Reduction-oxidation homeostasis of plastids is crucial for embryo development and heterotrophic metabolism in Arabidopsis.
Abstract
In illuminated chloroplasts, one mechanism involved in reduction-oxidation (redox) homeostasis is the malate-oxaloacetate (OAA) shuttle. Excess electrons from photosynthetic electron transport in the form of nicotinamide adenine dinucleotide phosphate, reduced are used by NADP-dependent malate dehydrogenase (MDH) to reduce OAA to malate, thus regenerating the electron acceptor NADP. NADP-MDH is a strictly redox-regulated, light-activated enzyme that is inactive in the dark. In the dark or in nonphotosynthetic tissues, the malate-OAA shuttle was proposed to be mediated by the constitutively active plastidial NAD-specific MDH isoform (pdNAD-MDH), but evidence is scarce. Here, we reveal the critical role of pdNAD-MDH in Arabidopsis (Arabidopsis thaliana) plants. A pdnad-mdh null mutation is embryo lethal. Plants with reduced pdNAD-MDH levels by means of artificial microRNA (miR-mdh-1) are viable, but dark metabolism is altered as reflected by increased nighttime malate, starch, and glutathione levels and a reduced respiration rate. In addition, miR-mdh-1 plants exhibit strong pleiotropic effects, including dwarfism, reductions in chlorophyll levels, photosynthetic rate, and daytime carbohydrate levels, and disordered chloroplast ultrastructure, particularly in developing leaves, compared with the wild type. pdNAD-MDH deficiency in miR-mdh-1 can be functionally complemented by expression of a microRNA-insensitive pdNAD-MDH but not NADP-MDH, confirming distinct roles for NAD- and NADP-linked redox homeostasis.
Reduction-oxidation (redox) reactions play pivotal roles for most metabolic processes and occur in all cellular compartments. The origin of all reducing power in plants is the chloroplast thylakoid membrane system, where light-driven photosynthetic electron transport leads to the coupled formation of ATP and the reducing equivalent NADPH (Dietz and Pfannschmidt, 2011). Sudden changes in light intensity and withdrawal of ATP and NADPH for biosynthetic processes in varying amounts can potentially disturb the ATP:NADPH ratio. Maintaining this ratio within certain limits, however, is crucial for plant metabolism, because it avoids the accumulation of excess electrons and the production of cytotoxic reactive oxygen species and allows for the continued production of ATP (Apel and Hirt, 2004; Logan, 2006; Scheibe and Dietz, 2012). Accordingly, plants have several mechanisms to dissipate excess electrons, avoid damage to cellular components, and maintain redox homeostasis. These mechanisms include nonphotochemical energy quenching, chlororespiration, cyclic electron transport, and the Mehler reaction (Scheibe et al., 2005).
Reducing equivalents in the form of dedicated electron carriers or reduced cofactors (e.g. ferredoxin and NADH) are not generally transported directly across membranes; however, they can be shuttled indirectly as malate in exchange for oxaloacetic acid (OAA). This redox-poising mechanism is known as the malate valve in illuminated plastids or more generally, the malate-OAA shuttle (Heber, 1974; Scheibe, 2004; Taniguchi and Miyake, 2012). The key enzyme of the malate-OAA shuttle is malate dehydrogenase (MDH), which catalyses the reversible interconversion of malate and OAA. Isoforms of MDH are present in various cell compartments (Gietl, 1992), and each isoform is specific to either cosubstrate NAD (NAD-MDH; EC 1.1.1.37) or NADP (NADP-MDH; EC 1.1.1.82). The Arabidopsis genome encodes eight putative NAD-MDH isoforms: two isoforms are peroxisomal MDH (PMDH; PMDH1 and PMDH2; Pracharoenwattana et al., 2007; Eubel et al., 2008), two isoforms are mitochondrial MDH (MMDH; MMDH1 and MMDH2; Millar et al., 2001; Lee et al., 2008; Tomaz et al., 2010), and one isoform is plastidial MDH (plastid-localized NAD-dependent MDH [pdNAD-MDH]; Berkemeyer et al., 1998). The remaining three isoforms have no detectable target sequence and are thought to be cytosolic MDH (CMDH; CMDH1, CMDH2, and CMDH3). The Arabidopsis genome also encodes an additional NADP-dependent isoform of MDH, which is localized to the plastid (Hebbelmann et al., 2012).
The physiological role of the different isoforms depends on the subcellular localization and the different metabolic pathways occurring there. For instance, MMDH was reported to be involved in two processes that are at least partly mitochondrial: leaf respiration and photorespiration (Tomaz et al., 2010). An MMDH null mutant (mmdh1 mmdh2) was slow growing and showed elevated leaf respiration in the dark and the light, although photosynthetic capacity was not affected. Tomaz et al. (2010) proposed that MMDH uses NADH to reduce OAA to malate, which is then shuttled to the cytosol, rather than generate NADH to fuel mitochondrial respiration (Tomaz et al., 2010). PMDH might serve at least two different functions. First, during fatty acid β-oxidation, which generates NADH, PMDH is proposed to regenerate the electron acceptor NAD by reducing OAA to malate, which is then shuttled to the cytosol in exchange for OAA (Pracharoenwattana et al., 2007). Second, PMDH is thought to generate NADH during photorespiration by oxidation of malate imported from the cytosol (Reumann and Weber, 2006; Cousins et al., 2008). Arabidopsis mutants lacking PMDH (pmdh1 pmdh2) are severely impaired in β-oxidation, and seedling establishment is strongly impaired and dependent on the supply of exogenous sugar (Pracharoenwattana et al., 2007), a phenotype characteristic of β-oxidation mutants (Pinfield-Wells et al., 2005; Baker et al., 2006). However, after transfer of established pmdh1 pmdh2 seedlings to compost, they grew only slightly slower than wild-type plants (Pracharoenwattana et al., 2007).
Until recently, genetic evidence for the roles of the plastidial MDH isoforms was scarce. In most C4 plants, NADP-MDH is directly involved in CO2 fixation, catalyzing the formation of the stable CO2 carrier malate from the primary CO2 fixation product OAA (Scheibe, 1987). However, in C3 plants, NADP-MDH has long been proposed to have its major function in the malate valve, leading to shuttling of reducing power (as malate) from the chloroplast to the cytosol during the day and thereby regenerating the electron acceptor NADP inside the chloroplasts (Heber, 1974; Lance and Rustin, 1984; Scheibe, 1987). NADP-MDH is redox activated by thioredoxins in the light and essentially inactive in the dark (Scheibe, 1987; Buchanan and Balmer, 2005). The widely accepted belief that chloroplasts only possess this one strictly light-/redox-activated NADP-MDH temporarily led to the conclusion that the malate valve only works in illuminated chloroplasts (Berkemeyer et al., 1998; Scheibe, 2004). However, a recent study showed that Arabidopsis plants lacking NADP-MDH (nadp-mdh) were indistinguishable from wild-type plants, even under conditions that are supposed to provoke the accumulation of excess electrons and the production of cytotoxic reactive oxygen species (high light and short days; Hebbelmann et al., 2012). This finding indicates that NADP-MDH is not crucial for providing electron acceptors in chloroplasts, but it rather suggests that other mechanisms can counteract or prevent overreduction of the chloroplast.
The existence of a second MDH isoform in plastids, which uses NAD as cofactor, has been questioned, because it could not be ruled out that NAD-MDH activity detected in isolated chloroplasts was caused by contamination from other organelles (Siebke et al., 1991; Backhausen et al., 1998). In 1998, Berkemeyer et al. (1998) reported the cloning, heterologous expression, and in vitro characterization of a pdNAD-MDH from Arabidopsis (At3g47520). In contrast to NADP-MDH, pdNAD-MDH is active under both light and dark conditions in isolated chloroplasts, and the activities of both enzymes are within the same range in the light (Backhausen et al., 1998; Berkemeyer et al., 1998). However, up to now, genetic evidence for the in vivo function of pdNAD-MDH is missing, and experimental data are scarce. Backhausen et al. (1998) showed that chloroplasts and heterotrophic chromoplasts isolated from different sources followed by incubation in the dark concomitantly produced 3-phosphoglycerate and malate on addition of dihydroxyacetone phosphate and OAA to the medium. It was proposed that 3-phosphoglycerate production was in a glycolytic step involving glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and that pdNAD-MDH regenerates the electron acceptor NAD required by GAPDH through reduction of OAA to malate, thus operating the malate-OAA shuttle in the dark and in nongreen tissues (Scheibe, 2004; Taniguchi and Miyake, 2012).
Here, we aimed to evaluate the function of this MDH isoform in plastid metabolism by analyzing Arabidopsis plants with a transposon insertion in the pdNAD-MDH gene and Arabidopsis plants with reduced pdNAD-MDH by means of artificial microRNA silencing.
RESULTS
A Transposon Insertion at the pdNAD-MDH Locus Results in Embryo Death
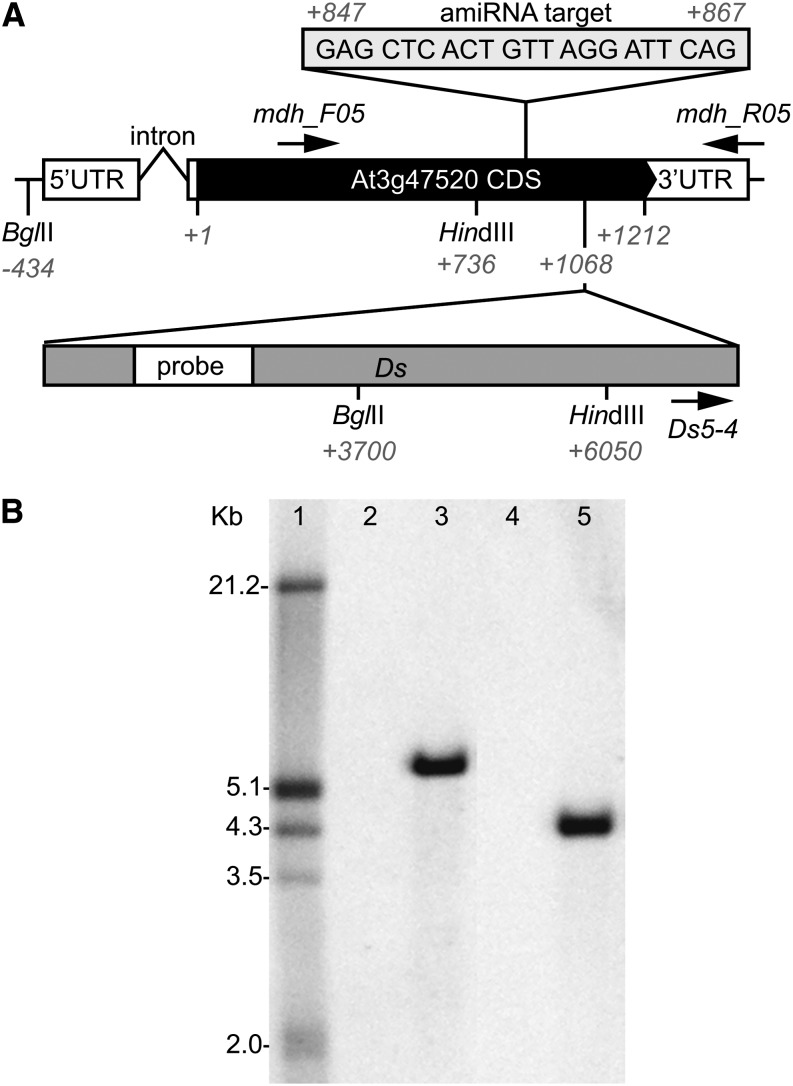
To determine the function of pdNAD-MDH in Arabidopsis, we searched for publicly available lines with altered pdNAD-MDH expression (www.arabidopsis.org). We found only one line with a transposon insertion in the pdNAD-MDH coding sequence: ET8629, which is an enhancer trap line from the Trapper collection (genetrap.cshl.org; Sundaresan et al., 1995). PCR with gene- and insert-specific primers followed by sequencing of the product revealed that the insertion of the dissociator (Ds) element is at position 1,068 of 1,212 nucleotides of the coding sequence (Fig. 1A). Using Southern blot analysis, we confirmed that it is the sole Ds element insertion in the ET8629 genome (Fig. 1B).
Figure 1.
Gene structure of pdNAD-MDH in the wild type and the transposon insertion line ET8629 (pdnad-mdh). A, In wild-type plants, the pdNAD-MDH gene (At3g47520) consists of one intron in the 5′ untranslated region (UTR) and one exon representing the coding sequence (CDS) followed by the 3′ UTR. Numbers represent nucleotide positions relative to the translational start +1. The position of the Ds element in the enhancer trap line pdnad-mdh and the relative position of the probe for Southern blotting are shown. Sequence and position of the target for silencing by amiRNA in the miR-mdh lines and position of restriction enzymes used in Southern blotting are indicated. Locations of primers used for genotyping are depicted as arrows. B, Southern blot analysis showed that pdnad-mdh has only one Ds insertion; 10 µg of genomic DNA per lane from wild-type (lanes 2 and 4) and heterozygous pdnad-mdh (lanes 3 and 5) plants digested with HindIII (lanes 2 and 3) or BglII (lanes 4 and 5) are shown. Position of the digoxigenin (DIG)-labeled probe on the Ds element is as shown in A. Sizes (in kilobase pairs) of the DIG-labeled molecular weight marker III (lane 1; Roche) are indicated on the left of the membrane.
In plants heterozygous for the Ds insertion, pdNAD-MDH expression was slightly, but not significantly, lower than in wild-type plants as determined by quantitative reverse transcription PCR (RT-PCR; Supplemental Table S1), and these plants were lacking any visual phenotype. Therefore, we tried to isolate homozygous lines from the progeny of the heterozygous line after self-pollination. Of 262 genotyped plants, 89 plants lacked a Ds insertion, 173 plants were heterozygous for the insertion, and none were homozygous. This segregation ratio is not significantly different (χ2 analysis, 1 degree of freedom, P = 0.01, χ2 = 0.048) from the expected 1:2:0 (wild type:Ds heterozygous:Ds homozygous) ratio, suggesting that a homozygous insertion affects plant viability.
To test this proposition, mature green siliques of heterozygous ET8629 plants were inspected for aborting seeds. From a total of 1,161 seeds from 20 siliques of individual plants, we observed 295 white and 866 normal green seeds, representing an abortion frequency that is not significantly different from the expected 25% for an embryo-lethal effect (χ2 analysis, 1 degree of freedom, P = 0.01, χ2 = 0.104; Fig. 2A). During later stages of seed development, the white seeds turned dark brown and collapsed. Abortion was not caused by a gametophyte defect, because genotyping the progeny of crosses of heterozygous ET8629 and wild-type plants revealed a transmission rate of the insertion of 47% (71 heterozygous and 79 wild-type plants), which is not significantly different from the 50% value expected for intact gametophytes (χ2 analysis, 1 degree of freedom, P = 0.01, χ2 = 0.427).
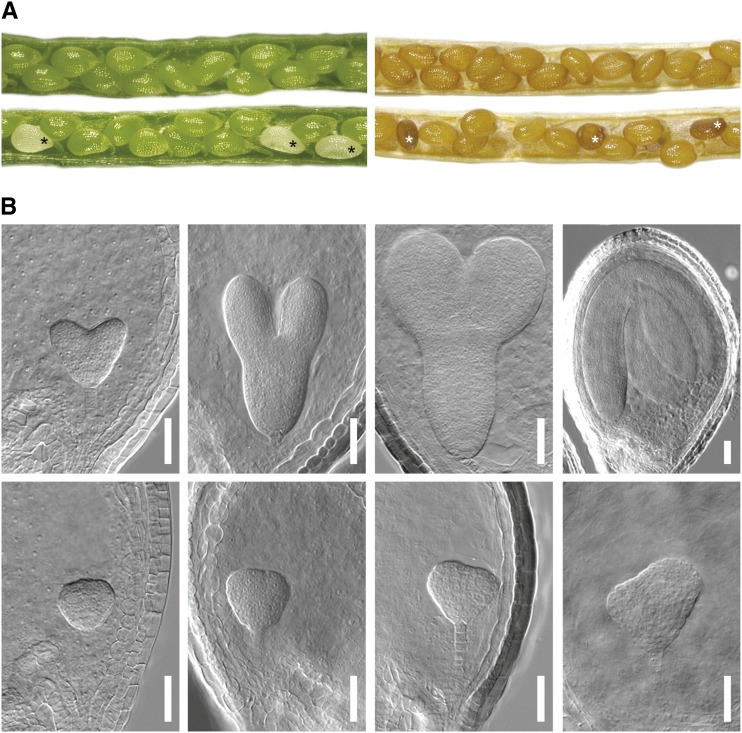
Figure 2.
Homozygous transposon insertion in pdNAD-MDH causes seed abortion and embryo arrest. A, Pictures of immature (left) and mature (right) siliques of wild-type (top) and heterozygous pdnad-mdh (bottom) plants. Asterisks denote aborted seeds. B, Green (top) and white (bottom) seeds from young to old (from left to right) siliques of heterozygous pdnad-mdh plants were cleared and investigated by DIC microscopy. Embryos in the same column were from the same silique. Embryos from green seeds were at heart, early and late torpedo, and late walking stick stages (from left to right). Embryos from white seeds did not reach heart stage but arrested in the globular-to-heart transition stage. Bar = 50 µm.
These observations indicate that ET8629 is a pdnad-mdh mutant and that the Ds insertion in ET8629 is embryo lethal in the homozygous but not the heterozygous state. To prove this proposition, we performed a complementation analysis. We generated heterozygous pdnad-mdh lines expressing a fusion of pdNAD-MDH and enhanced yellow fluorescent protein (eYFP) under control of the endogenous promoter (P) of pdNAD-MDH (PpdNAD-MDH::pdNAD-MDH::eYFP), selected for the presence of the Ds element and PpdNAD-MDH::pdNAD-MDH::eYFP in the first generation after transformation (T1) and genotyped the T2 offspring for homozygous Ds insertions. Of 44 T2 plants analyzed, we identified 22 plants carrying homozygous Ds insertions, and all of them expressed pdNAD-MDH-eYFP (judged by fluorescence microscopy). Altogether, these data show that the pdNAD-MDH-eYFP fusion protein is functional and that it complemented embryo lethality.
Embryos from Aborting Seeds Do Not Complete Morphogenesis
To investigate at which stage embryo development was affected, we inspected cleared seeds at most developmental stages by differential interference contrast (DIC) microscopy. During early stages of embryo development, up to the midglobular stage, we could not observe significant morphological differences between embryos from wild-type and heterozygous pdnad-mdh plants. At this time, it was also not yet possible to distinguish between green and white seeds of heterozygous pdnad-mdh plants macroscopically. However, in older siliques, the morphological differences became apparent: approximately three-quarters of the embryos developed normally to maturity, whereas the rest arrested in the globular-to-heart transition stage (Fig. 2B). To test if this arrest correlated with seed abortion, we separated normal green from white or collapsed seeds 9–12 d after fertilization followed by clearing and inspection by DIC microscopy. All embryos from white or collapsed seeds were arrested in the globular-to-heart transition stage. However, all embryos from green seeds had already progressed to the walking stick stage or beyond. We never observed arrested embryos in normal green seeds. Altogether, these observations suggest that pdNAD-MDH serves a crucial function during embryo development.
pdNAD-MDH Is Expressed in Green and Nongreen Tissues Throughout Development and the Diurnal Cycle
To check whether pdNAD-MDH is expressed during embryo development, we analyzed eYFP expression in embryos dissected from green seeds of heterozygous pdnad-mdh plants carrying PpdNAD-MDH::pdNAD-MDH::eYFP by fluorescence microscopy. We observed eYFP fluorescence at the globular stage and all subsequent stages of embryo development; representative pictures are shown in Figure 3A. Expression was not restricted to distinct areas of the embryo at any developmental stage but rather, was distributed over the whole embryo proper and the suspensor. We saw slightly higher eYFP fluorescence in the embryonic shoot apical meristem and at the bases of the cotyledons at torpedo stage, but chlorophyll autofluorescence was also higher in these regions. In subsequent stages, eYFP expression increased in the cotyledons, the hypocotyl, and the root. Chlorophyll autofluorescence was detected earliest at the late heart stage and from then on, in all subsequent stages. Chlorophyll and eYFP fluorescence largely colocalized, but in general, eYFP fluorescence preceded chlorophyll autofluorescence.
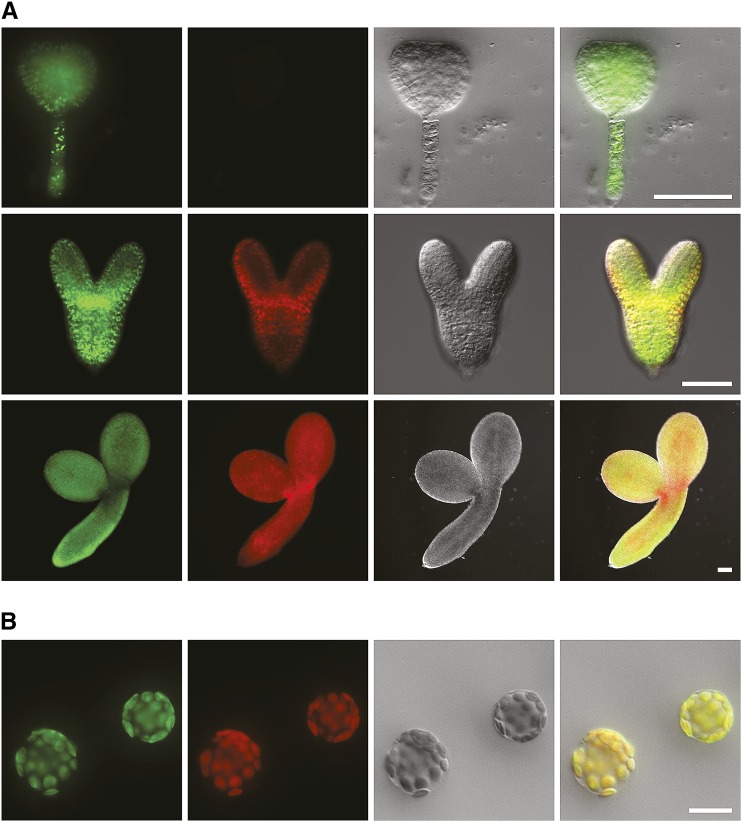
Figure 3.
pdNAD-MDH is a plastidial protein expressed during embryogenesis. Dissected embryos (A) and isolated leaf mesophyll protoplasts (B) of plants expressing a pdNAD-MDH-eYFP fusion protein under control of the endogenous pdNAD-MDH promoter. Left to right, eYFP fluorescence, chlorophyll autofluorescence, DIC, and merged pictures. Embryos at globular-to-heart transition (A, top), torpedo (A, middle), and mature (A, bottom) stages. eYFP fluorescence is visible at all these stages, and chlorophyll fluorescence appears only after the globular-to-heart transition stage. Chlorophyll and eYFP fluorescence colocalize in chloroplasts (B). Protoplasts isolated from leaves 4 h after dawn. Bar in A = 50 µm; bar in B = 20 µm.
Another line of evidence for embryonic expression of pdNAD-MDH comes from an independently produced set of Arabidopsis plants expressing a transcriptional fusion of the pdNAD-MDH promoter and the GUS coding sequence. GUS activity was detected from the globular stage on and throughout all subsequent stages (Supplemental Fig. S1). Additionally, we detected GUS activity in most of the tissues investigated. More specifically, we observed GUS expression in young and old rosette leaves, cotyledons, roots, stems, flowers (low activity in sepals, petals, and carpels; high activity in anthers, pollen, filaments, and stigmata) and carpels as siliques mature (Supplemental Fig. S1). Publicly available microarray data analyzed with Genevestigator (Hruz et al., 2008) confirmed the global and embryonic expression patterns that we observed with the eYFP and GUS lines (Supplemental Fig. S2). Microarray data from two studies analyzing diurnal gene expression (Smith et al., 2004; Bläsing et al., 2005) also showed that pdNAD-MDH expression was constantly high throughout the day-night cycle and did not reveal a diurnal expression pattern (Supplemental Fig. S3).
In summary, these data show that pdNAD-MDH is expressed at the stages of embryo development at which homozygous pdnad-mdh embryos arrest as well as in other heterotrophic and autotrophic tissues throughout the diurnal cycle.
pdNAD-MDH Is Localized to Plastids
To date, in vivo data on the subcellular localization of pdNAD-MDH are limited to large-scale subcellular proteomics datasets. The SUBA3 Web site currently lists 16 proteomics studies that identified pdNAD-MDH, 11 of which propose plastidial localization (http://suba.plantenergy.uwa.edu.au; Tanz et al., 2013). Here, we analyzed subcellular localization of pdNAD-MDH-eYFP in protoplasts isolated from heterozygous pdnad-mdh plants transformed with PpdNAD-MDH::pdNAD-MDH::eYFP (T1; see above). Chlorophyll and eYFP fluorescence clearly colocalized, thus confirming plastidial localization of pdNAD-MDH (Fig. 3B).
pdNAD-MDH Silencing Results in Small and Pale Green Plants
In addition to investigating the role of pdNAD-MDH for heterotrophic metabolism during embryo development, we also studied its role in photosynthetic metabolism. To perform this study, we generated Arabidopsis lines with reduced pdNAD-MDH levels using the artificial microRNA (amiRNA) technology (Schwab et al., 2006; Ossowski et al., 2008). The silencing construct specifically targets regions 847–867 of the pdNAD-MDH coding sequence (Fig. 1A). Expression of the silencing construct in these lines with reduced pdNAD-MDH levels by means of amiRNA (miR-mdh) was under control of the cauliflower mosaic virus 35S promoter (PCaMV 35S). This promoter is normally considered to allow for constitutive expression, but several studies have shown that it is hardly active during early embryogenesis (Benfey and Chua, 1989; Custers et al., 1999; Völker et al., 2001; Sunilkumar et al., 2002; Huanca-Mamani et al., 2005). Therefore, it seems likely that pdNAD-MDH will be silenced only at later stages of development, hence avoiding detrimental effects in embryos.
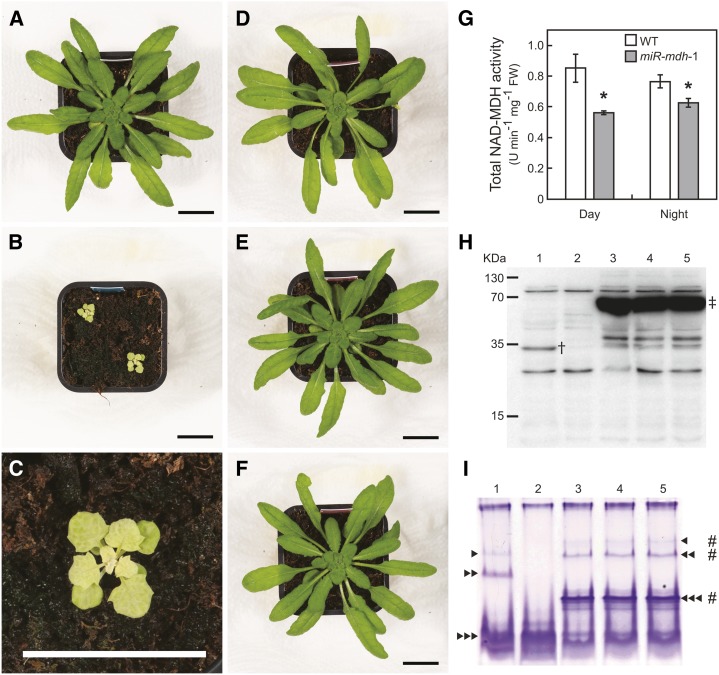
We analyzed phenotypes of the offspring of 20 primary transformants in the T2 generation and found 16 lines displaying apparent differences to the wild type. These lines had noticeably pale, flat, and jagged leaves and showed obvious growth retardation and reduction in seed yield compared with wild-type plants (Supplemental Fig. S4). No pdNAD-MDH protein was detectable by immunoblot analysis using a polyclonal antibody raised against a peptide of pdNAD-MDH (VESSLRALDGDGDVC; α-pdNAD-MDH1) in any of these lines (Fig. 4, representative line; Supplemental Fig. S5, antibody specificity). All additional investigations were on one homozygous line (T4) with a single insertion of the silencing construct, which was designated as miR-mdh-1. The visible phenotype of 5-week-old miR-mdh-1 plants is illustrated in Figure 4, B and C. At this stage, the projected leaf area of miR-mdh-1 plants was 100 times lower compared with the wild type (43 ± 4 and 4,318 ± 225 mm2, respectively; mean ± se of four plants). Expression of pdNAD-MDH in miR-mdh-1 plants was reduced by 82% compared with the wild type (Supplemental Table S1), and as mentioned before, pdNAD-MDH protein was not detectable (Fig. 4H). Likewise, total NAD-MDH activity was reduced by 34% at midday and 18% at night in miR-mdh-1 plants compared with the wild type (Fig. 4G). Given that total NAD-MDH activity represents the sum of all eight NAD-specific isoforms, we specifically investigated pdNAD-MDH activity by native gel electrophoresis followed by activity staining (Fig. 4I). Because of the presence of the different isozymes, NAD-MDH activity was apparent as multiple bands in the wild type, but three bands of activity were lost in miR-mdh-1 (Fig. 4I). To check if the loss of these three bands is specifically ascribable to pdNAD-MDH silencing, we performed additional experiments. First, we measured the transcript levels of MDH isoforms (NADP-MDH, PMDH1 and PMDH2, MMDH1 and MMDH2, and CMDH1 and CMDH2) by quantitative RT-PCR. Transcription of none of the isoforms was significantly changed in miR-mdh-1 compared with the wild type (Supplemental Table S2). Second, we compared the activity band profile of miR-mdh-1 with the profile of mutants lacking the peroxisomal (Pracharoenwattana et al., 2007) or mitochondrial (Tomaz et al., 2010) NAD-MDH isozymes. None of the three activity bands missing in miR-mdh-1 were altered in the other mutants (Supplemental Fig. S6A). Third, immunoblot analysis of native gels with α-pdNAD-MDH2 antibodies revealed signals that colocalized with the three activity bands, indicating that they are attributable to pdNAD-MDH (Supplemental Fig. S6, B and C).
Figure 4.
Silencing of pdNAD-MDH leads to reductions in MDH activity and pale green plants with stunted growth. A to F, Pictures of 40-d-old plants grown under standard conditions. Plants expressing an amiRNA silencing construct targeting pdNAD-MDH under control of PCaMV 35S (miR-mdh-1; B and C) exhibit pale green leaves and stunted growth compared with the wild type (A). Expression of amiRNA-insensitive pdNAD-MDH-eYFP-HA in the miR-mdh-1 background (i.e. miRins-mdh-1), also under control of PCaMV 35S, complements the above-mentioned phenotype. Three complemented primary transformants (T1) are shown: miRins-mdh-1-6 (D), miRins-mdh-1-7 (E), and miRins-mdh-1-8 (F). Bars = 20 mm. G, Total NAD-dependent MDH activity of 4- and 8-week-old plants (wild type and miR-mdh-1, respectively) was determined in the middle of the day and the middle of the night. Asterisks denote significant differences (Student’s t test; P < 0.05) between the genotypes (n = 5). FW, Fresh weight; WT, wild type. Immunoblot (H) and activity gel (I) analyses show reductions in pdNAD-MDH protein and activity levels in miR-mdh-1 (lane 2) compared with the wild type (lane 1). The complemented lines miRins-mdh-1-6, miRins-mdh-1-7, and miRins-mdh-1-8 (lanes 3–5) show high levels of the pdNAD-MDH-eYFP-HA fusion protein (H), which displays NAD-MDH activity (I). Primary antibody was α-pdNAD-MDH1. Numbers to the left of the blot indicate the size of the molecular mass marker (kD). Arrowheads indicate the positions of three different pdNAD-MDH activities. Hash marks indicate the positions of the pdNAD-MDH-eYFP-HA fusion protein activities. Dagger indicates the position of endogenous pdNAD-MDH (34 kD) protein. Double-dagger indicates the position of pdNAD-MDH-eYFP-HA-fusion (74 kD) protein.
To analyze silencing specificity more rigorously, we generated an amiRNA-insensitive construct encoding a pdNAD-MDH-eYFP fused to a hemagglutinin (HA) tag (pdNAD-MDH-eYFP-HA) and expressed it in the miR-mdh-1 background under control of PCaMV 35S (pdNAD-MDH construct that is insensitive to amiRNA silencing [miRins-mdh]). All 16 BASTA-resistant primary transformants were complemented for the pale and dwarf phenotype to some extent, and five of these plants were indistinguishable from the wild-type controls by eye. Three apparently complemented lines were analyzed further for protein content and enzyme activity (miRins-mdh-1-6, miRins-mdh-1-7, and miRins-mdh-1-8; Fig. 4). All three plants showed high levels of the fusion protein, and as indicated by activity staining of native gels, the fusion protein was active. Its activity was again detected as three distinct bands, but each band migrated more slowly, presumably because of the additional eYFP-HA tag. Essentially, the same results were obtained when we complemented another independent silencing line (miR-mdh-8; Supplemental Fig. S4) with the same approach as shown in Supplemental Figure S7. Altogether, these data show that paleness and stunted growth of miR-mdh are specifically caused by reduced pdNAD-MDH levels.
pdNAD-MDH Influences Chloroplast Ultrastructure and Photosynthetic Metabolism
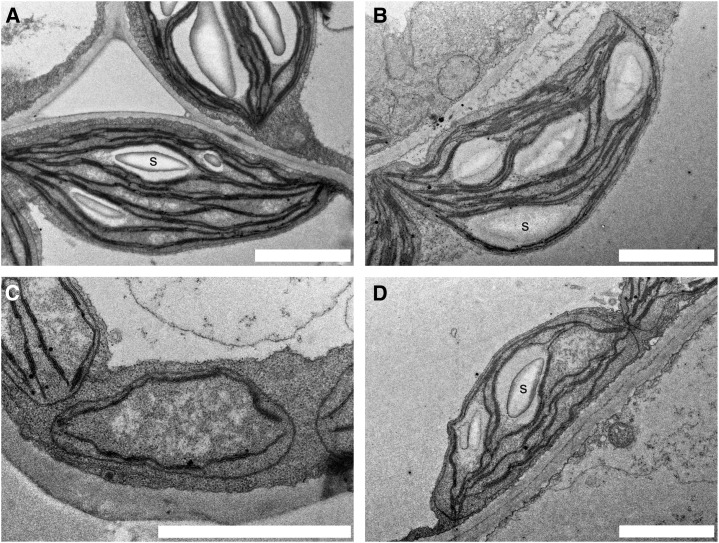
The pale phenotype observed with miR-mdh-1 plants led us to investigate chloroplast ultrastructure by transmission electron microscopy. Because we noticed that young, developing miR-mdh-1 leaves were paler than older, mature leaves, we examined leaf mesophyll chloroplasts from both young and old leaves of miR-mdh-1 and wild-type plants (Fig. 5). Chloroplast ultrastructure was altered in miR-mdh-1 compared with the wild type. The differences were most obvious in young leaves when miR-mdh-1 chloroplasts had far fewer thylakoids, had no starch granules, had an unusual, roundish shape, and at the most, were one-half the length of wild-type chloroplasts. In older leaves, these differences become less apparent. In general, miR-mdh-1 chloroplasts in older leaves still had fewer thylakoids and starch granules than wild-type chloroplasts, but the disparities in size and shape were negligible.
Figure 5.
Chloroplast ultrastructure is altered in miR-mdh-1 plants. Transmission electron micrographs of wild-type (A and B) and miR-mdh-1 (C and D) mesophyll chloroplasts from young developing (A and C) and mature (B and D) leaves. Chloroplasts from young miR-mdh-1 leaves (C) have no starch (S), have fewer thylakoids, and are reduced in length compared with the young wild type (A). Mature miR-mdh-1 chloroplasts (D) still seem to have fewer thylakoids and less starch than mature wild-type chloroplasts (B), but they now exhibit a similar length. Bars = 2 µm.
The observations of disturbed chloroplast ultrastructure, pale phenotype, and growth retardation of miR-mdh-1 plants led us to investigate photosynthesis-related parameters more closely. Although the chlorophyll a:b ratio was not significantly altered, miR-mdh-1 plants had only 24% of wild-type chlorophylla+b levels per leaf area, clearly reflecting the pale phenotype (Table I; Supplemental Fig. S8, plant images). However, chlorophyll fluorescence measurements revealed no significant differences between miR-mdh-1 and the wild type concerning photochemical (ΦPSII and variable fluorescence [Fv]/maximum fluorescence yield [Fm]) and nonphotochemical quenching, indicating that PSII efficiency is not altered (Supplemental Table S3). At the same time, ground state fluorescence (F0), Fm, and Fv are significantly lower in miR-mdh-1 compared with wild type, reflecting the reduced chlorophyll levels in the silencing line.
Table I. Silencing of pdNAD-MDH causes reductions in chlorophyll levels.
Plants were grown under standard conditions for 3–4 weeks (wild type) and 8–9 weeks (miR-mdh-1). Chlorophyll (Chl) levels, projected leaf area, and fresh weight of whole rosettes were determined in the middle of the light period. Values represent the mean ± se (n = 8).
| Chla | Chlb | Chla+b | Chla:Chlb | Leaf Area | Fresh Weight | |
|---|---|---|---|---|---|---|
| µmol m−2 | mm2 | mg | ||||
| Wild type | 311.3 ± 9.3 | 72.3 ± 4.2 | 383.5 ± 11.2 | 4.4 ± 0.3 | 677.6 ± 60.8 | 220.6 ± 33.4 |
| miR-mdh-1 | 76.5 ± 4.0a | 14.1 ± 1.7a | 90.6 ± 5.5a | 6.0 ± 0.6 | 817.6 ± 29.4 | 198.1 ± 8.6 |
miR-mdh-1 values statistically different from the respective wild-type values (Student’s t test, P < 0.05).
To test whether reduced chlorophyll levels influenced photosynthetic carbon fixation, we did gas exchange measurements of miR-mdh-1 and wild-type plants under ambient growth conditions using custom-built chambers coupled to an LI-7000 infrared gas analyzer (Licor). For technical reasons, we compared wild-type and miR-mdh-1 plants with similar projected leaf areas rather than plants of the same age (miR-mdh-1, 8 to 9 weeks old; wild type, 3 to 4 weeks old; images in Supplemental Fig. S8). The transpiration rates (Es) of wild-type and miR-mdh-1 plants were not significantly different (Table II). However, the net photosynthetic rate (A) of miR-mdh-1 plants was reduced by 49% during the day and 89% at night (i.e. the respiration rate) compared with the wild type (Table II).
Table II. Silencing of pdNAD-MDH entails reductions in photosynthetic rate.
Plants were grown under standard conditions for 3–4 weeks (wild type) and 8–9 weeks (miR-mdh-1) before transfer to gas exchange chambers (Supplemental Fig. S8, plant images). After a 2-d adaptation period, A and E were measured in custom-made gas exchange chambers over one complete 12-h-light/12-h-dark cycle at standard growth conditions. A and E were averaged over day and night, respectively. Projected leaf area was determined at the end of the light period. Values represent the mean ± se of two independent experiments with four plants per genotype each (n = 8 plants).
| A |
E |
|||
|---|---|---|---|---|
| Day | Night | Day | Night | |
| µmol m−2 s−1 | mmol m−2 s−1 | |||
| Wild type | 6.93 ± 0.25 | −0.49 ± 0.08 | 1.39 ± 0.07 | 0.96 ± 0.04 |
| miR-mdh-1 | 3.53 ± 0.12a | −0.05 ± 0.02a | 1.19 ± 0.09 | 0.91 ± 0.02 |
| Ratio miR-mdh-1:wild type (%) | 51.0 ± 1.7 | 10.7 ± 4.9 | 81.6 ± 7.0 | 91.1 ± 5.3 |
Values that are statistically different from the respective wild-type values (Student’s t test, P < 0.01).
Similar results were obtained with another set of plants grown at high light intensities (600 µmol m−2 s−1) under otherwise identical conditions. However, this time, we observed a small difference in photochemical quenching. Although nonphotochemical quenching and the maximum quantum efficiency of PSII photochemistry (Fv/Fm) was still unchanged, the quantum yield of PSII photochemistry (ΦPSII) was slightly but significantly decreased in miR-mdh-1 compared with the wild type (Supplemental Table S4). Furthermore, the photosynthetic rate of miR-mdh-1 was now decreased to 39% of the wild-type rate during the day, and nighttime respiration in miR-mdh-1 was below the detection limit.
pdNAD-MDH Silencing Causes Changes in Metabolite Levels
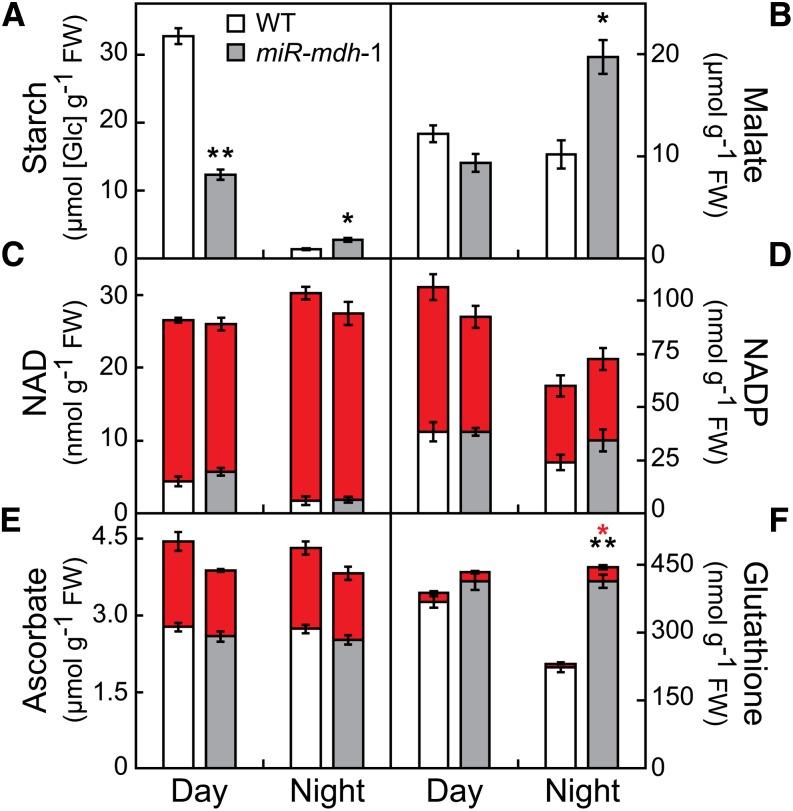
Levels of leaf starch, a major photosynthetic product in Arabidopsis, were reduced by 62% at the end of the day in miR-mdh-1 plants compared with the wild type (Fig. 6A), which likely is a consequence of the reduced photosynthetic rate during the day. At the end of the night, starch levels remained two times as high in miR-mdh-1 as in the wild type, indicating that nighttime catabolism is equally affected. Next, we measured levels of other MDH-related metabolites as well as key players in maintaining the plant’s redox status at midday and in the middle of the night (Fig. 6). Although malate was unchanged between both genotypes at midday, it increased to double the wild-type levels in miR-mdh-1 at night (Fig. 6B). NAD(H) levels did not change accordingly. The NAD(H) pool was more reduced in the light than the dark, but we did not observe differences between the genotypes (Fig. 6C). Similarly, both the total amount and the ratio of reduced to oxidized forms did not change with two other redox-related compounds: NADP(H) and ascorbate (Fig. 6, D and E). However, the cellular thiol-disulfide buffer, glutathione, was significantly affected in the night but not during the day (Fig. 6F). Although total glutathione at night decreased to 60% of the daytime value in the wild type, it remained high in miR-mdh-1, which resulted in a 92% increase in total glutathione in miR-mdh-1 compared with the wild type at night. In addition, the reduced glutathione/total glutathione ratio at night was significantly lower in miR-mdh-1 than the wild type (86.8 ± 1.8% and 93.5 ± 0.3%, respectively; P < 0.02, Student’s t test), indicating that the thiol-disulfide buffer is slightly more oxidized in miR-mdh-1. Taken together, the metabolite data indicate that miR-mdh-1 metabolism is mainly affected during the night.
Figure 6.
Metabolite levels in miR-mdh-1 and wild-type plants at day and night. Gray bars, miR-mdh-1; white bars, wild type. C to F, Red blocks indicate the oxidized form of the respective compound (NAD+ for NAD, NADP+ for NADP, dehydroascorbate for ascorbate, and oxidized glutathione for glutathione), whereas gray and white bars represent the reduced forms (NADH for NAD, NADPH for NADP, ascorbate for ascorbate, and reduced glutathione for glutathione). Plants were grown under standard conditions for 4 (wild type) and 8 (miR-mdh-1) weeks. Harvest for starch measurements (A) was at the end of day and night for wild type and miR-mdh-1 (n = 10 plants each). For determination of malate (B) and all other metabolites, plants were harvested after 6 h of light and dark (n = 5 plants each). Values represent mean ± se. Differences between the genotypes were analyzed by Student’s t test. Significant differences are indicated by black asterisks (starch, malate, and reduced forms in C to F) and red asterisks (oxidized forms in C to F)(*P < 0.005; **P < 0.001).
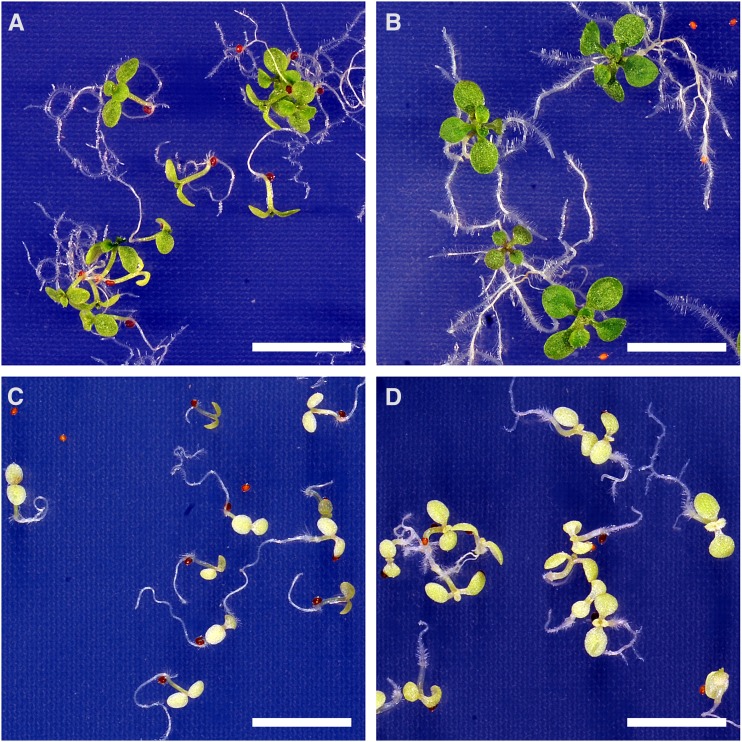
Although NAD(H) levels are unaffected in miR-mdh-1, we cannot exclude the fact that the flux through the NAD(H) pool is reduced in the mutant. The proposed role for pdNAD-MDH was the regeneration of the electron acceptor NAD in the dark and nongreen tissues (Scheibe, 2004). Another enzyme, which is present in chloroplasts and plastids of heterotrophic tissues and generates NAD, is NADH-dependent Gln-2-oxoglutarate aminotransferase (NADH-GOGAT; Lancien et al., 2002). To test if increased NAD production by NADH-GOGAT can complement the growth phenotype of miR-mdh-1, we analyzed growth of wild-type and miR-mdh-1 seedlings on agar medium with and without the NADH-GOGAT substrates Gln and 2-oxoglutarate (2-OG). As shown in Figure 7, miR-mdh-1 seedlings grew only slightly better on the supplemented medium, where the primary leaves had just appeared 3 weeks after germination, than on the control medium, where primary leaves were still absent. In contrast, wild-type seedlings developed much farther than miR-mdh-1 on both media, and the NADH-GOGAT substrates stimulated growth more than in miR-mdh-1. This result indicates that NADH-GOGAT cannot complement the miR-mdh-1 growth phenotype and that this phenotype is, thus, not a result of a decreased rate of plastidial NAD generation.
Figure 7.
Autotrophic growth of miR-mdh-1 and wild-type seedlings on medium with and without NADH-GOGAT substrates. Wild-type (A and B) and miR-mdh-1 (C and D) seeds were sown on agar medium containing (B and D) and lacking (A and C) the NADH-GOGAT substrates l-Gln (10 mm) and 2-OG (2.5 mm), stratified for 3 days, and then cultivated at standard growth conditions. Pictures were taken 3 weeks after germination. Bars = 1 cm.
DISCUSSION
The Role of pdNAD-MDH in Embryo Development and Heterotrophic Metabolism
pdNAD-MDH is crucial during embryo development, which is indicated by the facts that null mutant embryos arrested at the globular-to-heart transition stage and that the seeds were subsequently aborted (Fig. 2). Our data from plants carrying pdNAD-MDH promoter fusions with either pdNAD-MDH-eYFP or the GUS gene show that pdNAD-MDH is expressed in embryos at all examined stages of development (Fig. 3; Supplemental Fig. S1). Analysis of publicly available transcriptional data with the Genevestigator tool (www.genevestigator.com; Hruz et al., 2008) confirmed high expression levels of pdNAD-MDH in the embryo (Supplemental Fig. S2).
The absence of pdNAD-MDH in gametophytes did not affect fertility, which was indicated by the transmission rates of 50% in the progeny of reciprocal crosses between wild-type and heterozygous pdnad-mdh plants. However, it is not a rare phenomenon. Many of the embryo-defective mutants listed in the SeedGenes database (www.seedgenes.org; Tzafrir et al., 2003) are not defective in gametophyte development (Muralla et al., 2011). Two different explanations have been proposed. First, gene products originating from diploid, heterozygous sporocytes could be sufficient to complement the loss of gene expression in mutant gametophytes. As a consequence, the lethal phenotype would only appear after exhaustion of the parental gene products. Second, the mutation could cause a defect in a cellular process that is only essential at or after a certain developmental stage. In fact, proplastids have been shown to have significant metabolic activity and play a vital role in development of meristematic tissues (Aach et al., 1997).
Several central processes characterize the globular-to-heart transition stage of embryo development. A prominent example is the transition of proplastids to chloroplasts and thus, the acquisition of an active photosynthetic apparatus (Mansfield and Briarty, 1991; West and Harada, 1993; Devic, 2008). Concomitant with the transition from proplastids to chloroplasts are processes, such as increased plastidial fatty acid biosynthesis for thylakoid membrane production, that require a high turnover of ATP and the reducing equivalents NADH and NADPH (Mudd et al., 1987; Rawsthorne, 2002; Dörmann, 2006). Given that this stage is the stage of developmental arrest in the mutant, it is reasonable to propose a key role for the NAD-linked malate-OAA shuttle enzyme in chloroplast development in these heterotrophic tissues. In fact, functional chloroplasts are a prerequisite for embryo development (Uwer et al., 1998; Despres et al., 2001; Apuya et al., 2002; Tzafrir et al., 2004).
Previous reports indicate that, in vitro, pdNAD-MDH preferably catalyzes the reduction of OAA to malate (Scheibe, 2004; Cvetić et al., 2008). Thus, regeneration of the electron acceptor NAD inside the plastid was proposed as the most likely role for pdNAD-MDH. However, the reaction direction will depend on the metabolic fluxes in vivo, and we cannot rule out the possibility that pdNAD-MDH catalyzes the reverse reaction generating NADH to be used for cellular processes, like fatty acid synthesis. Indeed, it was shown that indirect import into plastids of reducing equivalents from the cytosol is an additional source of NADH for fatty acid biosynthesis in nonphotosynthetic tissues (for review, see Rawsthorne, 2002 and Dörmann, 2006). Malate, generated by MMDH during leaf respiration, could, thus, indirectly deliver reducing equivalents to plastids. Notably, the increased leaf malate content of miR-mdh-1 in the dark (Fig. 6B) now questions the originally proposed reaction direction. A straightforward explanation for this increase seems to be that, in the wild type at night, pdNAD-MDH catalyzed the oxidation of malate to OAA, thus generating NADH. However, because the amount and the oxidation state of the NAD(H) pool were unchanged between wild type and miR-mdh-1 (Fig. 6C), this proposition is dubious. It is possible, however, that the flux through the NAD(H) pool is altered in miR-mdh-1 without causing an actual change in the oxidation state.
We tried to address the question of the actual role of pdNAD-MDH by analyzing seedling growth on agar medium with and without GOGAT substrates. NADH-GOGAT is one of the enzymes known to oxidize NADH to NAD inside the plastid by converting Gln and 2-OG to two molecules of Glu. If pdNAD-MDH is, as proposed previously (Scheibe, 2004), primarily needed to supply the electron acceptor NAD for GAPDH to allow for continued production of ATP, NAD supplied by NADH-GOGAT could possibly complement the anticipated NAD shortage in miR-mdh-1. As shown in Figure 7, miR-mdh-1 seedlings on agar medium containing Gln and 2-OG developed only slightly further than seedlings on agar medium without supplement, whereas wild-type seedlings apparently grew better on the supplemented media. Assuming that the addition of Gln and 2-OG led to increased NADH-GOGAT activity, we propose that additional plastidial NAD supply cannot rescue the miR-mdh-1 growth phenotype. Together with the increased nocturnal malate levels in miR-mdh-1, this result suggests that the key role of pdNAD-MDH in the dark and in heterotrophic tissues is the supply of NADH for central plastidial processes (for example, fatty acid biosynthesis). However, while our manuscript was in revision, another study was published that deviates from this proposition (Selinski et al., 2014). This study shows that transcript levels of plastidial GAPDH were decreased by 50% in heterozygous pdnad-mdh plants and conversely, that pdNAD-MDH expression was reduced in gapdh knockout plants (Selinski et al., 2014). Hence, Selinski et al. (2014) reasoned that pdNAD-MDH is the proposed malate valve enzyme providing NAD for GAPDH in the dark, thus ensuring continued ATP supply in the plastid. The divergent conclusions from the two studies show that the exact pdNAD-MDH reaction has not unambiguously been resolved, indicating the need for additional research. The other data presented in the work by Selinski et al. (2014) are partly confirmative of our work (e.g. embryo lethality of homozygous pdnad-mdh, plastidic localization, and tissue-specific expression of pdNAD-MDH) and partly complementary, because they focus on transcriptional rather than biochemical or physiological analyses.
To analyze the impact of pdNAD-MDH deficiency on redox homeostasis more closely, we measured the levels of the key metabolites determining the intracellular redox state (glutathione and pyridine nucleotides) and ascorbate, a major antioxidant in plant cells, in leaves of miR-mdh-1 compared with the wild type. Similar to the invariant NAD(H) pool mentioned above, we could not detect significant changes in abundance and oxidation states of the NADP(H) and ascorbate pools (Fig. 6). A drastic change in redox maintenance is, therefore, unlikely to be the ultimate cause for the miR-mdh-1 phenotype. However, nocturnal glutathione increased in abundance and was more oxidized in miR-mdh-1 compared with the wild type (Fig. 6F). Glutathione is a vital cellular thiol-disulfide buffer, and depletion of glutathione synthesis causes embryo lethality in Arabidopsis (Cairns et al., 2006). Our metabolite measurements represent the average concentrations through all cellular compartments. For instance, in Arabidopsis mesophyll leaf cells, approximately 30% of the total cellular glutathione was estimated to localize to chloroplasts (Queval et al., 2011). It is, thus, reasonable to assume that local concentrations within chloroplasts are not appropriately reflected by our data. The actual redox state inside chloroplasts might, therefore, be more compromised than it seems and contribute significantly to the observed phenotype of miR-mdh-1. Glutathione reduction is catalyzed by glutathione reductase in an NADPH-dependent reaction; therefore, pdNAD-MDH is not directly involved in glutathione reduction. However, there is close interplay between glutathione, NAD(P), and ascorbate in plant metabolism (Noctor, 2006), indicating that pdNAD-MDH effects glutathione levels indirectly.
The Role of pdNAD-MDH in Photosynthetic Metabolism
In addition to its essential function for heterotrophic metabolism in embryos, we showed that silencing of pdNAD-MDH by amiRNA affected autotrophic metabolism as well. miR-mdh-1 plants were severely dwarfed and pale green compared with wild-type plants (Fig. 4; Table I). Chloroplast ultrastructure reflected the differences in chlorophyll levels: miR-mdh-1 chloroplasts have fewer thylakoid membranes and grana stacks, especially in young leaves, where the plastids are significantly smaller than in the wild type (Fig. 5). The fact that chloroplast malformation (and thus, the pale phenotype) is more severe in young, developing tissue is remarkable. Together with our observation that pdnad-mdh embryos arrest during the proplastid-to-chloroplast transition phase, they strongly suggest that pdNAD-MDH has a crucial function for chloroplast establishment. To date, the factors contributing to the attenuation of the pale phenotype as the leaves age were not known. From the expression data, there is no evidence that pdNAD-MDH activity would be more important at earlier stages of development, because expression is high during development of all the tissues analyzed (Supplemental Figs. S1 and S2). Other unknown factors must be involved, and additional research is needed to identify these factors.
The photosynthetic rate under standard growth conditions was decreased by 49% in miR-mdh-1 compared with the wild type (Table II). Chlorophyll fluorescence parameters indicate that this reduction is not caused by altered PSII efficiency (Supplemental Table S3): F0, Fm, and Fv were significantly reduced in miR-mdh-1, reflecting the low chlorophyll levels, but photochemical and nonphotochemical quenching was unchanged. When we repeated the experiments under high-light conditions, the results were very similar, although the differences between wild type and miR-mdh-1 were more pronounced. The photosynthetic rates increased compared with standard growth conditions by 54% and 18% in wild type and miR-mdh-1, respectively (Table II; Supplemental Table S4), indicating that light intensity at standard conditions was not saturating. Chlorophyll fluorescence data in high light indicate a tendency to reduced photochemical quenching in miR-mdh-1. Quantum yield of PSII photochemistry (ΦPSII), one of the two parameters for photochemical quenching, was significantly reduced in miR-mdh-1 (Supplemental Table S3). However, nonphotochemical quenching and the second parameter for photochemical quenching (Fv/Fm) were unchanged. Altogether, it seems unlikely that the mild reduction in photochemical quenching could account for the reduced photosynthetic rate in miR-mdh-1 at high light. We propose that the photosynthetic rate is, rather, reduced as a direct result of the low chlorophyll levels in miR-mdh-1. Moreover, pdNAD-MDH does not seem to play a crucial role in maintaining plastidial redox homeostasis in the light, hence confirming the previous proposition that pdNAD-MDH is not involved in the classical malate valve during photosynthesis (Scheibe, 2004). One has to keep in mind, however, that the wild-type and miR-mdh-1 plants used for these experiments were of different ages, making it difficult to draw ultimate conclusions.
The lower levels of starch, a major product of photosynthesis, in miR-mdh-1 at the end of the day (Fig. 6) also reflect the reduced photosynthetic rate and thus, are most likely a result of the reduced chlorophyll levels as well. Increased starch levels at the end of the night indicate a lower rate of starch mobilization in miR-mdh-1. The reduced respiration rate of miR-mdh-1 at night (Table II) is likely a consequence of this lack of storage carbohydrates. Mutants of Arabidopsis that cannot synthesize or degrade starch (phosphoglucomutase or starch-excess1, respectively) show a similar reduction in nighttime respiration (Paparelli et al., 2013).
The severities of the phenotypes reported here indicate that pdNAD-MDH loss cannot be rescued by other plastidial redox-poising mechanisms. All five Arabidopsis mdh single mutants published so far (nadp-mdh, mmdh1, mmdh2, pmdh1, and pmdh2; Pracharoenwattana et al., 2007; Tomaz et al., 2010; Hebbelmann et al., 2012) were viable and showed essentially no phenotypic differences to the wild type. This finding is at least partially caused by overlapping gene functions, which are indicated by the fact that double mutant plants lacking both mitochondrial or both peroxisomal isoforms exhibited distinct phenotypic differences to the wild type. However, in the plastid, only one NAD-dependent isoform is present, and NADP-MDH cannot compensate for the loss of pdNAD-MDH, confirming that NAD/NADH and NADP/NADPH pools have distinct functions.
CONCLUSION
In summary, we postulate that pdNAD-MDH fundamentally contributes to plastidial redox homeostasis in the dark and in heterotrophic tissues. We also confirm the previous assumption that pdNAD-MDH does not significantly contribute to the classical malate valve in the light. The fact that another recent publication also comes to the conclusion that pdNAD-MDH plays a crucial role in plastidial energy poise (Selinski et al., 2014) underlines the significance of our work. However, additional studies are needed to provide more insight into which of the metabolic or signaling processes linked to the redox state of the plastid are impaired, leading to the severe phenotypes that we have presented here. Inducible silencing of the pdNAD-MDH gene seems a particularly promising approach. Another interesting aspect is the fact that pdNAD-MDH contributes to three distinct activities on native gels (Fig. 4; Supplemental Fig. S6), which could indicate that pdNAD-MDH forms active complexes with other proteins. Testing this hypothesis and identifying potential interaction partners provide interesting prospects to further characterize pdNAD-MDH functions.
MATERIALS AND METHODS
Primer Sequences and Chemicals
Sequences of oligonucleotide primers used in this work are found in Supplemental Table S5. Unless stated otherwise, all chemicals used were from Sigma-Aldrich.
Plants and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study were grown in nutrient-rich, peat-based compost (Substrat 2; Klasmann-Deilmann) in controlled environment chambers (AR-95L; Percival Scientific) in a 12-h light/12-h dark regime with a uniform illumination of 150 µmol photons m−2 s−1, a constant temperature of 20°C, and constant 60% relative humidity (standard growth conditions). High-light experiments were done under standard growth conditions, except that illumination was 600 µmol photons m−2 s−1. The Arabidopsis enhancer trap line ET8629 was obtained from the Martienssen group at Cold Spring Harbor Laboratory (genetrap.cshl.org; Sundaresan et al., 1995). Arabidopsis mutants with T-DNA insertions in the mitochondrial (Tomaz et al., 2010) and peroxisomal (Pracharoenwattana et al., 2007) isoforms (mmdh1-2, mmdh2, mmdh1-2 mmdh2, pmdh1, pmdh2, and pmdh1 pmdh2) were a gift from Steven M. Smith (University of Western Australia). The Arabidopsis wild-type references used were Landsberg erecta for ET8629 (pdnad-mdh) and Columbia-0 (Col-0) for all other lines. Seedlings for GUS analyses were cultured on 0.8% (w/v) agar medium containing half-strength Murashige and Skoog medium (Duchefa Biochemie BV) under standard growth conditions, except that the photoperiod was 24 h. For growth analyses, seeds were sown on medium containing 0.8% (w/v) agar, 2.5 mm KCl, 1.25 mm KH2PO4, 1.25 mm (NH4)2SO4, 1 mm MgSO4, 1 mm CaCl2, 24 µm H3BO3, 20 µm Fe-EDTA, 4 µm MnCl2, 0.6 µm Na2MoO4, 0.4 µm ZnSO4, and 0.2 µm CuSO4. The pH was adjusted to 5.7 with KOH, and where indicated, the medium was supplemented with 10 mm l-Gln and 2.5 mm 2-oxoglutarate. Generation of transgenic Arabidopsis plants was by Agrobacterium tumefaciens-mediated transformation by floral dipping (Clough and Bent, 1998).
Southern Blot
The Ds insertion in ET8629 (pdnad-mdh) was analyzed by Southern blotting as described (Sambrook et al., 1989). The 859-bp probe was generated by PCR using the primers GUS_probe_fw and GUS_probe_rv, pGWB533 (Nakagawa et al., 2007) as the template, and DIG labeling was with DIG DNA Labeling Mix (Roche Applied Science). Chemiluminescence was detected using CDP-Star substrate (Roche Applied Science) and a FluorChem SP imaging system (ProteinSimple).
Generation of Silencing Lines and Silencing-Insensitive Lines
Selection of the silencing target sequence (GAG CTC ACT GTT AGG ATT CAG; positions 847–867 of the pdNAD-MDH coding sequence) and design of the oligonucleotide primers were done according to the instructions of the Web MicroRNA Designer tool (WMD3; wmd3.weigelworld.org; Ossowski et al., 2008). Cloning of the silencing construct was performed exactly as described in the standard protocol from the WMD3 Web site using the primers amiRNA oligo I, II, III, and IV. A 424-bp EcoRI-BamHI fragment of the resulting construct was ligated into pHannibal (Wesley et al., 2001). A 2,567-bp NotI fragment from the resulting vector, consisting of the CaMV 35S promoter, the amiRNA silencing construct, and the octopine synthase terminator, was ligated into the NotI site of the binary vector pART27 (Gleave, 1992), giving rise to the plant expression vector used for transformation of Arabidopsis Col-0 plants.
miRins-MDH was generated by silent exchanges of 6 nucleotides of the amiRNA target sequence (exchanges are underlined: GAA CTT ACA GTG AGA ATC CAG) by PCR. After addition of attachment sites (attB) by PCR (primers attB MDH fwd and attB MDHnoStop rev), miRins-MDH was first recombined into pDONR221 (Invitrogen) and finally, recombined into the binary vector pEARLEY101 (Earley et al., 2006) for PCaMV 35S-mediated expression after transformation of miR-mdh-1 by floral dipping (Clough and Bent, 1998).
Generation of PpdNAD-MDH::GUS Lines
The binary vector pEX/B4/PMDH/B1/GUS/B2 for expression of a transcriptional fusion of the pdNAD-MDH promoter and the GUS coding sequence in Arabidopsis was generated using the MultiSite Gateway Technology (Invitrogen). The 1.8-kb genomic DNA region upstream of the pdNAD-MDH ATG was amplified by PCR using attB linker primers (pMDH_attB4_fw and pMDH-1_attB1r_rv). The following recombination reaction with pDONR P4-1r (Invitrogen) yielded the entry clone pEN/L4/PMDH/R1. The 1.85-kb GUS coding sequence was amplified by PCR using primers gusA_fw and gusA_rv, and the vector pGWB533 (Nakagawa et al., 2007) as the template, and it was subcloned into pJet1.2 (Thermo Fisher Scientific). After attachment of attB sites by PCR (primers gusA_attB1_fw and gusA_attB2_rv), the BP reaction with pDONR 221 yielded the entry clone pEN/L1/GUS/L2. Recombination of pEN/L4/PMDH/R1 and pEN/L1/GUS/L2 with the destination vector pB7m24GW,3 (Karimi et al., 2005; gateway.psb.ugent.be) in the following recombination reaction yielded the final binary vector pEX/B4/PMDH/B1/GUS/B2 used for transformation of heterozygous pdnad-mdh.
Generation of PpdNAD-MDH::pdNAD-MDH::eYFP Lines
The binary vector for in planta expression of a translational fusion of pdNAD-MDH and eYFP under control of the endogenous pdNAD-MDH promoter, pEX/B4/PMDH/B1/pdNAD-MDH/B2/eYFP/B3, was generated using the MultiSite Gateway Technology. Because the pdNAD-MDH gene has only one intron in the 5′ UTR, the coding sequence was amplified from genomic DNA of Arabidopsis Col-0 using the primers GenChlMDH up and GenChlMDH down 2 and subcloned into pJet1.2. After the addition of attB linkers by PCR (primers attB MDH forward and attB MDHnoStop rev), the BP reaction with destination vector pDONR 221 yielded the entry clone pEN/L1/MDHnoSTOP/L2. The eYFP coding sequence was amplified by PCR using primers eYFP_fw and eYFP_STOP_rv, and the vector p2YGW7 (Karimi et al., 2005; gateway.psb.ugent.be) as a template; attB sites were attached by PCR (primers eYFP_attB1_fw and eYFP_attB2_rv), and the following recombination reaction with the destination vector pDONR P2r-P3 (Invitrogen) resulted in the entry clone pEN/R2/eYFPstop/L3. The final binary vector for in planta expression, pEX/B4/PMDH/B1/pdNAD-MDH/B2/eYFP/B3, resulted from a recombination reaction with the entry clones pEN/L4/PMDH/R1, pEN/L1/MDHnoSTOP/L2 and pEN/R2/eYFPstop/L3, and the destination vector pB7m34GW,0 (Karimi et al., 2005; gateway.psb.ugent.be).
Photosynthetic Measurements
Gas exchange of whole Arabidopsis rosettes was measured using a custom-built system consisting of eight chambers connected in parallel to an infrared gas analyzer (LI-7000; Licor). For each measurement, four plants each of the wild type and the pdNAD-MDH silencing line miR-mdh-1 were introduced into the chambers, and after an adaptation period of 2 d, gas exchange was measured for a complete 12-h-light/12-h-dark cycle. Air with 380 μL L−1 CO2 and 65% relative humidity was channeled through the chambers with a constant flow of 200 µmol s−1. CO2 concentration and relative humidity of in- and outgoing air were recorded every 30 s. The eight chambers were measured consecutively for a period of 6 min each. The projected leaf area was determined by taking pictures at the end of the light period followed by pixel counting using ImageJ software (Schneider et al., 2012). A and E were calculated according to the work by von Caemmerer and Farquhar (1981) on the basis of ΔCO2, ΔH2O, and the projected leaf area. Ultimately, both A and E of individual rosettes were averaged over day and night, respectively. For fresh weight determination, the rosettes were harvested 4 h after dawn, balanced, immediately frozen in liquid nitrogen, and stored at −80°C.
Chlorophyll and Chlorophyll Fluorescence Determinations
Chlorophyll measurements were performed on the plants used for photosynthetic measurements. Approximately 50 mg of pulverized plant material was extracted four times for 30 min on ice in the dark in 1 mL 80% (v/v) buffered (2.5 mm Tris, pH 7.8) acetone each. The four extractions were combined; chlorophyll a and b were measured on a Lambda 25 UV/Vis spectrophotometer (Perkin Elmer) and calculated as described (Porra et al., 1989). Chlorophyll fluorescence parameters were measured using a FluorCam system (Photon Systems Instruments) as described (Stettler et al., 2009).
Metabolite Measurements
Metabolite levels were determined as described in the works by Kötting et al. (2009; starch), Nunes-Nesi et al. (2007; malate), and Queval and Noctor (2007; NADH, NAD+, NADPH, NADP+, ascorbate, dehydroascorbate, reduced glutathione, and oxidized glutathione).
Enzyme Activity Measurements
NAD-dependent MDH activity was determined in two ways. First, total NAD-MDH activity was measured spectrophotometrically as described (Tomaz et al., 2010). Second, gel-based assays were performed as follows. Arabidopsis leaves were extracted in all-glass homogenizers in ice cold 100 mm 3-(N-Morpholino)-2-hydroxypropanesulfonic acid (pH 7.2), 1 mm EDTA, 10% (v/v) ethylene glycol, and 2 mm dithiothreitol at a ratio of 5 µL buffer/mg fresh weight, adjusted to the same protein concentration with extraction buffer, mixed with 2× loading dye (50% [v/v] glycerol and 0.05% [w/v] bromophenol blue), and separated on 8% (w/v) nondenaturing polyacrylamide gels at 4°C and 15 mA per gel (minigel, 1 mm; Bio-Rad) until the blue dye eluted followed by additional separation for 15 min (total: ∼300–400 Vh). Gels were incubated in 100 mm Tris (pH 8.0), 13.3 mm MgCl2, 1 mm l-malate, 0.5 mm NAD (β-NAD hydrate), 0.4 mm nitrotetrazolium blue chloride, and 27 µm N-methylphenazonium methyl sulfate at 25°C until blue formazan precipitate appeared (20–60 min). Reaction was stopped by washing the gels three times in water.
Antibodies and Western Blots
First antibodies α-pdNAD-MDH1 and α-pdNAD-MDH2 were raised against two pdNAD-MDH peptides (VESSLRALDGDGDVC and CVLKKKGVYDPKKLF, respectively) in rabbits (GenScript Corporation) and immunopurified using immobilized recombinant pdNAD-MDH (Berkemeyer et al., 1998). Second antibody was goat α-rabbit IgG-horseradish peroxidase (Bio-Rad). Working concentrations of α-pdNAD-MDH1, α-pdNAD-MDH2, and α-rabbit IgG-horseradish peroxidase were 1:2,000, 1:2,000, and 1:10,000, respectively.
For standard immunoblot analyses, equal amounts of proteins were separated by SDS-PAGE, electroblotted onto polyvinylidene fluoride membrane, and probed with first and second antibodies. Chemiluminescence was detected using the ChemiGlow West Kit on a FluorChem SP imaging system (ProteinSimple). Immunoblot analyses of nondenaturing gels were essentially the same, except that, after electrophoresis, gels were incubated in 20 mm Tris, 150 mm Gly, and 1% (w/v) SDS at 75°C for 5 min and briefly rinsed with blotting buffer before electroblotting. Antibody specificity is shown in Supplemental Figure S5.
Histochemical Detection of GUS Activity
Plant material was stained for GUS activity as described (Santelia et al., 2011), except that incubation in staining solution was for 1, 2, 6, or 24 h at 25°C. Images were taken as described below, and they represent GUS staining of at least three independent transgenic lines.
Microscopy
Dissection of embryos for fluorescence microscopy was done according to the instructions on the SeedGenes Web site (www.seedgenes.org/Tutorial; Tzafrir et al., 2003). Protoplasts for pdNAD-MDH-eYFP localization studies were isolated as described (Kötting et al., 2005). Fluorescence of eYFP (excitation: 500 ± 10 nm; emission: 535 ± 15 nm) and chlorophyll (excitation: 565 ± 15 nm; emission: 620 ± 30 nm) were recorded on an Axio Imager 2 microscope and analyzed with ZEN pro 2011 software (both Carl Zeiss). DIC pictures were taken with the same microscope and processed alike. Seeds were cleared with Hoyer medium before DIC microscopy as described (Stangeland and Salehian, 2002). Procedures for transmission electron microscopy were as described (Stettler et al., 2009), except that analyses were performed on a Morgagni 268 transmission electron microscope (FEI). GUS activity in tissues other than seeds and embryos was recorded with a VHX-1000D Digital Microscope (Keyence) or a D700 digital camera (Nikon).
Quantitative RT-PCR
RNA extraction, cDNA synthesis, and quantitative PCR were performed as described (Santelia et al., 2011). Relative transcript levels of the following genes were successfully determined using the oligonucleotide primers listed in Supplemental Table S5: pdNAD-MDH (At3g47520), NADP-MDH (At5g58330), MMDH1 (At1g53240), MMDH2 (At3g15020), PMDH1 (At2g22780), PMDH2 (At5g09660), CMDH1 (At1g04410), CMDH2 (At5g43330), and protein phosphatase 2A (At1g13320). Transcript of CMDH3 (At5g56720) could not unequivocally be detected.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_114620/AY128281.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Spatial and temporal expression patterns of pdNAD-MDH.
Supplemental Figure S2. Expression levels of pdNAD-MDH in different tissue types.
Supplemental Figure S3. Diurnal expression levels of pdNAD-MDH.
Supplemental Figure S4. Phenotypic appearance of different miR-mdh silencing lines.
Supplemental Figure S5. Specificities of two anti-pdNAD-MDH antibodies.
Supplemental Figure S6. Identification of distinct NAD-MDH activities.
Supplemental Figure S7. Complementation of the miR-mdh-8 silencing line.
Supplemental Figure S8. Images of plants used for photosynthetic measurements.
Supplemental Table S1. Relative expression levels of pdNAD-MDH (At3g47520).
Supplemental Table S2. Relative expression levels of MDH genes in miR-mdh-1 and wild-type plants.
Supplemental Table S3. Chlorophyll fluorescence parameters of miR-mdh-1 and wild-type plants.
Supplemental Table S4. Gas exchange measurement of miR-mdh-1 and wild type in high-light conditions.
Supplemental Table S5. Nucleotide sequences of primers used in this work.
Acknowledgments
We thank John Bussel and Steven M. Smith (University of Western Australia) for providing seeds of mitochondrial and peroxisomal MDH mutants, Renate Scheibe (University of Osnabrück) for providing the pdNAD-MDH expression clone, and our colleagues at Eidgenössiche Technische Hochschule Zürich for help with plant maintenance (Andrea Ruckle), gas exchange measurements (Katharina Kölling), MultiSite Gateway cloning (Nathan Pumplin), and embryo dissection (Pawel Roszak).
Glossary
- DIC
differential interference contrast
- EC
European Community
- F0
initial (minimum) PSII fluorescence in the dark-adapted state
- Fm
maximum PSII fluorescence in the dark-adapted state
- Fv
variable PSII fluorescence in the dark-adapted state
- GOGAT
glutamate synthase
- MDH
malate dehydrogenase
- 2-OG
2-oxoglutarate
- RT-PCR
reverse transcription PCR
- Col-0
Columbia-0
- UTR
untranslated region
- amiRNA
artificial mircoRNA
Footnotes
This work was supported by Eidgenössische Technische Hochschule Zürich (S.B., M.S., T.S., S.E., S.C.Z., and O.K.), the Swiss-South African Joint Research Program (grant no. IZ LS Z3122916 to S.B., S.C.Z., and O.K.), the National Science Council (grant no. 100-2311-B001-006), and Academia Sinica, Taiwan (H.-C.L., W.-L.L, and J.C.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aach H, Bode H, Robinson DG, Graebe JE. (1997) ent-Kaurene synthase is located in proplastids of meristematic shoot tissues. Planta 202: 211–219 [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Goldberg RB. (2002) RASPBERRY3 gene encodes a novel protein important for embryo development. Plant Physiol 129: 691–705; erratum Apuya NR, Yadegari R, Fischer RL, Harada JJ, Goldberg RB. (2002) Plant Physiol 130: 1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhausen JE, Vetter S, Baalmann E, Kitzmann C, Scheibe R. (1998) NAD-dependent malate dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta 205: 359–366 [Google Scholar]
- Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. (2006) Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci 11: 124–132 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Chua NH. (1989) Regulated genes in transgenic plants. Science 244: 174–181 [DOI] [PubMed] [Google Scholar]
- Berkemeyer M, Scheibe R, Ocheretina O. (1998) A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. J Biol Chem 273: 27927–27933 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. (2006) Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cousins AB, Pracharoenwattana I, Zhou W, Smith SM, Badger MR. (2008) Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol 148: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custers JBM, Snepvangers SCHJ, Jansen HJ, Zhang L, van Lookeren Campagne MM. (1999) The 35S-CaMV promoter is silent during early embryogenesis but activated during nonembryogenic sporophytic development in microspore culture. Protoplasma 208: 257–264 [Google Scholar]
- Cvetić T, Veljović-Jovanović S, Vucinić Ž. (2008) Characterization of NAD-dependent malate dehydrogenases from spinach leaves. Protoplasma 232: 247–253 [DOI] [PubMed] [Google Scholar]
- Despres B, Delseny M, Devic M. (2001) Partial complementation of embryo defective mutations: a general strategy to elucidate gene function. Plant J 27: 149–159 [DOI] [PubMed] [Google Scholar]
- Devic M. (2008) The importance of being essential: EMBRYO-DEFECTIVE genes in Arabidopsis. C R Biol 331: 726–736 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Pfannschmidt T. (2011) Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 155: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P. (2006) Lipid synthesis, metabolism and transport. In Wise RR, Hoober JK, eds, The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 335–353 [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH. (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C. (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim Biophys Acta 1100: 217–234 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hebbelmann I, Selinski J, Wehmeyer C, Goss T, Voss I, Mulo P, Kangasjärvi S, Aro EM, Oelze ML, Dietz KJ, et al. (2012) Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J Exp Bot 63: 1445–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber U. (1974) Metabolite exchange between chloroplasts and cytoplasm. Annu Rev Plant Physiol 25: 393–421 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huanca-Mamani W, Garcia-Aguilar M, León-Martínez G, Grossniklaus U, Vielle-Calzada JP. (2005) CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 17231–17236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, et al. (2009) STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance C, Rustin P. (1984) The central role of malate in plant metabolism. Physiol Veg 22: 625–641 [Google Scholar]
- Lancien M, Martin M, Hsieh MH, Leustek T, Goodman H, Coruzzi GM. (2002) Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J 29: 347–358 [DOI] [PubMed] [Google Scholar]
- Lee CP, Eubel H, O’Toole N, Millar AH. (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Logan BA. (2006) Oxygen metabolism and stress physiology. In Wise RR, Hoober JK, eds, The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 539–553 [Google Scholar]
- Mansfield SG, Briarty LG. (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Mudd JB, Bishop DG, Sanchez J, Kleppinger-Sparace KF, Sparace SA, Andrews J, Thomas S. (1987) Biosynthesis of chloroplast glycerolipids. In Fuller G, Nes WD, eds, Ecology and Metabolism of Plant Lipids. ACS Symposium Series, Vol 325 ACS, Washington, DC, pp 10–24 [Google Scholar]
- Muralla R, Lloyd J, Meinke D. (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Noctor G. (2006) Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ 29: 409–425 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Paparelli E, Parlanti S, Gonzali S, Novi G, Mariotti L, Ceccarelli N, van Dongen JT, Kölling K, Zeeman SC, Perata P. (2013) Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell 25: 3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson TR, Graham IA. (2005) Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J 43: 861–872 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. (2007) Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Queval G, Jaillard D, Zechmann B, Noctor G. (2011) Increased intracellular H₂O₂ availability preferentially drives glutathione accumulation in vacuoles and chloroplasts. Plant Cell Environ 34: 21–32 [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Rawsthorne S. (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41: 182–196 [DOI] [PubMed] [Google Scholar]
- Reumann S, Weber AP. (2006) Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim Biophys Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- Santelia D, Kötting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, et al. (2011) The phosphoglucan phosphatase Like Sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell 23: 4096–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R. (1987) NADP+-malate dehydrogenase in C3-plants: regulation and role of a light-activated enzyme. Physiol Plant 71: 393–400 [Google Scholar]
- Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Scheibe R, Backhausen JE, Emmerlich V, Holtgrefe S. (2005) Strategies to maintain redox homeostasis during photosynthesis under changing conditions. J Exp Bot 56: 1481–1489 [DOI] [PubMed] [Google Scholar]
- Scheibe R, Dietz KJ. (2012) Reduction-oxidation network for flexible adjustment of cellular metabolism in photoautotrophic cells. Plant Cell Environ 35: 202–216 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J, König N, Wellmeyer B, Hanke GT, Linke V, Neuhaus HE, Scheibe R. (2014) The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol Plant 7: 170–186 [DOI] [PubMed] [Google Scholar]
- Siebke K, Laisk A, Neimanis S, Heber U. (1991) Regulation of chloroplast metabolism in leaves: evidence that NADP-dependent glyceraldehydephosphate dehydrogenase, but not ferredoxin-NADP reductase, controls electron flow to phosphoglycerate in the dark-light transition. Planta 185: 337–343 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z. (2002) An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol Biol Rep 20: 107–114 [Google Scholar]
- Stettler M, Eicke S, Mettler T, Messerli G, Hörtensteiner S, Zeeman SC. (2009) Blocking the metabolism of starch breakdown products in Arabidopsis leaves triggers chloroplast degradation. Mol Plant 2: 1233–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS. (2002) Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol Biol 50: 463–474 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Miyake H. (2012) Redox-shuttling between chloroplast and cytosol: integration of intra-chloroplast and extra-chloroplast metabolism. Curr Opin Plant Biol 15: 252–260 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA. (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaz T, Bagard M, Pracharoenwattana I, Lindén P, Lee CP, Carroll AJ, Ströher E, Smith SM, Gardeström P, Millar AH. (2010) Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol 154: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T. (1998) Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10: 1277–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker A, Stierhof YD, Jürgens G. (2001) Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci 114: 3001–3012 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD. (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- West M, Harada JJ. (1993) Embryogenesis in higher plants: an overview. Plant Cell 5: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]