A new family of oil body associated proteins present in plants, fungi, and bacteria are involved in determining oil body size and shape.
Abstract
A transcriptomic approach has been used to identify genes predominantly expressed in maize (Zea mays) scutellum during maturation. One of the identified genes is oil body associated protein1 (obap1), which is transcribed during seed maturation predominantly in the scutellum, and its expression decreases rapidly after germination. Proteins similar to OBAP1 are present in all plants, including primitive plants and mosses, and in some fungi and bacteria. In plants, obap genes are divided in two subfamilies. Arabidopsis (Arabidopsis thaliana) genome contains five genes coding for OBAP proteins. Arabidopsis OBAP1a protein is accumulated during seed maturation and disappears after germination. Agroinfiltration of tobacco (Nicotiana benthamiana) epidermal leaf cells with fusions of OBAP1 to yellow fluorescent protein and immunogold labeling of embryo transmission electron microscopy sections showed that OBAP1 protein is mainly localized in the surface of the oil bodies. OBAP1 protein was detected in the oil body cellular fraction of Arabidopsis embryos. Deletion analyses demonstrate that the most hydrophilic part of the protein is responsible for the oil body localization, which suggests an indirect interaction of OBAP1 with other proteins in the oil body surface. An Arabidopsis mutant with a transfer DNA inserted in the second exon of the obap1a gene and an RNA interference line against the same gene showed a decrease in the germination rate, a decrease in seed oil content, and changes in fatty acid composition, and their embryos have few, big, and irregular oil bodies compared with the wild type. Taken together, our findings suggest that OBAP1 protein is involved in the stability of oil bodies.
The scutellum is a shield-like structure surrounding the embryo axis that occurs in all grass species, including early diverging taxa, and is generally considered as functionally equivalent to the cotyledon (Negby, 1984; Vernoud et al., 2005). The scutellum plays an important role in the hydrolysis and transport of endosperm-stored substrates during germination, but the scutellum itself also accumulates part of the seed reserves in special lipids (Tzen and Huang, 1992; Subbarao et al., 1998; White and Weber, 2003). Maize (Zea mays) scutellum only represents 11% of the mass of the kernel but accumulates 90% of the oil, 20% of the storage proteins, 10% of the sugars, and 90% of the phytate (Watson, 2003) and is the main source of vitamins and minerals of the kernel (Mazzolini et al., 1985; Lombi et al., 2011).
Triacylglycerols (TAGs) in seeds accumulate in special cytoplasmatic organelles called oil bodies (OBs; Murphy, 2001), which consist in a hydrophobic central core of neutral lipids, such as TAGs, surrounded by a monolayer of amphipathic phospholipids, glycolipids, and/or sterols, with a series of proteins bound to the surface of the OB (Purkrtova et al., 2008). OBs are also present in animals, fungi, and prokaryotes (Yang et al., 2012). Although the main role of OBs is to accumulate nutrients, increasing data indicate that in eukaryotic cells, OBs are also involved in other roles as lipid and protein trafficking between organelles (Raposo and Stenmark, 2008). Proteomic analyses suggest that OBs can serve as transient storage depots for proteins that lack appropriate binding partners in the cell and may provide a general cellular strategy for handling excess proteins (Cermelli et al., 2006). OBs also seem to be involved in the protection of plant embryos against freeze (Shimada et al., 2008).
A limited number of proteins are associated with OBs. Each type of organisms contains a specific set of them, for example, perilipins in mammal cytosolic OBs (Kimmel et al., 2010), 19 proteins in fungi (Grillitsch et al., 2011), and phasins in bacterial OBs (Yang et al., 2012). The most abundant plant proteins associated with OBs are oleosins (Capuano et al., 2007; Huang et al., 2009). Oleosins are tightly associated with OBs due to the presence of a highly hydrophobic central “core” composed of 70 to 80 hydrophobic and nonpolar amino acids (Murphy, 2001). In the center of this hydrophobic stretch lies a conserved motif containing three Pro residues that are crucial for the OB localization of oleosins (Abell et al., 1997). Oleosins maintain the integrity of the OBs and regulate their size by preventing them from coalescence (Tzen and Huang, 1992). Caleosins are a second group of plant proteins associated with OBs that also contain a hydrophobic core of about 30 amino acids and an E-F hand calcium-binding motif in its C-terminal end (Chen et al., 1999; Frandsen et al., 2001). Caleosins seem to have a role in OB degradation during germination (Poxleitner et al., 2006). Other plant OB-associated proteins are steroleosins, characterized by the presence of a NADPH-binding region and a soluble sterol-binding dehydrogenase domain (Lin et al., 2002). Oleosins are not present in green algae, but a lipid droplet surface protein was identified in the algae Nannochloropsis sp. with structural similarities to other OB-associated proteins (Vieler et al., 2012), and a major lipid droplet protein was also identified in Chlamydomonas reinhardtii and other green algae species (Moellering and Benning, 2010; Peled et al., 2011). Proteomic analysis in different plants and algae identified a wide variety of proteins associated with OBs (Katavic et al., 2006; Jolivet et al., 2009; Nguyen et al., 2011; Tnani et al., 2011, 2012). Different roles have been proposed to the OB-associated proteins, including OB biogenesis, stability, trafficking, and mobilization.
In this study, we identified the genes predominantly expressed in the scutellum during seed maturation compared with other parts of the seed. Among them, we identified a gene coding for a protein of unknown function we called OBAP1. Homologous genes are present in all plant species and in some fungi and bacteria. Here, we demonstrate that OBAP1 is associated with the OBs. Our results demonstrate that Oil Body Associated Protein1 (OBAP1) is necessary to maintain the structure of the OBs and for seed germination in Arabidopsis (Arabidopsis thaliana).
RESULTS
Identification of Maize Genes Predominantly Expressed in Scutellum
Scutellum samples were dissected from immature embryos 30 d after pollination (dap), which corresponds to the embryo maturation stage marked by the growth of the embryo and the accumulation of reserve substances (Vernoud et al., 2005). RNA was extracted and a complementary DNA (cDNA) library constructed. EST clones from the unamplified library were randomly picked, and those with inserts larger than 130 bp were sequenced from their 5′ ends. Supplemental Tables S1 and S2 list the 1,553 sequenced ESTs and their corresponding 789 genes and include accession numbers, functional category, and best BLASTP match. Fifty-five ESTs corresponded to transposable elements (3.5%). Sixty-seven percent of the ESTs corresponding to genes were singletons, and the rest assembled into contigs of two to 37 sequences. Among the 18 largest contigs composed of more than seven ESTs, the majority corresponded to zeins. Considering all genes with an identified function, 30% are involved in metabolism (25 in lipid metabolism) and 30% in protein synthesis and processing (Supplemental Table S3). To identify genes specifically or predominantly expressed in the immature scutellum, 618 clones corresponding to different genes were selected from the scutellum library (Supplemental Table S4). The inserts of the cDNA clones were PCR amplified and spotted onto nylon membranes. Macroarray hybridization experiments were carried out in triplicate with radio-labeled retrotranscribed probes synthesized with RNA extracted from dissected embryo axis, endosperm, and scutellum (30 dap). A gene was designated as being predominantly expressed in scutellum when, after normalization, the average signal intensity with scutellum probes was greater than 2.5 times that of embryo axis and of endosperm samples. Four genes showed a significantly higher expression in scutellum (Supplemental Table S4): two genes coding oleosins (oleosin I and II), a gene involved in defense against pathogens (antimicrobial maize basic peptide1 [MBP1]), and a gene of undetermined function (GRMZM2G044627). This last gene is represented by three clones in the scutellum library, which indicates a moderately high level of expression. We named it obap1 because, as we will show later, it encodes an Oil Body Associated Protein.
OBAP1 Belongs to an Evolutionary Conserved Family of Proteins
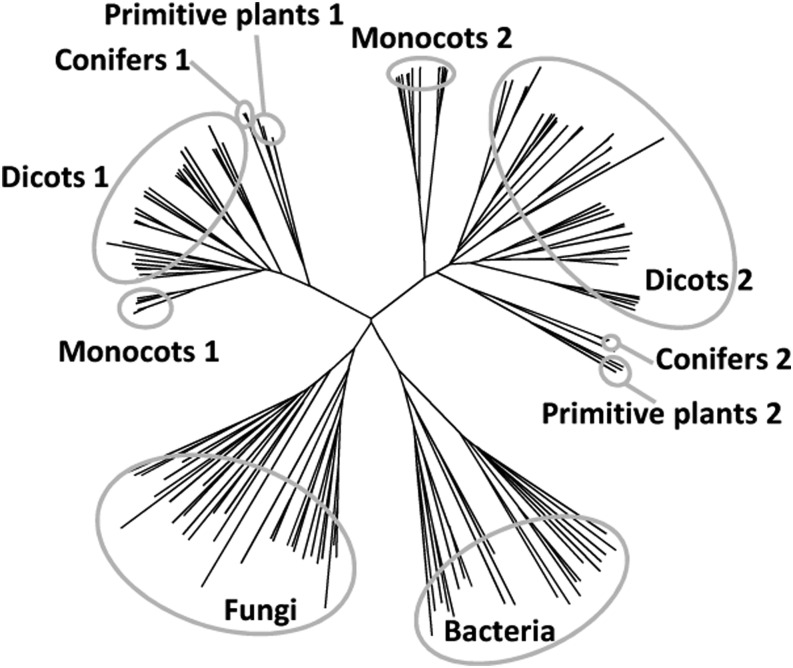
The obap1 gene is located in chromosome 4 (155907185–155909704) and contains one intron. Transcript sequence databases describe only one transcript variant, and the in silico expression database eFP Browser suggests that higher expression occurs in mature embryo (Sekhon et al., 2011). The maize genome contains two additional genes encoding proteins similar to OBAP1: obap2a (GRMZM2G043521), located in chromosome 3 and 33% similar, and obap2b (GRMZM2G107570), located in chromosome 8 and 36% similar. OBAP2A and OBAP2B are 63% similar between them. The genes obap2a and obap2b also contain one intron, and the in silico data indicate that their highest expression also occurs in mature embryo. A total of 235 protein sequences similar to maize OBAP1 were identified in databases corresponding to 153 species, 165 of them full-length sequences (Supplemental Table S5). They corresponded to several plant species including monocots, dicots, conifers, primitive plants, mosses, and algae (Liliopsida, Eudicotyledons, Coniferophyta, Filicophyta, Lycopodiophyta, Bryophyta, and Chlorophyta) but also to some fungi (Ascomycota, Basidiomycota, Oomycetes, and Zygomycetes) and some prokaryotes (Proteobacteria, Planctomycetes, Bacteroidetes, and Actinobacteria). Only one sequence corresponding to an animal species was found (accession no. FG621146). This sequence corresponds to an EST collection obtained from desiccated samples of the nematode Plectus murrayi (Adhikari et al., 2009). However, it is unlikely that this sequence really corresponds to a P. murrayi transcribed gene because similar genes are not present in the genomes of any other nematode species, including the fully sequenced genome of Caenorhabditis elegans, and because this sequence is 100% similar to a peanut (Arachis hypogaea) transcribed mRNA (accession no. EE126736), which strongly suggests that it corresponds to a contamination. In conclusion, according to the current available information, obap genes are not present in animals. Whereas in plants obap genes are present as small gene families, fungi and prokaryote genomes contain only one copy, although not in all species. For example, Escherichia coli and yeast (Saccharomyces cerevisiae) genomes do not contain obap genes. A phylogenetic tree based on all the available OBAP full-length sequences shows that there are two OBAP families in plants, one in fungi, and one in prokaryotes (Fig. 1; Supplemental Fig. S1). Both plant subfamilies contain genes from multiple species and representatives from monocotyledons, dicotyledons, conifers, and primitive plants. In general, monocot plants contain three copies of the gene, one from family 1 and two from family 2, and dicots, conifers, and primitive plants contain a variable number of genes distributed in the two subfamilies. For example, Arabidopsis genome contains two genes of subfamily 1 (obap1a and obap1b) and three genes of subfamily 2 (obap2a, obap2b, and obap2c). Inside each group, the sequences are distributed accordingly to the species phylogeny (Supplemental Fig. S2).
Figure 1.
Unrooted neighbor-joining tree based on aligned protein sequences of OBAP proteins from different species.
Expression Pattern of Maize obap1 Gene
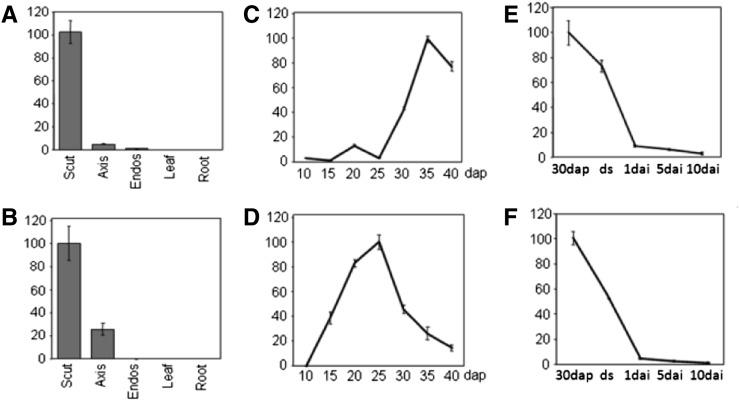
Quantitative reverse transcription-PCR experiments revealed that at 30 dap, maize obap1 mRNA is about 20-fold more abundant in the scutellum than in the embryo axis and about 100 times more in the scutellum than in the endosperm (Fig. 2A), confirming the macroarray results. No expression was detected in leaves or roots. A similar pattern was obtained for oleosin1 gene (Fig. 2B). Expression was also examined in embryos at different stages of development. In both genes, the highest expression was found in maturing embryos, but whereas obap1 has a maximum at about 35 dap (Figs. 2C), oleosin1 has a maximum about 10 d earlier (Fig. 2D). After imbibition, the mRNA accumulation in embryos in both cases decreases rapidly (Fig. 2, E and F).
Figure 2.
Relative abundance of maize obap1 and oleosin1 genes normalized to actin in different organs and developmental stages measured by quantitative reverse transcription-PCR. The sd of three repeats is shown. A, C, and E, obap1 mRNA accumulation. B, D, and F, oleosin1 mRNA accumulation. A and B, Total RNA from dissected parts of maize kernels 30 dap and dissected parts of 15-d-old seedlings (leaf and root). C and D, Total RNAs extracted from immature embryos at different dap. E and F, Total RNAs extracted from dissected scutellum of embryos at 30 dap, from dry seeds, and at different days after imbibition (dai). Scut, Scutellum; Axis, embryo axis; Endos, endosperm, ds, dry seeds.
The obap1a Gene in Brassicaceae
Five obap genes are present in the Arabidopsis genome. At1g05510 (AtObap1a) is the more similar to the maize obap1. According to Arabidopsis microarray databases (Arabidopsis eFP Browser), AtObap1a is highly expressed during seed maturation, its expression drops quickly after imbibition, and no expression was observed in other organs (Supplemental Fig. S3). An antibody was produced against Arabidopsis OBAP1a and used in western-blot analyses to study protein accumulation in different organs and during seed development and germination (Fig. 3). AtOBAP1 protein is not present in seeds at early stages of development but accumulated rapidly during seed maturation (Fig. 3A). Mature leaves and stems do not contain OBAP1a. Protein accumulation decreases rapidly after imbibition, becoming undetectable only 5 d after imbibition (Fig. 3B).
Figure 3.

Arabidopsis OBAP1a protein. A, Immunoblot of homogenates from Arabidopsis seeds at different stages of development (S1, globular stage, to S6, dry seeds) and from Arabidopsis mature leaves (Le) and stems (St). B, Immunoblot of homogenates from Arabidopsis seeds and seedlings at different days after imbibition.
Subcellular Localization of OBAP1 Protein
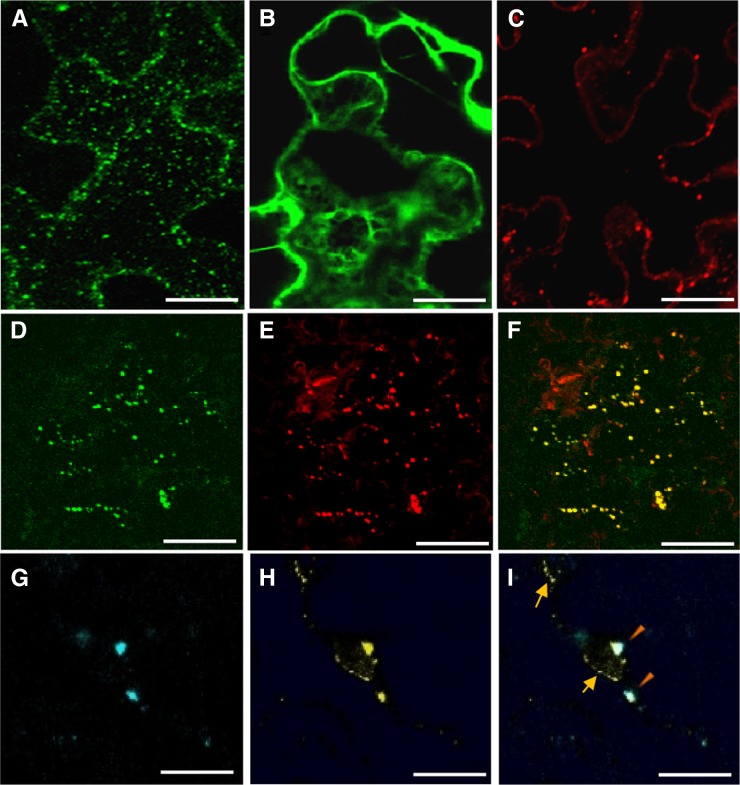
OBAP1 protein sequences do not contain any recognizable protein motif or transmembrane helix that could give us clues about its function. Maize OBAP1 (LOC 100193701) and Arabidopsis OBAP1a (At1g05510) proteins appeared in proteomic analysis of maize embryos and Arabidopsis seeds, and in both cases, their concentration increased during artificial ageing (Rajjou et al., 2008; Xin et al., 2011). Arabidopsis OBAP1a also appeared in a proteomic analysis focused in the identification of proteins whose Tyr phosphorylation varied significantly in seeds imbibed for 2 d in abscisic acid (Ghelis et al., 2008). These proteomic analysis did not provide any experimental data on the possible subcellular distribution of AtOBAP1a; however, the protein is defined by Ghelis et al. (2008) as a “putative lipoprotein.” Some of the homologous genes from prokaryote species are also described similarly in GeneBank, although no experimental data have been published for any of them. To clarify OBAP1 subcellular localization, the full-length ZmObap1 coding region was fused with the yellow fluorescent protein (YFP) coding region in C terminus under the control of 35S promoter and analyzed by confocal laser scanning microscopy after transient expression by agroinfiltration in tobacco (Nicotiana benthamiana) leaf epidermal cells. Confocal microscopy revealed YFP fluorescence in small organelles distributed in the cytoplasm (Fig. 4A). Fluorescence obtained with YFP alone shows a more uniform distribution in the cell (Fig. 4B). This punctuate distribution was similar to the distribution of OBs in tobacco cells as can be seen after Nile Red treatment that stains neutral lipids, and it has been used to localize OBs (Huang et al., 2009). Used in tobacco leaf epidermal cells, Nile Red stains spherical lipid-based particles distributed in the cytoplasm (Fig. 4C). When tobacco leaf epidermal cells agroinfiltrated with OBAP-YFP construct (Fig. 4D) were stained with Nile Red (Fig. 4E), both signals mainly colocalized (Fig. 4F), indicating that the punctuate structures in which OBAP1-YFP localizes correspond to OBs. Finally, we carried out coexpression experiments to verify the colocalization of OBAP1 with the well-known OB-associated protein OLEOSIN2 (Fig. 4, G–I). As expected, OLEOSIN2 fused to cyan fluorescent protein (OLEOSIN2-CFP) was localized in spherical bodies (Fig. 4G), and part of the OBAP-YFP protein colocalized with OLEOSIN2 (Fig. 4H). However, the ectopic expression of OLEOSIN2-CFP induces that part of the OBAP1-YFP protein that appeared to be located in aligned small spots (Fig. 4I) that do not contain OLEOSIN2-CFP (Fig. 4I).
Figure 4.
Subcellular localization of maize OBAP1 in tobacco cells. Confocal projection images of transiently transformed tobacco leaf epidermal cells. A, Tobacco cell transformed with OBAP1-YFP. B, Tobacco cell transformed with YFP. C, Nontransformed tobacco cell stained with Nile Red. D, Tobacco cell transformed with OBAP1-YFP. E, the same transformed tobacco cell stained with Nile Red. F, colocalization of OBAP1-YFP and Nile Red. G, Tobacco cell transformed with Oleosin2-CFP. H, The same tobacco cell transformed with OBAP1-YFP. I, Colocalization of Oleosin2-CFP and OBAP1-YFP. Arrows indicate endoplasmic reticulum-like structures, and triangles indicate OB-like structures. Bars = 10 μm (A–F) and 2 μm (G–I).
We used the anti-OBAP1 antibody to obtain further evidence of the association of OBAP1a to OBs. First, immunogold transmission electron microscopy was used to determine AtOBAP1a subcellular localization in mature rapeseed (Brassica napus L.) embryos. Gold labeling was observed predominantly in the surface of the OBs (Fig. 5). Second, we used the flotation centrifugation method (Katavic et al., 2006) to isolate the OB-enriched protein fraction from dry rapeseed seeds. The OB layer, the total lysate, and the cytosolic fraction were delipidated, precipitated, separated by SDS-PAGE, and blotted onto a nitrocellulose membrane for immunodetection (Fig. 6). Although the antibody raised against Arabidopsis OBAP1a recognized proteins present in the OB protein fraction, demonstrating the association of OBAP with the OBs, it also recognized proteins present in the cytosolic fraction.
Figure 5.
Ultrastructural localization of OBAP1 in rapeseed embryos. Immunolabeling of OBs with anti-OBAP1 serum. The position of OBs is indicated (ob). The black or white arrows indicate the position of the gold particles. A, B, C, and D images correspond to different rapeseed embryo samples treated similarly.
Figure 6.

Western-blot analysis of the total protein (TOT), OB fraction, and soluble protein fraction (SOL) of Arabidopsis mature seed extract using an anti-AtOBAP1a antibody. The arrow indicates the expected position of AtOBAP1a.
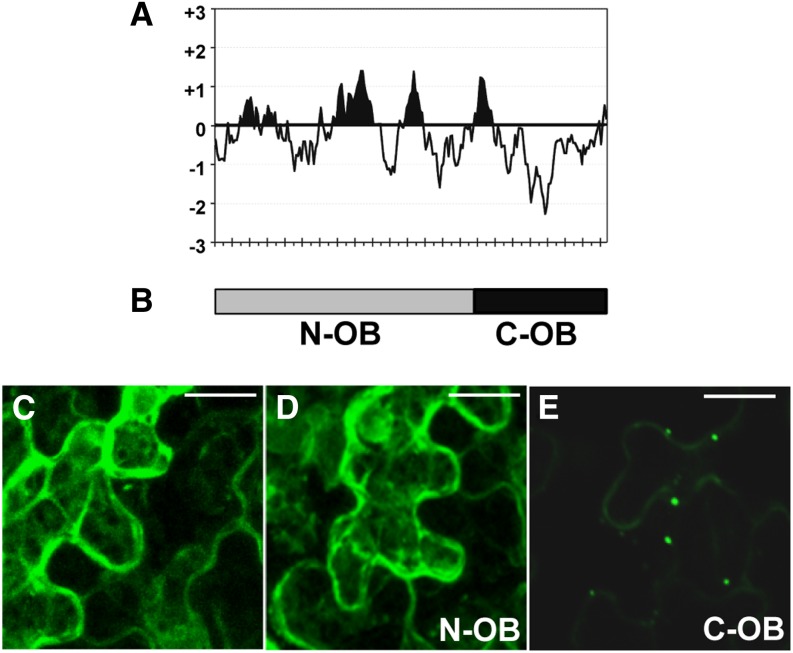
Confocal and transmission electron microscopy experiments indicated that OBAP1a is mainly located in the OBs. However, subcellular fractioning experiment raised some doubts in respect to this subcellular distribution. The association to OBs of other plant proteins such as oleosins and caleosins are known to be due to the presence of a long, central highly hydrophobic domain. Oleosins have a central hydrophobic domain of about 70 residues (Supplemental Fig. S4A; Sarmiento et al., 1997), caleosins of about 30 residues (Supplemental Fig. S4B; Naested et al., 2000), and steroleosins of about 40 residues (Supplemental Fig. S4C; Lin et al., 2002). These proteins also contain a characteristic Prorich motif. OBAP1 proteins do not contain a Prorich motif, and although hydropathy plots show some hydrophobic regions, they are short (less than 20 residues) and not as hydrophobic as in the other three plant OB-associated proteins (Fig. 7A). To determine if these short hydrophobic regions are the responsibility of the OB localization of OBAP1, we divided the obap1 coding region in two parts and fused them to the yfp coding region: the N-terminal part (N-OB) contains the hydrophobic regions, and the C-terminal part (C-OB) contains the most hydrophilic part of the protein (Fig. 7B). When introduced into tobacco leaf epidermal cells, the N-terminal part shows a subcellular distribution similar to YFP alone (Fig. 7, C and D), whereas the C-terminal part shows a punctuate distribution characteristic of the OB distribution (Fig. 7E). These results suggest that OBAP1 does not interact with the central lipidic domain of the OBs and that OB localization is due to another type of interaction as, for example, the interaction with another OB-associated protein.
Figure 7.
Mechanism of OB association of OBAP1. A, Kyte-Doolittle hydropathy index plot for maize OBAP1 protein calculated with window size of 15 amino acids. Each signal in the x axis represents 10 amino acids. B, Schematic representation of the two fragments of OBAP1 protein fused to YFP. C to E, Confocal projection images of transiently transformed tobacco leaf epidermal cells. C, Tobacco cell transformed with YFP. D, Tobacco cell transformed with N-OBAP1-YFP (N-OB). E, Tobacco cell transformed with C-OBAP1-YFP (C-OB). Bar = 8 μm.
Insertion Mutants and RNAi Lines for AtObap1a Gene
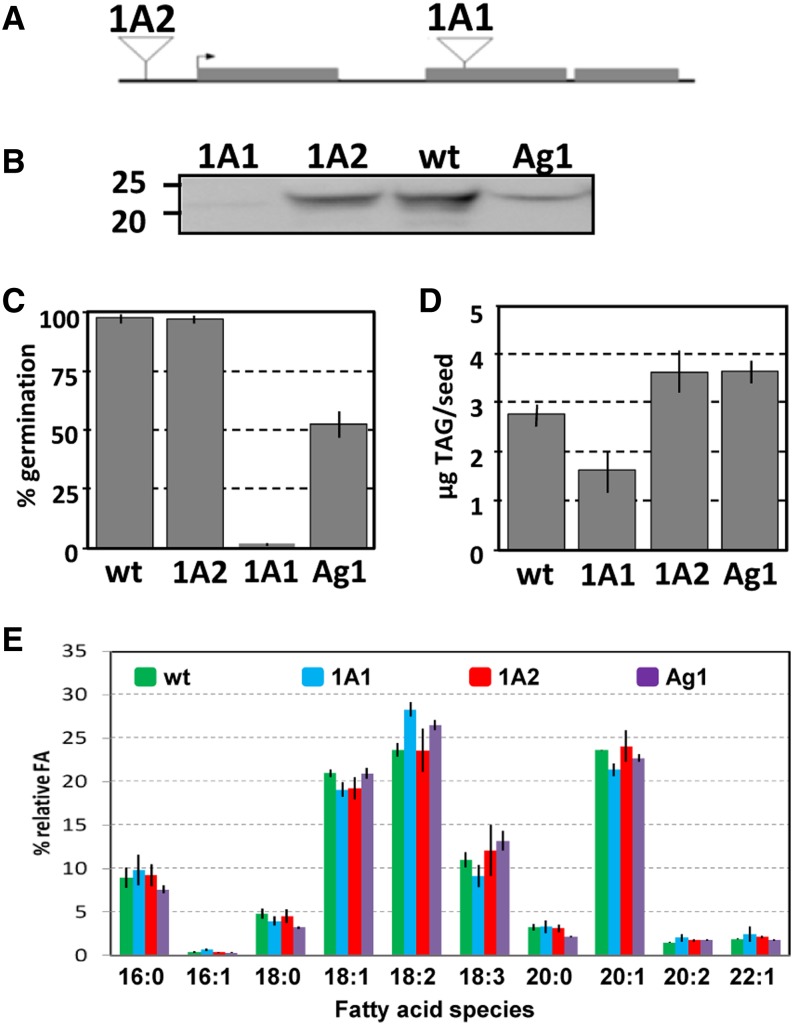
We used insertional mutagenesis and RNA interference (RNAi) approaches to determine the biological function of Arabidopsis OBAP1a. Public collections of transfer DNA (T-DNA) insertion in Arabidopsis were screened, and we have found two lines containing T-DNA insertions in AtObap1a gene. Seeds of these homozygous mutant lines were obtained from Nottingham Arabidopsis Stock Centre (NASC) T-DNA collection. One insertion was located in the second exon (Fig. 8A), and we designated it AtObap1a-1 (1A1). The second insertion was located about 500 bp upstream of the ATG, and we called it AtObap1a-2 (1A2). We have also used the RNAi method to reduce AtOBAP1a expression. We have produced an RNAi line specific to AtObap1a gene we called Ag1. Immunoblot analyses using anti-AtOBAP1a antibody showed that, whereas the abundance of OBAP1a protein in 1A2 seeds is not altered, the seeds of the 1A1 line do not contain detectable amounts of OBAP1a protein and the seeds from Ag1 line contain a reduced quantity (Fig. 8B). Whereas 1A2 seeds germinated similarly to the wild type, Ag1 seeds germinated poorly (about 50% compared with the wild type) and mutant 1A1 seeds germinated less than 2% compared with the wild type (Fig. 8C). After germination, no difference in seedling development was observed between Ag1 or 1A1 lines and the wild type. No significant differences were observed in the weight of the seeds of the different lines.
Figure 8.
Arabidopsis AtOBAP1a mutants and interference lines. A, Schematic representation of the position of the T-DNA insertions in the mutant lines 1A1 and 1A2 with respect to the Arabidopsis obap1a gene. The arrow indicates the ATG start codon. B, Immunoblot of homogenates from mature seeds of the Arabidopsis wild type (wt; ecotype Columbia) and insertion mutants 1A1 and 1A2 and from the RNAi line Ag1, with anti-OBAP1a antibody. C, Seed germination rates of the different lines. sds are indicated. D, TAG content of seeds of the different lines. E, Fatty acid composition of the TAGs of the seeds in the different lines. sds is indicated.
To investigate whether alterations in AtOBAP1a protein deposition would affect the synthesis of storage lipids, we performed biochemical analyses of seed TAG content and fatty acids (FA) composition. When compared with the wild type, we observed no differences in the total amount of TAG in 1A2 and Ag1 seeds but a significant reduction in 1A1 seeds, which contain about 60% TAG of the wild-type amount (Fig. 8D). Although there are quantitative differences in the accumulation of TAG, qualitative analyses of FA composition showed only minor statistically significant differences when compared with the wild type (P < 0.05 by Student’s t test; Fig. 8E). Whereas 1A2 seeds did not show any significant differences in overall FA profiles in TAG compared with the wild type, the seeds of the 1A1 and Ag1 lines contain significantly more linoleic acid (18:2; 20% increase in 1A1 and 12% increase in Ag1) and less eicosenoic acid (20:1; 10% reduction in 1A1 and 4% reduction in Ag1).
We conducted microscopy analyses to investigate the possible effects of different AtOBAP1a accumulation on the morphology of OBs. First, we extracted embryos from mature seeds of each line, stained them with Nile Red (which specifically stains neutral lipids), and observed them in the confocal microscope (Fig. 9, A–C). Whereas wild-type embryos showed a uniform distribution of relatively small OBs (Fig. 9A), embryos from 1A1 (Fig. 9B) and Ag1 (Fig. 9C) contained bigger OBs with an irregular shape and located in the central part of the cell. To obtain a more precise image of these changes in OB structure, the embryos were also observed using electron microscopy (Fig. 9, D–I). Compared with the wild type (Fig. 9, D and E), the OBs in embryos of 1A1 mutant (Fig. 9, F and G) and Ag1 RNAi line are enlarged and irregularly shaped (Fig. 9, H and I). The effect is more intense in the 1A1 insertion line compared with Ag1.
Figure 9.
OBAP1 deficiency causes enlargement of OBs in embryos. A to C, Confocal projection images of Arabidopsis mature embryos stained with Nile Red. A, Ecotype Columbia. B, T-DNA insertion mutant 1A1. C, RNAi line Ag1. D to I, Electron micrographs of Arabidopsis mature seeds. D to E, Ecotype Columbia. F and G, T-DNA insertion mutant 1A1. H to I, RNAi line Ag1. Some of the OBs are indicated by red arrows. Bars = 2 μm (D–G and I) and 1 μm (H).
DISCUSSION
Maize kernels contain about 4.5% oil, and about 90% is accumulated in the OBs of the scutellar parenchymal cells (Watson, 2003). In consequence, the maize scutellum could be considered as a good source to identify new genes and proteins involved in the synthesis and accumulation of storage lipids in plants. In 30-dap kernels, the scutellar cells did not further divide, and they only increase in size and accumulate storage molecules, including oil (Bommert and Werr, 2001). The peak of oleosin gene expression occurs about 25 dap (Fig. 2D). Functional categories of the expressed genes in 30-dap scutellum showed a predominance of metabolic functions and a low representation of genes involved in cell division and development. Genes coding for storage proteins and for proteins involved in protein synthesis and processing are also highly represented, reflecting the fact that scutellum also accumulates storage proteins. A transcriptomic analysis of dissected barley (Hordeum vulgare) embryos at a similar developmental stage also showed that ribosomal proteins and other components of the translation machinery are highly expressed in the scutellum (Potokina et al., 2002). We identified four genes predominantly expressed in scutellum and two of them encode oleosins, which is coherent with the oil storage function of this organ. Another gene encodes an antimicrobial peptide MBP1. Pathogens could try entering into the embryo through the endosperm, and scutellum seems to play a protective role against them. Accordingly, some genes involved in pathogen defense have been described to be expressed in scutellum: a barley gene coding for a β-1,-3-glucanase, a maize gene coding for a ribosome-inactivating protein, and the maize and wheat (Triticum aestivum) cystatin genes (Jutidamrongphan et al., 1991; Guo et al., 1999; Corr-Menguy et al., 2002).
No clues were available for the role of the protein encoded by the fourth identified gene. We have demonstrated here that the protein is associated with OBs, and we called it OBAP1. All the available expression data (generated by us or available in expression databases) indicate that obap genes are predominantly expressed during embryo development, and their expression pattern is very similar to the oleosin genes. Fusions of OBAP1 to fluorescent protein tags demonstrate that OBAP1 colocalizes with OBs in tobacco leaf epidermal cells, which contains bona fide OBs (Wahlroos et al., 2003). These results were confirmed by immunolocalization experiments of OBAP1a in electron microscopy samples of mature rapeseed embryos. Although oleosins are the most abundant plant OB-associated proteins, they are not the most unique. Caleosins (Chen et al., 1999) and steroleosins (Lin et al., 2002) are also localized in OBs, and the proteomic analyses on maize scutellum and Brassica napus and sunflower (Helianthus annus L.) seeds suggest that other less abundant OB-associated proteins must exist (Katavic et al., 2006; Hajduch et al., 2007; Tnani et al., 2012).
An apparently contradictory result was obtained in subcellular fractioning experiment in which OBAP1 proteins appear to be located in both the OB and the soluble fractions. However, something similar happens with oleosins, the more characteristic plant OB proteins, because about 5% to 10% of rapeseed oleosins were invariably targeted to the non-OB fraction in similar experiments (Sarmiento et al., 1997), and similarly, a proportion of Arabidopsis caleosin can be detected in the soluble fraction (Naested et al., 2000). Despite these, in situ experiments demonstrate that both proteins, as well as OBAP1 protein, are predominantly located in the surface of the OBs. The proportion of protein located in the OB fraction is much lower in the case of OBAP1 compared with oleosin and caleosin, and this may reflect that OBAP1 attachment to OBs is weaker. Oleosin and caleosin association to OBs is mediated by the presence of a long, central hydrophobic region; however, OBAP1 does not have long hydrophobic regions, and the fraction of the protein responsible for the interaction is the C terminus, corresponding to the most hydrophilic part of the protein. These data suggest that probably the interaction of OBAP1 with OBs is not mediated by a direct interaction of the protein with the central TAG core of the OB. Alternatively, OBAP1 may interact with OBs in a similar manner, as has been suggested with major lipid-droplet proteins in algae, that is, by the direct contact of its polar residues with the phospholipids or with other components of the surface of the OBs, like other proteins (Huang et al., 2013). Oleosins and caleosins contain a signal peptide and are directed to the endoplasmic reticulum and trafficked to the surface of OBs (Beaudoin and Napier 2002; Froissard et al., 2009). Unlike oleosins, OBAP1 does not have a signal peptide, which suggests that initially it accumulates in the cytoplasm rather than in the endoplasmic reticulum, and this is also coherent with the hypothesis of a superficial interaction of OBAP to the OBs once they have been formed. Coherently to this hypothesis, the pick of expression of OBAP1 in maize embryos occurs later than the pick of expression of oleosin.
When obap1 is coexpressed with oleosin2, part of the OBAP1 protein localizes in linear structures resembling endoplasmic reticulum. Interestingly, overexpression of oleosin has a similar effect on caleosin localization (DeDomenico et al., 2011). These results suggest the possibility that OBAP1 interacts with caleosins and not with oleosins. Homology searches indicate that obap genes are present in all plant species, including primitive plants, and in some fungi and prokaryotes. Oleosins are only present in plants (Huang et al., 2009), but caleosins are present in plants and some fungi (Murphy, 2005). Caleosin and OBAP are both present in, for example, Aspergillus niger, Ustilago maydis, Magnaporthe grisea, Neurospora crassa, or Chaetomium globosum but not in yeast. Further analyses will be necessary to determine this possible interaction.
Irregularly expanded oil-containing structures appear when OBAP1 is not present and seed germination rate and oil content decreases, whereas fatty acid composition is altered. Oleosin mutants produce similar changes (Siloto et al., 2006; Shimada et al., 2008), and authors suggested that OB expansion push the nuclei to the cell periphery and may prevent them from playing their roles, inducing cell degeneration and death (Shimada et al., 2008). On the other hand, not only total oil content, but also the average size of the OBs determines the germination rate because large OBs reduce the accessibility of lipases to the TAGs during germination (Siloto et al., 2006). From a qualitative point of view, the suppression of OBAP1 has a significant effect on fatty acid preferences in TAG, increasing 18:2 (linoleic acid) at the expense of 20:1 (eicosanoic acid). Similar alterations in the oil content and fatty acid composition of TAGs were observed in an Arabidopsis diacylglycerol acyltransferase1 mutant in which TAG biosynthesis is reduced (Katavic et al., 1995). All these data suggest that, similar to oleosins, OBAP1 protein is necessary to maintain OB integrity and may also exert some influence on qualitative and quantitative aspects of TAG accumulation in seeds.
Different studies have revealed that OBs, previously considered static storage depots of cellular neutral lipids, may be more active organelles (Beller et al., 2006; Sato et al., 2006; Bartz et al., 2007; Wan et al., 2007). Animal lipid droplets interact with various cellular organelles (peroxisomes, glyoxisomes, mitochondria, etc.; Liu et al., 2007), and lipid droplets have also been considered a refugee of proteins destined for destruction, such as toxins or viral proteins, or proteins that will be used when conditions change, such as signaling proteins (Welte, 2007). An interesting possibility is that OBAP mediates some of these interactions. Supporting this idea, the Arabidopsis Interactome Mapping Project, based on experimental yeast two-hybrid data, found that Arabidopsis OBAP1a, OBAP1b, and OBAP2c interact with TEOSINTE-BRANCHED1/CYCLOIDEA/PCF14 (TCP14) transcription factor. Arabidopsis OBAP1b is also predicted to interact with other transcription factors of the TCP family (TCP11 and TCP15) and with MEDIATOR OF ABSCISIC ACID-REGULATED DORMANCY1 (Arabidopsis Interactome Mapping Consortium, 2011). If confirmed, these interactions open several new possible functions for OBs and OBAP proteins in plants.
MATERIALS AND METHODS
Plant Materials and Plant Growth Conditions
Maize (Zea mays ‘W64A’) was grown under standard conditions (16 h/8 h at 26°C; 150 μmol m–2 s–1). Tobacco (Nicotiana benthamiana) was grown in soil under long-day conditions (16 h/8 h at 22°C; 150 μmol m–2 s–1). Seeds from Brassica carinata (N2-7399) and Glycine max (N85-2124) were obtained from the U.S. Department of Agriculture. The following SALK Arabidopsis (Arabidopsis thaliana) insertion mutants were acquired from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/): 1A1 (SALK_011689) and 1A2 (SALK_017397). The RNAi line Ag1 was requested to AGRIKOLA project (systematic RNAi knockouts in Arabidopsis; www.agrikola.org; Hilson et al., 2003).
For Arabidopsis germination assays, seeds were surface sterilized for 2 min with 75% ethanol in water (v/v), followed by 5 min in 1% NaClO solution, washed five times in sterile distilled water, and then placed onto 0.8% (w/v) agar in Murashige and Skoog medium, pH 5.7. Plates were kept for 2 d in the dark at 4°C to break dormancy (stratification) and transferred to 16-h/8-h photoperiod at 120 mol m–2 s–1 at 21°C. After 1 week, germination rate was evaluated. Four batches of 100 seeds were analyzed per line.
RNA Extraction, cDNA Library Construction, and Sequencing
Total RNA was extracted with TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions, treated with DNAse I and purified with RNAeasy Plant Mini Kit (Qiagen). A cDNA library was constructed using 1 μg of total RNA and the SMART cDNA Library Construction Kit (Clontech) with oligo(dT) priming. PCR-amplified cDNAs were ligated into pCRII-TOPO (Invitrogen) and introduced into Escherichia coli. Insert amplification was performed by PCR using the primers 5′-GGAAACAGCTATGACCATGATTACG-3′ and 5′-GTCACGACGTTGTTAAACGACGGC-3′.
Single-pass sequencing was performed from the 5′ end of the selected PCR products using the primer 5′-GTATCAACGCAGAGTCG-3′ at the Sequencing Service (Centre for Research in Agricultural Genomics). The EST sequences (accession nos. AM937535–AM939368) were compared against the public DNA and protein databases using BLASTN, BLASTX, and TBLASTX (Altschul et al., 1997) at National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and CoGeBLAST (http://genomevolution.org/CoGe/CoGeBlast.pl). The functional category classification was derived from the BLAST results based in Gene Ontology database (http://geneontology.org/).
Construction, Hybridization, and Data Analysis of cDNA Macroarrays
cDNA inserts were amplified by PCR using the primers 5′-ACTAGTTAATACGACTCACTATAGGGGGCCGAGGCGGCCGACATGTT-3′ and 5′-CCATGGTATTAGGTGACACTATAGAACGCAGAGTGGCCATTACGGCC-3′ in 96-well plates. Their concentration was adjusted to 100 ng µL–1 and denatured with 0.2 n NaOH. Spotting was done with a 96-pin Multi-Print Arrayer (V&P Scientific) using arrays of 36 × 24 spots and 200-nL hanging pins. Spots were printed onto Hybond-N+ nylon membranes (Amersham). After air drying, membranes were cross linked by UV radiation at 120 μJ sec–1 for 1 min using Stratalinker (Stratagene) and stored at 4°C until usage. Hybridization probes were prepared by the retrotranscription of 10 µg total RNA in presence of [α-32P]dCTP annealed with 1 µg oligo(dT)12-18 (Invitrogen), using SuperScript II RT (Invitrogen) for 70 min at 42°C. The RNA strand was hydrolyzed by adding 1 μL 5 m NaOH (37°C for 15 min), and the reaction was neutralized by adding 1 μL 5 m HCl and 5 μL 1 m Tris-HCl (pH 7.0). Probes were purified using Bio-Spin P-30 columns (Amersham). Prehybridization was done with MicroHyb hybridization buffer (Invitrogen), 10% dextran sulfate, and 100 μg mL–1 denatured salmon sperm DNA (42°C for 2 h). Hybridizations were done in the same buffer (42°C for 18 h). Washing was done in 2× SSC/0.1% SDS twice for 30 min at 65°C. Washed membranes were exposed to a phosphoimager screen (GE Healthcare) for 3 d, and the screens were scanned in a phosphoImager (Molecular Imager, Bio-Rad). Hybridizations were done in triplicate. Image files were imported into the Quantity-One program (Bio-Rad) for spot quantification. Normalization was done based on the average intensities of seven housekeeping genes: actin (AM937802), heat shock protein Hsp70 (AM939105), elongation factor 1-α (AM938565), translation initiation factor 5A (AM937959), polyubiquitin containing 7 ubiquitin monomer (AM938744), α-7 tubulin (AM938025), and ubiquitin extension protein 2 (AM938244). Statistical analysis was done using the Student’s t test (P < 0.05).
In Silico Analyses
Expression data on maize genes was obtained in Maize Genetics and Genomics Database (http://www.maizegdb.org/expression/expr_tools/expr_tools.php) and maize eFP Browser (http://bar.utoronto.ca/efp_maize/cgi-bin/efpWeb.cgi?dataSource=Sekhon_et_al; Sekhon et al., 2011). Expression data on Arabidopsis genes was obtained in the Arabidopsis eFP Browser (http://bbc.botany.utoronto.ca/efp; Winter et al., 2007), Genevestigator (https://www.genevestigator.ethz.ch/; Zimmermann et al., 2004), and AtGeneExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp; Schmid et al., 2005). Protein motif analyses were performed in SMART (http://smart.embl-heidelberg.de/; Letunic et al., 2012), TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000), ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999), SignalP (http://www.cbs.dtu.dk/services/SignalP/; Petersen et a., 2011), and TMpred (prediction of transmembrane regions and orientation; http://www.ch.embnet.org/software/TMPRED_form.html; Hofmann and Stoffel, 1993). Calculation of the hydropathy was done using ProtScale (http://www.expasy.ch/tools/protscale.html; Gasteiger et al., 2005) based on the Kyte and Doolittle index (Kyte and Doolittle, 1982).
The sequence of the maize OBAP1 protein was used as a query for the BLASP and TBLASTN programs against several sequence databases including GeneBank (http://blast.ncbi.nlm.nih.gov/), U.S. Department of Energy Joint Genome Institute (http://www.phytozome.net/search.php?show=blast), Plant Genomics Database (http://www.plantgdb.org/cgi-bin/blast/PlantGDBblast), GrainGenes (http://wheat.pw.usda.gov/GG2/index.shtml), the Gene Index Project (http://compbio.dfci.harvard.edu/tgi/), Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), Dana Farber Cancer Institute Maize Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=maize), The Institute for Genomic Research Rice Genome Annotation Project (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1), the Maize Genome Browser (http://www.maizesequence.org/Multi/blastview), and The Arabidopsis Information Resource (http://www.arabidopsis.org/). Taxonomical classification was determined according to the National Center for Biotechnology Information taxonomy homepage (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/). Amino acid sequences were aligned using ClustalW (Pole Bioinformatique Lyonnais: http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html). Phylogenetic tree was constructed using neighbor-joining method using Treecon program (Van de Peer and De Wachter, 1994).
Quantitative Real-Time Reverse Transcription-PCR
Total RNA was extracted from different tissues using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. From 2 µg of total RNA, DNA was synthesized by the Omniscript RT Kit (Qiagen) using an oligo(dT) primer according to the manufacturer’s instructions. cDNA samples were normalized by optical density 260, and PCR reactions were set up in a LightCycler 480 system (Roche Diagnostics) using SYBR Premix Ex Taq (TaKaRa) using 200 nm of the appropriate primers pairs and 2 µL of cDNA template up to a volume of 20 μL. Reactions included an initial denaturation step at 95°C for 30 s, followed by 50 cycles of 95°C for 5 s, 60°C for 20 s, and 72°C for 20 s. Melting-curve analysis was performed on each sample to ensure single-amplicon specificity. Actin was used as reference endogenous control for normalization purposes (primers 5′-TACCCAACTAAGCGCATGCC-3′ and 5′-GCATCTGAATCACGAAGCAGG-3′). Relative quantification was performed using standard curves from serial dilutions of cDNA. The efficiency for all genes tested was greater than 99%. All PCR reactions were performed in triplicate for each RNA sample. Relative quantification analyses were performed using the LightCycler 480 software. Gene-specific primers were designed using the Primer Express software (Applied Biosystems; http://www.appliedbiosystems.com/). Specific primers used were: for Obap1, 5′-CCGGTGTTCCTGATCTTCA-3′, 5′-GCACGTGAGCGAGGACAG-3′, and for oleosin1, 5′-TCCCGCAGATCATGATGGT-3′, 5′-CGAACTTCTTCTCCACACAGTCA-3′.
Construction of Transient Expression Clones and Subcellular Localization
DNA sequences encoding the complete coding regions of maize OBAP1 and Oleosin2 genes were amplified by PCR using the following primers: 5′-CACCATGGCGTCGTCGTGCCAGAAC-3′ and 5′-TTTAGTGAAGACCCTCCCGCC-3′ for OBAP1 and 5′-CACCATGGCGGACCGTGACCGCAGC-3′ and 5′-TTTCGAGGAAGCCCTGCCGCCGCCC-3′ for oleosin2. The resulting coding fragments were cloned into the Gateway binary vectors pYL-CFPct and pYL-YFPct, respectively, to be driven by a 35S promoter of Cauliflower mosaic virus (Rubio et al., 2005). Partial obap1 coding regions were obtained using the following primers: 5′-CACCATGGCGTCGTCGTGC-3′ and 5′-GTCCACCTGCCAGAAGTGGATGGT-3′ for N-OB, and 5′-CACCATGGCGTCGTCGTGC-3′ and 5′-GTCCACCTGCCAGAAGTGGATGGT-3′ for C-OB. The resulting fragments were cloned into the Gateway binary vector pYL-YFPct.
Plant Transformation
The floral dip method was used for Arabidopsis stable transformation (Clough and Bent, 1998). For transient transformation, tobacco leaves were agroinfiltrated by syringe method into the abbatial side of 3- to 5-week-old leaves (Voinnet et al., 2003). The binary plant vectors produced were mixed together with the HC-Pro silencing suppressor construct (Goytia et al., 2006). Three days after agroinfection, transformed leaves were observed.
Confocal Laser Scanning Microscopy
Confocal laser scanning microscopy images were acquired using an Olympus Fluoview FV10i inverted confocal laser scanning microscope using water immersion 60× objective. Fluorophore emissions were collected sequentially in double-labeling experiments. Single-labeling experiments showed no detectable crossover at the settings used for data collection. CFP was excited at 405 nm and was detected with the 460- to 500-nm filter set. YFP was excited at 515 nm and was detected with the 530- to 560-nm filter set. Nile Red was excited at 559 nm and was detected with the 570- to 670-nm filter set. For Arabidopsis embryo OB staining, Arabidopsis embryos were prepared from freshly harvested seeds, and OBs were stained with Nile Red as previously described (Greenspan et al., 1985).
Immunodetection of OBAP1a
The peptide antibody against Arabidopsis OBAP1a was produced by Abyntek Biopharma S.L. using the C-terminal peptide CEVDIKPVESVPRVFV. For immunodetection of OBAP1a, Arabidopsis tissues were homogenized in protein extraction buffer (50 mm Tris, pH 8.0, 2% SDS, and protease inhibitor cocktail). Aliquots of the extracts were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Proteins were probed against the anti-OBAP1a antibody and detected by alkaline phosphatase.
Electron Microscopy
Isolated embryos were fixed for 2 h in 2% (w/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde in phosphate buffer 0.1 m, pH 7.4. Following fixation, the embryos were washed with phosphate buffer 0.1 m, pH 7.4, for 10 min, resuspended in 1% (w/v) paraformaldehyde for 2 h, and rinsed three times with phosphate buffer. Embryos were postfixed for 2 h with 1% (w/v) osmium tetroxide containing 0.8% (w/v) potassium hexacyanoferrate prepared in phosphate buffer, followed by four washes with deionized water and sequential dehydration in acetone. All procedures were performed at 4°C. Samples were embedded in Spurr’s resin and polymerized at 60°C for 48 h. Ultrathin sections (70 nm) were obtained with a diamond knife (45°, Diatome). Sections were stained with uranyl acetate and Reynolds lead citrate for 10 min and examined with a JEOL 1400 transmission electron microscope equipped with a Gatan Ultrascan ES1000 CCD Camera. Embryos from three individuals were used in each analysis.
For gold immunolabeling, ultrathin sections were incubated in phosphate buffer with anti-OBAP1a antibody at 4°C overnight in a humidified chamber. The grids were washed with 0.5% (v/v) Tween 20 in phosphate-buffered saline and incubated in the blocking solution with a secondary anti-rabbit antibody coupled to 10-nm gold particles for 1 h at room temperature. Grids were washed five times with 0.5% (v/v) Tween 20 in phosphate-buffered saline for 10 min and washed with phosphate-buffered saline and finally with deionized water. Grids were stained with uranyl acetate and Reynolds lead citrate solutions.
Subcellular Fractioning
Five grams of dry seeds were homogenized in buffer (0.4 m Suc, 10 mm KCl, 1 mm MgCl2, 1 mm EDTA, 100 mm HEPES, pH 7.5, and 0.1 mm phenylmethylsulfonyl fluoride) with a mortar and pestle at 4°C. Homogenates were filtered through Miracloth and centrifuged at 13,000 rpm for 2 min to remove debris. Crude homogenates were then centrifuged at 13,000 rpm for 10 min at 4°C. The floating OB fraction was removed and resuspended in 200 µL 50 mm Tris-HCl (pH 7.2) buffer containing 8 m urea, and the nonfloating fraction was stored as cytosolic fraction. The resuspended floating OB fraction was diluted with 1 mL of buffer and OBs were recovered by centrifugation as before. This step was repeated twice. Both cytosolic and OB fractions were delipidated with 5 volumen of acetone. Proteins were recovered by centrifugation at 13,000 rpm for 2 min. Dry pellets were resuspended in 200 µL 50 mm Tris-HCl (pH 7.2) and 0.1% SDS. Protein concentration was determined using protein assay from Bio-Rad. Ten micrograms of protein were loaded per lane on 12% SDS-PAGE gel.
TAG Analysis
Lipids were extracted from 50 Arabidopsis seeds as previously described by Burgal et al. (2008), not including 1,1,1-13C-triolein as an internal standard. TAG purification was carried out by one-dimensional thin-layer chromatography according to Hernández et al. (2008). Fatty acid methyl esters from TAGs were produced by acid-catalyzed transmethylation (Garcés and Mancha, 1993) and analyzed by gas chromatography using a 7890A (Agilent Technologies) fitted with a capillary column (30-m length; 0.25-mm i.d.; 0.20-µm film thickness) of fused silica (Supelco) and a flame ionization detector. Hydrogen was used as carrier gas with a linear flux of 1.34 mL min–1 and a split ratio of 1:50. The injector and detector temperature was 220°C, and the oven temperature was 170°C. Heptadecanoic acid was used as internal standard to calculate the lipid content of the samples.
Sequence data from this article can be found in the GenBank/EMBL data libraries. Accession numbers are available in Supplemental Table S5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Polypeptide sequence alignment of OBAP proteins.
Supplemental Figure S2. Neighbor-joining tree of OBAP proteins from different species.
Supplemental Figure S3. Expression analysis of Arabidopsis obap1a gene (At1g05510) based on the eFP Browser database information.
Supplemental Figure S4. Mechanism of OB association.
Supplemental Table S1. EST sequences from immature scutellum library.
Supplemental Table S2. Maize genes represented in the EST sequences from immature scutellum library.
Supplemental Table S3. Functional categories of the maize genes represented in the EST sequences from immature scutellum library.
Supplemental Table S4. Array hybridization results.
Supplemental Table S5. Species whose genomes contain genes coding for OBAP proteins.
Acknowledgments
We thank Dr. Juanjo Lopez-Moya (Centre for Research in Agricultural Genomics) for providing p35S:HcPro, Dr. Vicente Rubio and Dr. Salomé Prat (National Center for Biotechnology-Spanish National Research Council) for providing us with the pVR-eCFPCt and pVR-YFPCt vectors, Maria Coca (Centre for Research in Agricultural Genomics) for providing us with the oleosin2-CFP construct, and Montse Amenós (Core Microscopy Facility, Centre for Research in Agricultural Genomics) and Alejandro Sánchez (Electronic Microscopy Facility, Universitat Autònoma de Barcelona) for technical assistance.
Glossary
- TAG
triacylglycerol
- OB
oil body
- dap
d after pollination
- cDNA
complementary DNA
- RNAi
RNA interference
- T-DNA
transfer DNA
- FA
fatty acids
- YFP
yellow fluorescent protein
Footnotes
This work was supported by the Spanish Ministry of Science and Innovation (grant nos. BIO2007–64791 and AGL2009–09151), the CONSOLIDER-INGENIO program (grant no. CSD2007–00036), the Xarxa de Referencia en Biotecnologia of the Generalitat de Catalunya, and the Consejo Superior de Investigaciones Científicas (JAE–Doc contract to M.L.H.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. (1997) Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9: 1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari BN, Wall DH, Adams BJ. (2009) Desiccation survival in an Antarctic nematode: molecular analysis using expressed sequenced tags. BMC Genomics 10: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. (2007) Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res 6: 3256–3265 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Napier JA. (2002) Targeting and membrane-insertion of a sunflower oleosin in vitro and in Saccharomyces cerevisiae: the central hydrophobic domain contains more than one signal sequence, and directs oleosin insertion into the endoplasmic reticulum membrane using a signal anchor sequence mechanism. Planta 215: 293–303 [DOI] [PubMed] [Google Scholar]
- Beller M, Riedel D, Jänsch L, Dieterich G, Wehland J, Jäckle H, Kühnlein RP. (2006) Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics 5: 1082–1094 [DOI] [PubMed] [Google Scholar]
- Bommert P, Werr W. (2001) Gene expression patterns in the maize caryopsis: clues to decisions in embryo and endosperm development. Gene 271: 131–142 [DOI] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J. (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano F, Beaudoin F, Napier JA, Shewry PR. (2007) Properties and exploitation of oleosins. Biotechnol Adv 25: 203–206 [DOI] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. (2006) The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol 16: 1783–1795 [DOI] [PubMed] [Google Scholar]
- Chen JC, Tsai CC, Tzen JT. (1999) Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol 40: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corr-Menguy F, Cejudo FJ, Mazubert C, Vidal J, Lelandais-Brière C, Torres G, Rode A, Hartmann C. (2002) Characterization of the expression of a wheat cystatin gene during caryopsis development. Plant Mol Biol 50: 687–698 [DOI] [PubMed] [Google Scholar]
- De Domenico S, Bonsegna S, Lenucci MS, Poltronieri P, Di Sansebastiano GP, Santino A. (2011) Localization of seed oil body proteins in tobacco protoplasts reveals specific mechanisms of protein targeting to leaf lipid droplets. J Integr Plant Biol 53: 858–868 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen GI, Mundy J, Tzen JT. (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112: 301–307 [DOI] [PubMed] [Google Scholar]
- Froissard M, D’andréa S, Boulard C, Chardot T. (2009) Heterologous expression of AtClo1, a plant oil body protein, induces lipid accumulation in yeast. FEMS Yeast Res 9: 428–438 [DOI] [PubMed] [Google Scholar]
- Garcés R, Mancha M. (1993) One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal Biochem 211: 139–143 [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In JM Walker, ed, The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp 571–607 [Google Scholar]
- Ghelis T, Bolbach G, Clodic G, Habricot Y, Miginiac E, Sotta B, Jeannette E. (2008) Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol 148: 1668–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytia E, Fernández-Calvino L, Martínez-García B, López-Abella D, López-Moya JJ. (2006) Production of plum pox virus HC-Pro functionally active for aphid transmission in a transient-expression system. J Gen Virol 87: 3413–3423 [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillitsch K, Connerth M, Köfeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. (2011) Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta 1811: 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BZ, Cleveland TE, Brown RL, Widstrom NW, Lynch RE, Russin JS. (1999) Distribution of antifungal proteins in maize kernel tissues using immunochemistry. J Food Prot 62: 295–299 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Casteel JE, Tang S, Hearne LB, Knapp S, Thelen JJ. (2007) Proteomic analysis of near-isogenic sunflower varieties differing in seed oil traits. J Proteome Res 6: 3232–3241 [DOI] [PubMed] [Google Scholar]
- Hernández ML, Guschina IA, Martínez-Rivas JM, Mancha M, Harwood JL. (2008) The utilization and desaturation of oleate and linoleate during glycerolipid biosynthesis in olive (Olea europaea L.) callus cultures. J Exp Bot 59: 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Small I, Kuiper MT. (2003) European consortia building integrated resources for Arabidopsis functional genomics. Curr Opin Plant Biol 6: 426–429 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. (1993) TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374: 166 [Google Scholar]
- Huang CY, Chung CI, Lin YC, Hsing YI, Huang AH. (2009) Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol 150: 1192–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NL, Huang MD, Chen TL, Huang AH. (2013) Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol 161: 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet P, Boulard C, Bellamy A, Larré C, Barre M, Rogniaux H, d’Andréa S, Chardot T, Nesi N. (2009) Protein composition of oil bodies from mature Brassica napus seeds. Proteomics 9: 3268–3284 [DOI] [PubMed] [Google Scholar]
- Jutidamrongphan W, Andersen JB, Mackinnon G, Manners JM, Simpson RS, Scott KJ. (1991) Induction of β-1,3-glucanase in barley in response to infection by fungal pathogens. Mol Plant Microbe Interact 4: 234–238 [PubMed] [Google Scholar]
- Katavic V, Agrawal GK, Hajduch M, Harris SL, Thelen JJ. (2006) Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L. (1995) Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. (2010) Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res 51: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LJ, Tai SS, Peng CC, Tzen JT. (2002) Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol 128: 1200–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Bartz R, Zehmer JK, Ying YS, Zhu M, Serrero G, Anderson RG. (2007) Rab-regulated interaction of early endosomes with lipid droplets. Biochim Biophys Acta 1773: 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombi E, Smith E, Hansen TH, Paterson D, de Jonge MD, Howard DL, Persson DP, Husted S, Ryan C, Schjoerring JK. (2011) Megapixel imaging of (micro)nutrients in mature barley grains. J Exp Bot 62: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzolini AP, Pallaghy CK, Legge GJF. (1985) Quantitative microanalysis of Mn, Zn and other elements in mature wheat seed. New Phytol 100: 483–509 [Google Scholar]
- Moellering ER, Benning C. (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. (2001) The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res 40: 325–438 [DOI] [PubMed] [Google Scholar]
- Murphy DJ (2005) Lipid-associated proteins. In DJ Murphy, ed, Plant Lipids: Biology, Utilisation and Manipulation. Blackwell, Oxford, pp 226–269 [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J. (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Negby FLSM. (1984) The structure and function of the scutellum of the Gramineae. Bot J Linn Soc 88: 205–222 [Google Scholar]
- Nguyen HM, Baudet M, Cuiné S, Adriano JM, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, et al. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11: 4266–4273 [DOI] [PubMed] [Google Scholar]
- Peled E, Leu S, Zarka A, Weiss M, Pick U, Khozin-Goldberg I, Boussiba S. (2011) Isolation of a novel oil globule protein from the green alga Haematococcus pluvialis (Chlorophyceae). Lipids 46: 851–861 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Potokina E, Sreenivasulu N, Altschmied L, Michalek W, Graner A. (2002) Differential gene expression during seed germination in barley (Hordeum vulgare L.). Funct Integr Genomics 2: 28–39 [DOI] [PubMed] [Google Scholar]
- Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC. (2006) A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J 47: 917–933 [DOI] [PubMed] [Google Scholar]
- Purkrtova Z, Jolivet P, Miquel M, Chardot T. (2008) Structure and function of seed lipid-body-associated proteins. C R Biol 331: 746–754 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SP, Belghazi M, Job C, Job D. (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 148: 620–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stenmark H. (2008) Membranes and organelles. Curr Opin Cell Biol 20: 357–359 [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Sarmiento C, Ross JH, Herman E, Murphy DJ. (1997) Expression and subcellular targeting of a soybean oleosin in transgenic rapeseed. Implications for the mechanism of oil-body formation in seeds. Plant J 11: 783–796 [DOI] [PubMed] [Google Scholar]
- Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. (2006) Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem 139: 921–930 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao KV, Datta R, Sharma R. (1998) Amylases synthesis in scutellum and aleurone layer of maize seeds. Phytochemistry 49: 657–666 [DOI] [PubMed] [Google Scholar]
- Tnani H, López I, Jouenne T, Vicient CM. (2011) Protein composition analysis of oil bodies from maize embryos during germination. J Plant Physiol 168: 510–513 [DOI] [PubMed] [Google Scholar]
- Tnani H, López I, Jouenne T, Vicient CM. (2012) Quantitative subproteomic analysis of germinating related changes in the scutellum oil bodies of Zea mays. Plant Sci 191-192: 1–7 [DOI] [PubMed] [Google Scholar]
- Tzen JT, Huang AH. (1992) Surface structure and properties of plant seed oil bodies. J Cell Biol 117: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Hajduch M, Khaled AS, Depège N, Rogowski P. (2005) Maize embryogenesis. Maydica 50: 469–483 [Google Scholar]
- Vieler A, Brubaker SB, Vick B, Benning C. (2012) A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol 158: 1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wahlroos T, Soukka J, Denesyuk A, Wahlroos R, Korpela T, Kilby NJ. (2003) Oleosin expression and trafficking during oil body biogenesis in tobacco leaf cells. Genesis 35: 125–132 [DOI] [PubMed] [Google Scholar]
- Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. (2007) Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J 21: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SA (2003) Description, development, structure and composition of the corn kernel. In PJ White, LA Johnson, eds, Corn: Chemistry and Technology. American Association of Cereal Chemists, Inc., St. Paul, pp 69–106 [Google Scholar]
- Welte MA. (2007) Proteins under new management: lipid droplets deliver. Trends Cell Biol 17: 363–369 [DOI] [PubMed] [Google Scholar]
- White PJ, Weber EJ (2003) Lipids of the kernel. In PJ White, LA Johnson, eds, Corn: Chemistry and Technology. American Association of Cereal Chemists, Inc., St. Paul, pp 356–406 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X, Lin XH, Zhou YC, Chen XL, Liu X, Lu XX. (2011) Proteome analysis of maize seeds: the effect of artificial ageing. Physiol Plant 143: 126–138 [DOI] [PubMed] [Google Scholar]
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, et al. (2012) The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res 53: 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]