagent (Invitrogen; http://www.invitrogen.com/), then split into two aliquots for microarray and qRT-PCR analyses. Two micrograms of total RNA treated with DNase (Ambion; http://www.ambion.com/) was used for qRT-PCR as described previously (Zhao et al., 2010). Gene-specific primers are listed in the Supplemental Material of our previous publication (Gallego-Giraldo et al., 2011b). For microarray analysis, the RNA was purified using the RNeasy MinElute Cleanup Kit (Qiagen; http://www.quiagen.com/), and 500 ng was hybridized with the Affymetrix Medicago GeneChip (http://www.affymetrix.com/). In this analysis, three biological replicates were used for each HCT (HCT1–HCT3) and wild-type (Wt1–Wt3) line. Each biological replicate consisted of a pool of six complete root systems. Experimental and statistical procedures were as described previously (Zhao et al., 2010). Differentially expressed genes between wild-type and HCT down-regulated lines were selected using associative analysis (Dozmorov and Centola, 2003). Functional annotation of differentially expressed genes was performed manually to the most similar Medicago truncatula and Arabidopsis proteins by BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Total RNA was also extracted from inoculated roots from both greenhouse and field experiments, following the same protocol described above.
Growth and Harvesting of Alfalfa in the Field
Transgenic HCT down-regulated lines 3a and 30a were crossed manually with the male-sterile line 208Ms-1 in the greenhouse. The resulting T1 seeds were planted in flats containing Metro-Mix 350 Growing Medium (Scotts-Sierra Horticultural Products) and inoculated with S. meliloti (Royal Peat Alfalfa/Clover inoculum; Becker Underwood). Before transferring plants to the field, confirmation of the presence (+) or absence (−) of the transgene in the T1 progeny from each HCT event, and of a single copy of the transgene, was performed as described (Bhattarai et al., 2012).
The field evaluations were performed at the Headquarters Farm of the Samuel Roberts Noble Foundation (Ardmore, OK) under U.S. Department of Agriculture-Animal and Plant Health Inspection Service permit 09-089-101-rm. Soil at the site is of the Normangee series (fine, smectitic, thermic Udertic Haplustalfs). The field was irrigated daily at 635 mm (0.25 inch) for the 10 d prior to root harvest performed on August 31, 2012. Roots were extracted using a Giddings Soil probe (model GSRTS; Giddings Machine) with a 20.32-cm (8-inch) diameter metal tube mounted on a tractor (John Deere 7420) at 20-cm depth. Excavated roots were collected in a 28-L bucket. Roots with soil were soaked in water for about 15 min before gently removing soil particles manually. Root fragments were hand collected. Root samples (100 mg) were frozen at −80°C for analysis of RNA. The remaining root samples were transported to the laboratory in an ice chest and double contained for further cleaning with a brush. Root samples were evaluated for root nodules, number of lateral roots, and overall root health. Root materials were then air dried in an oven at 50°C for 10 d to establish the dry matter yield of harvested samples. Dried roots were ground using a 1-mm-screen Wiley Mill (Thomas model 4; Thomas Scientific). Ground root samples were analyzed for total lignin by the acid detergent lignin procedure (Fukushima and Hatfield, 2004), for lignin monomer composition using the thioacidolysis method (Chen et al., 2006), and for total carbon and nitrogen as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript levels of lignin pathway genes in HCT-antisense and wild-type alfalfa roots as determined by qRT-PCR.
Supplemental Figure S2. Transcript levels of the plant leghemoglobin1 and bacterial nitrogenase genes following inoculation of wild-type and HCT down-regulated alfalfa roots with Rhizobia.
Supplemental Figure S3. Nitrogen fixation ability of roots from wild-type and HCT down-regulated alfalfa as assessed by acetylene reduction assay.
Supplemental Figure S4. Auxin transport in wild-type and HCT down-regulated alfalfa roots.
Supplemental Figure S5. Transcript levels of selected phenylpropanoid pathway genes in roots of alfalfa plants grown in the field for 3 years.
Supplemental Table S1. Genes that are differentially expressed in HCT-down-regulated roots compared to the wild type.
Supplemental Table S2. Functional categories of up-regulated genes in HCT down-regulated roots compared to the wild type.
Acknowledgments
We thank Drs. Rebecca Dickstein and Senjuti Sinharoy for critical reading of the manuscript.
Glossary
- qRT
quantitative real-time
- dpi
days post inoculation
- SA
salicylic acid
- PR
pathogenesis-related
Footnotes
This work was supported by the Oklahoma Center for the Advancement of Science and Technology and the Oklahoma Bioenergy Center (grants to R.A.D. and M.J.M.), by Forage Genetics International (to R.A.D.), by the Samuel Roberts Noble Foundation, and by the National Science Foundation (equipment grant no. DBI0400580).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Appleby CA. (1984) Leghemoglobin and rhizobium respiration. Annu Rev Plant Physiol Plant Mol Biol 35: 443–478 [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G. (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai K, Dixon RA, Monteros MJ (2012) Down-regulation of the HCT lignin gene in alfalfa: lignin reduction and transgene stability in the field. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America International Annual Meeting, Cincinnati, October 21–24, 2012. https://scisoc.confex.com/crops/2012am/webprogram/Paper73764.htm (Oct 22, 2012) [Google Scholar]
- Bright LJ, Liang Y, Mitchell DM, Harris JM. (2005) The LATD gene of Medicago truncatula is required for both nodule and root development. Mol Plant Microbe Interact 18: 521–532 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J. (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabannes M, Barakate A, Lapierre C, Marita JM, Ralph J, Pean M, Danoun S, Halpin C, Grima-Pettenati J, Boudet AM. (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J 28: 257–270 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen F, Duran AL, Blount JW, Sumner LW, Dixon RA. (2003) Profiling phenolic metabolites in transgenic alfalfa modified in lignin biosynthesis. Phytochemistry 64: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Chen F, Srinivasa Reddy MS, Temple S, Jackson L, Shadle G, Dixon RA. (2006) Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J 48: 113–124 [DOI] [PubMed] [Google Scholar]
- Dozmorov I, Centola M. (2003) An associative analysis of gene expression array data. Bioinformatics 19: 204–211 [DOI] [PubMed] [Google Scholar]
- Fagoaga C, Tadeo FR, Iglesias DJ, Huerta L, Lliso I, Vidal AM, Talon M, Navarro L, García-Martínez JL, Peña L. (2007) Engineering of gibberellin levels in citrus by sense and antisense overexpression of a GA 20-oxidase gene modifies plant architecture. J Exp Bot 58: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Foo E, Ross JJ, Reid JB. (2011) Relationship between gibberellin, ethylene and nodulation in Pisum sativum. New Phytol 189: 829–842 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61–76 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Ross JJ, Reid JB. (2005) Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol 138: 2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Chen F, Bouton JH, Foston M, Dixon RA, Wang ZY. (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima RS, Hatfield RD. (2004) Comparison of the acetyl bromide spectrophotometric method with other analytical lignin methods for determining lignin concentration in forage samples. J Agric Food Chem 52: 3713–3720 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA. (2011a) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. (2011b) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Guo D, Chen F, Wheeler J, Winder J, Selman S, Peterson M, Dixon RA. (2001) Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res 10: 457–464 [DOI] [PubMed] [Google Scholar]
- Hartwig UA, Maxwell CA, Joseph CM, Phillips DA. (1990) Chrysoeriol and luteolin released from alfalfa seeds induce nod genes in Rhizobium meliloti. Plant Physiol 92: 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Reid DE, Lorenc MT, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ. (2012) Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max [L.] Merr.) roots. Plant Biotechnol J 10: 995–1010 [DOI] [PubMed] [Google Scholar]
- Haynes JG, Czymmek KJ, Carlson CA, Veereshlingam H, Dickstein R, Sherrier DJ. (2004) Rapid analysis of legume root nodule development using confocal microscopy. New Phytol 163: 661–668 [DOI] [PubMed] [Google Scholar]
- Jeudy C, Ruffel S, Freixes S, Tillard P, Santoni AL, Morel S, Journet EP, Duc G, Gojon A, Lepetit M, et al. (2010) Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses. New Phytol 185: 817–828 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Ivashuta S, Bucciarelli B, Vance CP, Gantt JS, Vandenbosch KA. (2009) Knockdown of CELL DIVISION CYCLE16 reveals an inverse relationship between lateral root and nodule numbers and a link to auxin in Medicago truncatula. Plant Physiol 151: 1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffont C, Blanchet S, Lapierre C, Brocard L, Ratet P, Crespi M, Mathesius U, Frugier F. (2010) The compact root architecture1 gene regulates lignification, flavonoid production, and polar auxin transport in Medicago truncatula. Plant Physiol 153: 1597–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C. (1985) Thioacidolysis of lignin: comparison with acidolysis. J Wood Chem Technol 5: 277–292 [Google Scholar]
- Lapierre C, Pollet B, Rolando C. (1995) New insights into the molecular architecture of hardwood lignins by chemical degradative methods. Res Chem Intermed 21: 397–412 [Google Scholar]
- León-Barrios M, Dakora FD, Joseph CM, Phillips DA. (1993) Isolation of Rhizobium meliloti nod gene inducers from alfalfa rhizosphere soil. Appl Environ Microbiol 59: 636–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens S, Goormachtig S, Den Herder J, Capoen W, Mathis R, Hedden P, Holsters M. (2005) Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol 139: 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SR. (2001) Genes and signals in the rhizobium-legume symbiosis. Plant Physiol 125: 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Maekawa-Yoshikawa M, Takeda N, Imaizumi-Anraku H, Murooka Y, Hayashi M. (2009) Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J 58: 183–194 [DOI] [PubMed] [Google Scholar]
- Oke V, Long SR. (1999) Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol Microbiol 32: 837–849 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA. (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Peck MC, Fisher RF, Long SR. (2006) Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J Bacteriol 188: 5417–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Peoples MB, Brockwell J, Herridge DF, Rochester IJ, Alves BJR, Urquiaga S, Boddey RM, Dakora FD, Bhattarai S, Maskey SL, et al. (2009) The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48: 1–17 [Google Scholar]
- Peters NK, Frost JW, Long SR. (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233: 977–980 [DOI] [PubMed] [Google Scholar]
- Peters NK, Long SR. (1988) Alfalfa root exudates and compounds which promote or inhibit induction of Rhizobium meliloti nodulation genes. Plant Physiol 88: 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J, Cosson V, Muni RR, Wang M, Benedito VA, Andriankaja A, Cheng X, Jerez IT, et al. (2012) A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol 159: 1686–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Ott T. (2011) Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol 14: 458–467 [DOI] [PubMed] [Google Scholar]
- Prayitno J, Rolfe BG, Mathesius U. (2006) The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol 142: 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA. (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci USA 102: 16573–16578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel EL, Kassaw TK, Smith LS, Marsh JF, Oldroyd GE, Long SR, Frugoli JA. (2011) The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol 157: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA. (2007) Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68: 1521–1529 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Guo Z, Blancaflor EB, Masson PH, Chen R. (2005) Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J 42: 188–200 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Stacey G, Yu O. (2006) Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J 48: 261–273 [DOI] [PubMed] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. (2006) Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol 140: 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R. (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136: 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K. (2000) Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23: 97–114 [DOI] [PubMed] [Google Scholar]
- Yokota K, Li YY, Hisatomi M, Wang Y, Ishikawa K, Liu CT, Suzuki S, Aonuma K, Aono T, Nakamoto T, et al. (2009) Root-determined hypernodulation mutant of Lotus japonicus shows high-yielding characteristics. Biosci Biotechnol Biochem 73: 1690–1692 [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Stacey G, Yu O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J 57: 171–183 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Gallego-Giraldo L, Wang H, Zeng Y, Ding SY, Chen F, Dixon RA. (2010) An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J 63: 100–114 [DOI] [PubMed] [Google Scholar]