Multiprotein complexes are assembled and remodeled for the transport and localization of prolamine RNAs to the cortical endoplasmic reticulum in developing rice endosperm.
Abstract
RNAs for the storage proteins, glutelins and prolamines, contain zipcode sequences, which target them to specific subdomains of the cortical endoplasmic reticulum in developing rice (Oryza sativa) seeds. Fifteen RNA binding proteins (RBPs) specifically bind to the prolamine zipcode sequences and are likely to play an important role in the transport and localization of this storage protein RNA. To understand the underlying basis for the binding of multiple protein species to the prolamine zipcode sequences, the relationship of five of these RBPs, RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q, were studied. These five RBPs, which belong to the heterogeneous nuclear ribonucleoprotein class, bind specifically to the 5′ coding regions as well as to the 3′ untranslated region zipcode RNAs but not to a control RNA sequence. Coimmunoprecipitation-immunoblot analyses in the presence or absence of ribonuclease showed that these five RBPs are assembled into three multiprotein complexes to form at least two zipcode RNA-protein assemblies. One cytoplasmic-localized zipcode assembly contained two multiprotein complexes sharing a common core consisting of RBP-J and RBP-K and either RBP-A (A-J-K) or RBP-I (I-J-K). A second zipcode assembly of possibly nuclear origin consists of a multiprotein complex containing RBP-Q and modified forms of the other protein complexes. These results suggest that prolamine RNA transport is initiated in the nucleus to form a zipcode-protein assembly, which is remodeled in the cytoplasm to target the RNA to its proper location on the cortical endoplasmic reticulum.
RNA localization is recognized as a facile mechanism to target gene transcripts, and, in turn, their coded proteins, to specific intracellular locales within animals, plants, and microorganisms. It is especially conspicuous during early metazoan development, where it accounts for oocyte polarity and cell patterning in the early embryo (Palacios, 2007; Martin and Ephrussi, 2009). For example, 71% of 3,370 transcripts in Drosophila spp. embryos are targeted to specific cell compartments, supporting the role of mRNA localization as an essential process in controlling gene expression at the subcellular level (Lécuyer et al., 2007). Recently, several mRNAs were identified as targeting mRNAs to discrete regions within Escherichia coli. Therefore, mRNA localization is an important and general process to control cellular function in both prokaryotic and eukaryotic organisms (Nevo-Dinur et al., 2011).
In developing rice (Oryza sativa) endosperm, RNAs that code for the major storage proteins are sorted to specific subdomains of the cortical endoplasmic reticulum (ER). Prolamine RNAs are localized to the ER membranes (PB-ER) that delimit the prolamine intracisternal granules, while glutelin RNAs are sorted to adjacent cisternal ER (Li et al., 1993; Choi et al., 2000; Hamada et al., 2003a, 2003b; Washida et al., 2009a, 2009b). Although a third minor storage protein species, α-globulins, are copackaged with glutelins in the protein storage vacuoles, its mRNAs are targeted to and synthesized on the PB-ER, whereupon the newly synthesized protein is rapidly exported to the Golgi (Washida et al., 2012). Mislocalization of α-globulin RNA localization to the cisternal ER disrupts transport of storage proteins to the storage vacuole and their packaging. Hence, RNA localization not only facilitates the deposition of its coded proteins into specific endomembrane compartments but also prevents potential undesirable protein-to-protein interactions that may be detrimental in protein transport and packaging and possibly function (Washida et al., 2012).
mRNA localization requires one or more cis-localization elements or zipcodes on the targeted mRNA (Marchand et al., 2012). Cis-localization elements are normally located on the 3′ untranslated region (UTR) of the localized mRNA but are also frequently found in the 5′ UTR or coding regions (CDS; St Johnston, 2005). Cis-localization elements, responsible for RNA localization, have been identified for the rice storage protein RNAs (Hamada et al., 2003b; Washida et al., 2009a, 2012). Prolamine RNAs contain two cis-localization elements; one located 3′ to the signal peptide coding sequence and the other in the 3′ UTR (Hamada et al., 2003b). Both regulatory sequences, which share considerable sequence homology, are required for restricted localization of prolamine RNAs to the PB-ER. The presence of a single cis-determinant results in only partial localization, indicating that redundant signals are required for this process. Likewise, cis-localization elements have been identified for the rice glutelin (Washida et al., 2009a) and α-globulin RNAs (Washida et al., 2012).
Using the prolamine cis-localization element as bait in affinity chromatography studies, 15 unique proteins were identified, of which seven belonged to the heterogeneous nuclear ribonucleoprotein (hnRNP) class of RNA binding proteins (Crofts et al., 2010). HnRNPs possess multiple functions and participate in diverse cellular processes in the nucleus and cytosol, including mRNA biogenesis and mRNA export from nucleus (Dreyfuss et al., 2002; Stewart, 2007; Glisovic et al., 2008). HnRNPs have also been demonstrated to be essential for RNA localization in many organisms (St Johnston, 2005; Lewis and Mowry, 2007). Some notable examples are the transport of RNAs to the distal dentritic domain (Muslimov et al., 2011) by hnRNP A2 and the transport of myelin basic protein in oligodendrocytes by hnRNP A3 (Ma et al., 2002). Hence, it is likely that the identified rice hnRNPs play an important role in the movement of prolamine mRNAs (Crofts et al., 2010).
The molecular basis for the capture of a complex mixture of RNA binding proteins by the prolamine cis-localization elements remains unclear. The RNA binding specificity of the purified protein fraction could not be confirmed, as the eluted proteins bind very poorly to the immobilized cis-localization sequences. Moreover, recombinant forms of several of the hnRNPs also lacked significant specificity to the cis-localization sequences compared with control RNA sequences (Crofts et al., 2010). A likely cause for the failure to demonstrate RNA binding specificity is that these RNA binding proteins are assembled into one or more multicomplexes, which collectively confer recognition of and binding to the cis-localization element, but are unstable under the conditions used during affinity chromatography with the baited cis-localization sequences. To address the possible role of higher order ensembles of RNA binding proteins in prolamine RNA localization, five of the identified hnRNP homologs were selected and monospecific antibodies produced. Using affinity-purified forms as biochemical probes, we confirm the specific recognition of these hnRNPs to both the 5′ CDS and 3′ UTR cis-localization sequences. Coimmunoprecipitation (Co-IP) studies showed that these five proteins are organized into at least three multiprotein complexes with two of these oligomeric structures sharing common hnRNP species.
RESULTS
Structural Relationships of the Seven hnRNPs That Recognize the Prolamine Zipcode
An earlier study (Crofts et al., 2010) identified 15 RBPs, which were specifically captured under stringent binding conditions by biotinylated prolamine 5′ CDS zipcode sequences conjugated to streptavidin magnetic beads. An immediate question that arises from this result is why so many RBPs were captured by the prolamine 5′ CDS zipcode sequences. Among the various RBPs identified, seven were hnRNPs containing conserved tandem RNA recognition motifs (RRMs; Dreyfuss et al., 2002; Glisovic et al., 2008). As hnRNPs play a variety of roles in RNA transcription, transport, and stability, we initiated a study on the relationship of these captured hnRNPs.
Comparative sequence analysis indicated that several of the hnRNPs were closely related, resulting in the seven RBPs being resolved into five clades. RBP-A and RBP-G were distributed to the same clade while RBP-D and RBP-I belonged to the same hnRNP subfamily (Fig. 1). In view of the close structural relatedness of the RBP-A/RBP-G pair and RBP-D/RBP-I pair, only RBP-A and RBP-I were studied together with RBP-J, RBP-K, and RBP-Q.
Figure 1.
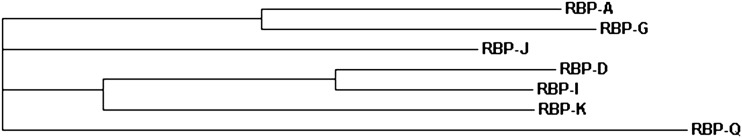
Phylogram of hnRNP homologs in identified prolamine zipcode binding proteins. The phylogenetic tree was generated by ClusterW2 with amino acid sequences of RBP (http://www.ebi.ac.uk/Tools).
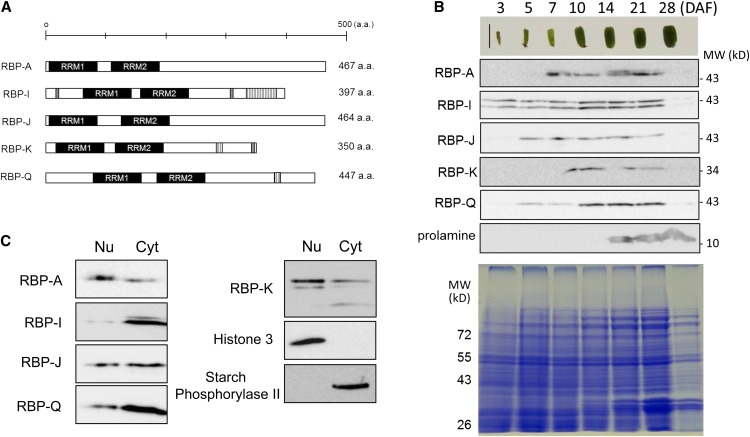
Schematic representations of the five hnRNPs are depicted in Figure 2A. Each hnRNP has two RRMs within the N-terminal half of the protein. All five RBPs have an enriched Gly content, which constitutes up to 17% in RBP-A, 28% in RBP-I, 28% in RBP-J, and 21% in RBP-Q, respectively. The biased Gly content is due to C-terminal (Gly)nX repeats. RBP-I has seven GAY (Gly-Ala-Tyr) repeats located at the C-terminal end, while RBP-K and RBP-Q have two of these repeats (Fig. 2A). RBP-I has three RGG (Arg-Gly-Gly) peptides, an RNA binding motif; one is found near the N terminus, while two are close to the C terminus. RBP-K also has a single RGG motif near the C terminus (Birney et al., 1993). Other than RBP-Q, which has a single Cys, these hnRNPs are devoid of Cys residues.
Figure 2.
Five selected hnRNPs that interact with the prolamine zipcode. A, Schematic structural representations of the hnRNPs RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q, which all contain a pair of RRMs (black rectangle). RBP-I and RBP-K contain RGG (Arg-Gly-Gly) motifs (gray rectangles) and GAY (Gly-Ala-Tyr) motifs (vertical line rectangles), while RBP-Q contains only the GAY motif. B, The temporal expression pattern of RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q as viewed by immunoblotting. Note that the RBPs are all readily detected when prolamine proteins begin to accumulate at the midstage of seed development. Only the 36-kD polypeptide band for RBP-K is shown, as the presence of the higher molecular-sized polypeptide bands at 45 and 41 kD in C was variable. C, Distribution of the RBPs in nuclear/chromatin and cytosolic fractions prepared from 10- to 13-DAF seed extracts. Histone3 and starch phosphorylase2 were used as markers of the nuclear/chromatin and cytosolic fractions, respectively. Nu, Nucleus; Cyt, cytosol. [See online article for color version of this figure.]
Monospecific antibodies were generated using bacterial-expressed recombinant proteins and affinity purified. Immunoblot analysis of seed extracts indicated that the antibodies for two of the hnRNPs, RBP-I and RBP-K, recognized multiple polypeptide forms. Antibodies raised against RBP-I recognized a polypeptide band at approximately 43 kD, close to the predicted molecular size of RBP-I based on coding sequence, and a slightly larger form at 45 kD (Fig. 2B). Three polypeptide bands were evident when using anti-RBP-K. In addition to its predicted molecular size at 36 kD, a prominent polypeptide band at 45 kD and a minor band at 43 kD were present (Figs. 2, B and C, and 3). RBP-A, RBP-J, and RBP-Q were detected as single polypeptide bands.
Figure 3.
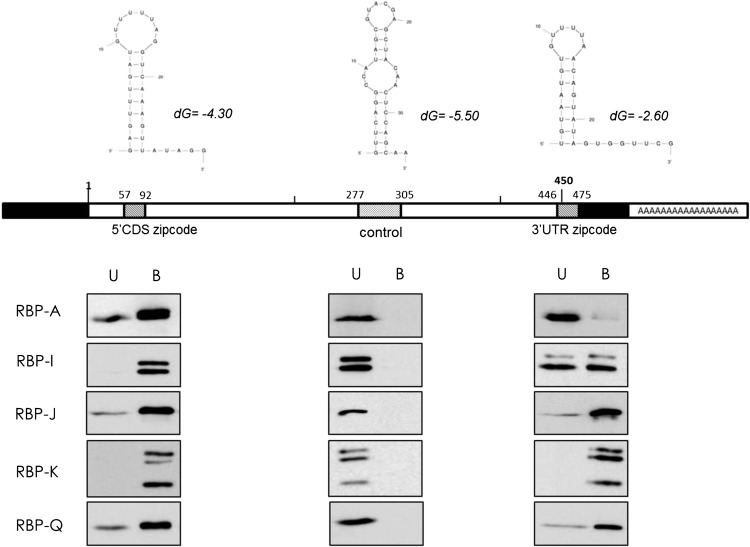
Recognition of 5′ CDS and 3′ UTR zipcodes by RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q. Rice seed extracts were incubated with biotinylated 5′ CDS and 3′ UTR zipcode or control sequences bound to streptavidin-conjugated magnetic beads. The bound (B) and unbound (U) fractions were then subjected to immunoblot analysis using affinity-purified antibodies raised against RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q. The top portion of the figure depicts the secondary structures of 5′ CDS and 3′ UTR zipcodes and control sequences as predicted by mfold web-based analysis tools (http://mfold.rna.albany.edu). Schematic structure of prolamine7 complementary DNA depicts the location of the 5′ CDS and 3′ UTR zipcodes and control sequence. The bottom portion of the figure depicts immunoblot results, which show that all five RBPs bind to 5′ CDS and 3′ UTR zipcodes but not to the control sequence.
The basis for the polymorphic polypeptide patterns seen for RBP-I and RBP-K on immunoblots containing seed extracts may be due to posttranslational modifications or cross reactivity to closely related hnRNPs. The 45- and 41-kD polypeptides may be posttranslational modified forms of the 36-kD RBP-K, as the amounts of these polypeptide bands were highly variable from experiment to experiment. Posttranslational modification is the most likely cause for the higher Mr polypeptide species at 45 kD for RBP-I. Mass spectrometry of this region of the SDS polyacrylamide gel readily identifies the presence of RBP-I (Supplemental Fig. S1), while the closely related RBP-D is absent.
Temporal Expression and Subcellular Distribution of hnRNPs during Seed Development
Immunoblotting was performed with total protein extracts isolated from developing seeds collected at different days after flowering (DAF) to monitor the distribution of these RBPs during seed development. RBP-I, which is detected as a doublet, is visible as early as 3 DAF and is expressed throughout the remainder of seed development with a maximum accumulation level at midstage from 10 to 21 DAF (Fig. 2B). The other hnRNPs are detected at later stages, but all attain their maximum levels at 10 DAF, a period preceding the synthesis of 13-kD prolamine polypeptides.
Figure 2C shows the distribution of the hnRNPs in nuclear/chromatin and soluble (cytosolic) fractions. The efficiency of subcellular fractionation was validated by assessing the distribution of marker proteins for nuclei/chromatin and cytosol. Histone 3, a chromatin-associated protein, is restricted to the nuclear/chromatin fraction, while phosphorylase II, a known cytosolic enzyme involved in starch degradation, is absent from nuclei and is present only in the cytosolic fraction. All five RBPs were detected in both nuclear/chromatin and cytosolic compartments, although their relative distribution levels varied depending on the hnRNP in question. RBP-A and the RBP-K species at 45 and 43 kD predominate in the nuclear fraction, while RBP-Q is preferentially present in the cytosolic fraction. By contrast, nearly all of RBP-I is distributed to the cytosol (Fig. 2C). Overall, these hnRNPs are nucleocytoplasmic proteins, although their relative distribution between these two different subcellular compartments are nonidentical, suggesting that they play different roles in RNA metabolism during seed development.
Biotinylated RNA Binding Assays
Prolamine mRNA has two zipcodes, one located 3′ to the signal peptide coding sequence (5′ CDS) and a second one within the 3′ UTR (Fig. 3). Both zipcodes are essential to maintain restricted prolamine mRNA transport to PB-ER, as the presence of a single zipcode results in partial mislocalization to the cisternal ER (Hamada et al., 2003b). The two zipcodes are partially redundant in sharing considerable sequences when gaps are introduced. To determine whether these RBPs recognized both zipcode sequences equally or preferred one zipcode sequence over the other, binding studies were conducted. Three different biotinylated RNAs were used, two containing the prolamine 5′ CDS and 3′ UTR zipcode sequences and a third nonzipcode RNA sequence from the central region of the prolamine CDS. All three RNA sequences were predicted to fold into stem-loop secondary structures (Fig. 3).
The binding assay shows that all five RBPs bind to both zipcode RNAs but not to the control RNA (Fig. 3). These results confirm our earlier binding studies to zipcode and control RNA sequences, which were performed under very stringent conditions (Crofts et al., 2010). Interestingly, RBP-A interacted weakly to the 3′ UTR zipcode, while binding by RBP-I to the 3′ UTR zipcode was considerably weaker than to the 5′ CDS one. RBP-K, RBP-J, and RBP- Q showed strong binding to both zipcode RNAs. Overall, all five RBPs specifically recognized the 5′ CDS and 3′ UTR zipcode RNAs compared with the control RNA sequence, although RBP-A and RBP- I showed preferential binding to the 5′ CDS zipcode.
Co-IP Studies Indicate the Assembly of RBPs into Several Multiprotein Complexes
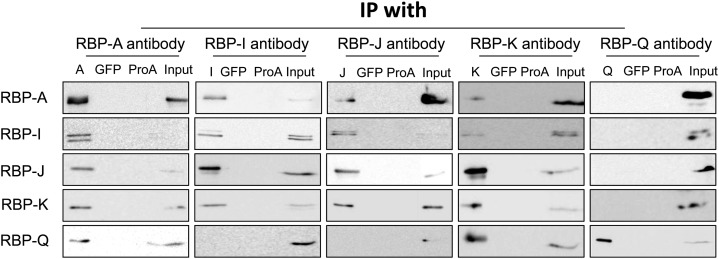
As all five RBPs display binding activity to the prolamine zipcode RNAs, we examined whether they recognized the RNA alone or together with other proteins as a multiprotein complex. Immunoprecipitation (IP) studies were carried out with each of the five antibodies at our disposal. After subjecting the IPs to SDS-PAGE, the presence of the various RBPs was examined by immunoblotting. All five RBPs were detected on immunoblots when incubated with their respective antibodies. By contrast, none of the RBPs were detected in control Co-IP studies with GFP antibody or with mock IP reactions containing antibody-free protein A agarose beads (Fig. 4).
Figure 4.
Interaction of the five RBPs as viewed by Co-IP analysis. Immunoprecipitates were generated by incubating rice seed extracts with protein A conjugated with various RBP antibodies. The immunoprecipitates were then subjected to immunoblot analysis for the presence of other RBPs. Mock IPs with anti-GFP and antibody-free protein A agarose beads (ProA) were performed as negative controls. Note that RBP-A, RBP-I, RBP-J, and RBP-K are found in reciprocal IPs, while RBP-Q is only found in IPs formed by anti-RBP-A and anti-RBP-K. Note also that RBP-A and RBP-K are not found in IPs generated with anti-RBP-Q, indicating that RBP-Q is present in multiple forms, with one form being masked from interacting with its antibody when associated with RBP-A and RBP-K. Only the 36-kD polypeptide band for RBP-K is shown, as the presence of the higher molecular weight forms seen in Figures 2 and 3 was highly variable and not reproducibly detected.
Figure 4 depicts the Co-IP results. Three different immunoblot patterns are evident, depending on the antibody used for the initial IP. IPs generated with antibodies raised against RBP-A and RBP-K not only contained their respective antigen, but the three other RBPs as well. By contrast, IPs formed using RBP-I and RBP-J antibodies contained only four of the RBPs, with RBP-Q being absent. Interestingly, IP generated by RBP-Q antibody only contained RBP-Q and not the other four RBPs. This observation indicates that the RBP-Q is sterically inaccessible from reacting to its antibody when associated with RBP-A and RBP-K. Overall, these results suggest that these RBPs are organized in at least two multiprotein complexes, one containing RBP-Q and the other devoid of this RBP.
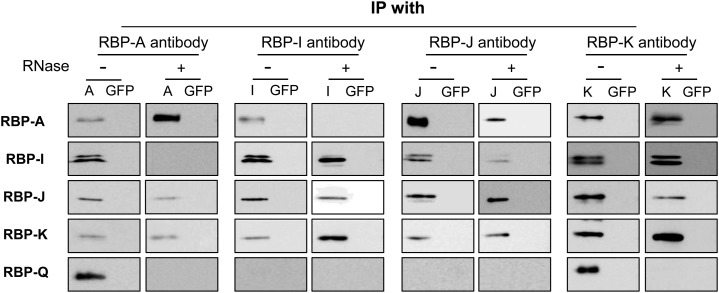
To gain further insights on the association of the RBPs on the prolamine zipcode, the Co-IP studies were repeated in the presence of ribonuclease (RNase; Fig. 5). Immunoblot analysis of IPs generated with antibodies raised against RBP-J or RBP-K showed the same polypeptide pattern as that seen in the absence of RNase, i.e. the presence of the other three RBPs (RBP-A, RBP-I, and, depending on the antibody used for IP, either RBP-K or RBP-J). IPs generated with antibodies to RBP-A and RBP-I contained different RBP compositions in the presence of RNase. IPs formed by RBP-A antibody in the presence of RNase was devoid of RBP-I and RBP-Q while retaining RBP-J and RBP-K. Likewise, IPs formed with RBP-I antibodies also contained RBP-J and RBP-K but lacked RBP-A and RBP-Q. Hence, the association of RBP-A to RBP-I or to RBP-Q is RNA dependent, and these RBPs are bound to the prolamine zipcode in different multiprotein complexes.
Figure 5.
The association of the five RBPs in Co-IPs treated with RNase. Immunoprecipitates formed using antibodies raised against RBP-A, RBP-I, RBP-J, RBP-K, or RBP-Q were treated with or without RNase and then subjected to immunoblot analysis. Protein A beads conjugated with anti-GFP were used as a negative control (GFP). Note that both RBP-A and RBP-I are found in RNase-resistant complexes with RBP-J and RBP-K but not with each other. The association of RBP-Q with RBP-A and RBP-K is RNase sensitive, indicating that the former does not directly interact with the two latter RBPs. Overall, these results support the existence of three RBP-containing multiprotein complexes.
The presence of RBP-J and RBP-K with both RBP-A and RBP-I indicates that these RBPs provide a common core of the RBP-A- and RBP-I-containing multiprotein complexes. The absence of RBP-Q in any IPs generated in the presence of RNase indicates that RBP-Q is bound to the prolamine zipcode as a third independent multiprotein complex and is sterically inaccessible from interacting with its own antibody. Taken together, the Co-IP results indicate that these five RBPs are bound to the prolamine zipcode as three independent multiprotein complexes, two of which share a common core containing RBP-J and RBP-K.
Yeast Two-Hybrid Analysis
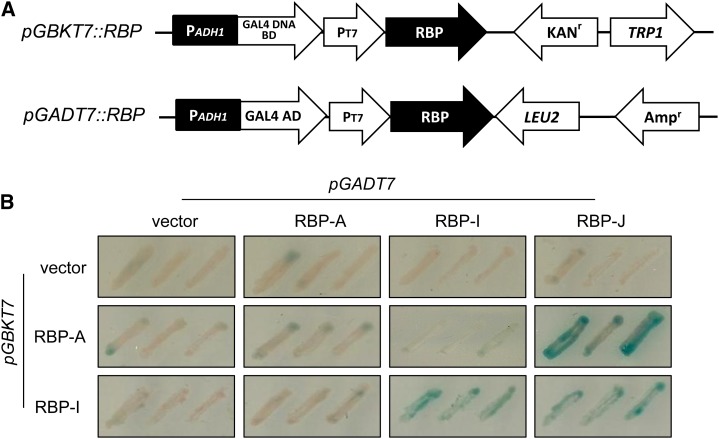
To validate the direct protein interaction in vivo, yeast (Saccharomyces cerevisiae) two-hybrid studies were carried out using the various RBP coding sequences as bait and prey (Fig. 6A). All RBP gene constructs were devoid of autotranscriptional activity. However, yeast cells harboring RBP-J as a bait construct and RBP-K as both bait and prey constructs exhibited high toxicity, and these gene constructs were excluded in these studies (data not shown). Mating conditions for each combination were determined by microscopic observation of the zygote (data not shown). RBP-A interacted with RBP-J in reciprocal combinations of bait and prey yeast cells. When RBP-I was used as bait, RBP-J as well as RBP-I showed positive signals, the latter result indicating that RBP-I can interact with itself and dimerize. Collectively, these results showed that RBP-A directly binds to RBP-J but not to RBP-I, while RBP-I interacts with RBP-J but not with RBP-A. These results are consistent with the conclusions made from the Co-IP studies.
Figure 6.
The interaction of RBP-A and RBP-I to other hnRNPs as viewed by yeast two-hybrid analysis. A, Schematic representation of gene constructs used for yeast two-hybrid analysis. Y187 yeast cells were transformed with pGBKT7 plasmids harboring RBP-A or RBP-I sequences or as an empty vector. The prey vector, pGADT7, was constructed with RBP-A, RBP-I, or RBP-J sequences and transformed into AH109 yeast strain. B, Yeast two-hybrid screening on Trp– and Leu– synthetic dextrose media containing X-α-gal. RBP-K sequences and the pGBKT7 containing RBP-J were toxic to yeast cells and, hence, were not further evaluated. Note that RBP-A and RBP-I interact with RBP-J, while RBP-I interacts with itself.
Biomolecular Fluorescence Complementation (BiFC) Test
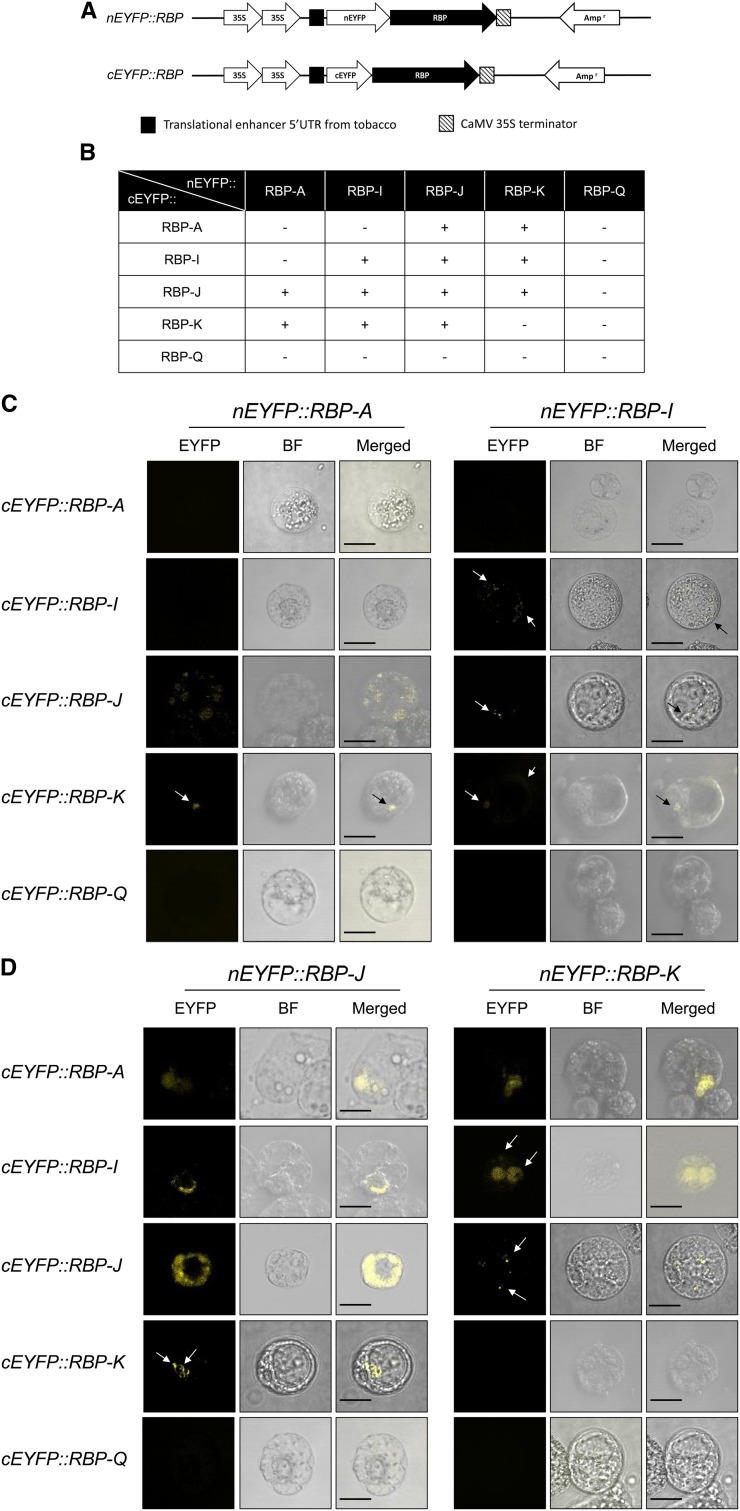
Yeast two-hybrid studies validated the interaction of RBP-A or RBP-I with RBP-J. The interaction with RBP-J was, however, restricted to using this RBP as a prey, while interactions with RBP-K were unable to be assessed, as expression of this RBP was toxic to yeast cells. To overcome this limitation, we employed BiFC to verify the in vivo interaction of these five RBPs in plant cells. BiFC has been popularly applied in living plant cells to assess potential protein-protein interaction by reconstruction of enhanced yellow fluorescent protein (EYFP; Ohad et al., 2007; Lee et al., 2012).
We designed the gene constructs where one RBP coding sequence was fused to the N-terminal EYFP fragment and the other RBP to the C-terminal of EYFP. The coding sequences of these translational fusions were under the control of a Cauliflower mosaic virus double 35S promoter (Fig. 7A). To ensure that the EYFP reconstruction reflected authentic interaction between the RBPs, the pair of RBP coding sequences was reciprocally exchanged between the two EYFP fragments. The pair of corresponding EYFP constructs were then transformed into tomato (Solanum lycopersicum) protoplasts by polyethylene glycol transformation method, incubated for 16 h, and then assessed for YFP fluorescence in protoplasts (Schütze et al., 2009). Empty vectors were transformed into protoplasts to check for background autofluorescence, which was removed by adjusting the gain on the confocal microscope (Supplemental Fig. S2).
Figure 7.
Interactions of RBPs in tomato protoplasts as assessed by BiFC. A, Schematic diagrams of RBP gene fusions to the N-terminal and C-terminal EYFP fragments. B, Summary of various protein-protein interactions among the five RBPs. C, BiFC confocal microscopic images of tomato protoplasts expressing nEYFP::RBP-A or nEYFP::RBP-I and cEYFP versions of RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q. D, BiFC confocal microscopic images of tomato protoplasts expressing nEYFP::RBP-J or nEYFP::RBP-K and cEYFP versions of RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q. The yellow fluorescence formed by direct interaction of RBPs was observed by confocal microscope. Individual panels for each BiFC pair of plasmids denote presence or absence of fluorescence (EYFP), bright field image (BF), and superimposed BF and EYFP images (Merged). Bar = 20 μm.
Figure 7, C and D, summarized the representative microscopic images observed in the cotransformed protoplasts as captured by confocal microscope. The results showed that nEYFP::RBP-J (nRBP-J) interacted with the cEYFP versions of RBP-A (cRBP-A), RBP-I (cRBP-I), and RBP-K (cRBP-K). Likewise, nEYFP::RBP-K (nRBP-K) was capable of forming functional EYFP with cRBP-A, cRBP-I, and cRBP-J in cotransformed protoplasts (Fig. 7D). These observations indicated that RBP-J and RBP-K interact directly with each other and also with RBP-A and RBP-I. In addition, cotransformed protoplasts expressing nRBP-A generated fluorescence signals with cRBP-J or cRBP-K (Fig. 7C). Fluorescence signals were also generated when nRBP-I was coexpressed with cRBP-J or with cRBP-K (Fig. 7C). However, there was no sign of interaction of cRBP-Q with the nEYFP versions of RBP-A, RBP-I, RBP-J, or RBP-K. Likewise, neither nRBP-A nor cRBP-A formed a stable interaction with cRBP-I or nRBP-I, respectively.
RBP-I forms a homodimer in the yeast two-hybrid assay, and this was verified in the BiFC studies. RBP-I homodimers are observed as small, faint speckles distributed throughout the protoplast cell (Fig. 7C). RBP-J also forms a homodimer, which is highly prevalent and distributed throughout the nonnuclear regions of the cell (Fig. 7D). By contrast, RBP-A and RBP-K do not self-assemble to form homodimers. Interactions of RBP-K with RBP-A or RBP-I were nuclear in location, with stronger expression seen when RBP-K were fused to nEYFP and weak expression when linked to cEYFP. The latter interaction with nRBP-I resulted in faint fluorescence being observed on the cell’s periphery in addition to a small signal in the nucleus.
The fluorescence signal intensity and cellular location of the RBP-A/RBP-J complex depend on the orientation of the interaction. Interactions of nRBP-J with cRBP-A and cRBP-I occur in the nucleus. Unlike the uniform distribution of the nRBP-J/cRBP-A complex in the nucleus, the nRBP-J/cRBP-I complexes were located toward the peripheral region of this organelle. Interestingly, while the nRBP-J/cRBP-A interaction was observed in the nucleus, the corresponding reverse pair (nRBP-A/cRBP-J) complexes were distributed as small foci throughout the cell.
Collectively, the results from BiFC studies corroborate and extend information on the interaction of the five RBPs as initially determined by Co-IP and yeast two-hybrid analysis. RBP-J and RBP-K directly interact with RBP-A or RBP-I as well as to each other to form two ternary complexes. The BiFC studies also confirm that RBP-A and RBP-I do not directly interact with each other and that RBP-Q does not interact with the other four RBPs.
DISCUSSION
Studies from this laboratory have established that prolamine and glutelin mRNAs are transported and localized to two distinct subdomains of the cortical ER, the PB-ER and cisternal ER, respectively (Li et al., 1993; Crofts et al., 2005). These RNAs are transported and targeted to these ER subdomains by a regulated process requiring cis-determinants or zipcodes (Hamada et al., 2003b; Washida et al., 2009a, 2012). Prolamine mRNAs contain two zipcode sequences, a 5′ CDS located downstream of the signal peptide coding sequence and a 3′ UTR localized signal (Fig. 3). Both zipcode sequences are required for restricted localization of prolamine mRNAs to the PB-ER.
Using the 5′ CDS zipcode sequences as bait, 15 proteins were identified that specifically interacted under stringent binding conditions with the 5′ CDS zipcode of prolamine mRNA (Crofts et al., 2010). Seven of the RBPs were identified as hnRNPs containing two RRM motifs (Fig. 2A). HnRNPs function in a variety of RNA processes, including transcription, splicing, nuclear export, cytoplasmic transport and localization of RNAs, translation, and RNA stability and turnover (Dreyfuss et al., 2002). Two of these hnRNPs, RBP-A and RBP-I, share significant homology with hnRNP40 (Hrp40; squid) and Hrp48, hnRNPs required for the localization of specific RNAs in Drosophila oocytes (Lall et al., 1999; Yano et al., 2004; St Johnston, 2005). These binding and sequence conservation properties support a role for these rice hnRNPs in RNA transport and localization during seed development.
Depending on their roles in RNA metabolism, the hnRNPs may be predominately distributed to the nucleus or cytoplasm although many are likely to be involved in processes occurring in both subcellular compartments, especially those involved in RNA transport and localization (Hachet and Ephrussi, 2004; Moore, 2005; Lewis and Mowry, 2007; Glisovic et al., 2008). Consistent with this view, the five hnRNPs studied are distributed to both the nucleus/chromatin and the cytosol fractions, although their relative distributions within these two compartments are not identical. RBP-A and the 45-kD RBP-K were preferentially located in the nucleus/chromatin fraction, while RBP-I and RBP-Q were distributed predominantly to the cytoplasm. By contrast, RBP-J was found in about equal amounts in these compartments.
Two of the antibodies used to in this study cross reacted with more than one polypeptide band as viewed by immunoblot analysis. RBP-I antibody reacted with a polypeptide at 43 kD, somewhat larger than the predicted molecular size (40 kD) of RBP-I based on coding sequence, and a slightly larger form at 45 kD. Although this larger band corresponded to the predicted size of the closely related RBP-D (Fig. 1), mass spectrometry analysis of the 45-kD gel region only detected RBP-I (Supplemental Fig. S1). Moreover, antibodies raised against RBP-D only react with a single polypeptide band at 45 kD.
RBP-K exhibited multiple bands at 45 and 36 kD (the predicted size) as well as a minor band at 42 kD. The larger polypeptide bands were not consistently detected, and, when seen, their amounts varied in relationship to the 36-kD band (results not shown). The basis for these polymorphic properties of RBP-I and RBP-K is not understood and was not explored, although posttranslational modifications are a likely reason for appearance of the larger polypeptides.
The five hnRNP homolog RBPs showed different patterns of accumulation during seed development. RBP-I was readily detected in 3-d-old developing seeds, while RBP-A and RBP-K were not detected until about 7 d. Despite the temporal differences, all five RBPs were readily present when prolamine polypeptides were being actively accumulated. Hence, the temporal accumulation patterns of these hnRNPs are consistent with their suggested role in prolamine RNA transport and localization.
Our studies show that the five selected RBPs bind to both the 5′ CDS and 3′ UTR zipcode sequences but not to a control RNA sequence (Fig. 3). This finding supports our earlier observations that the polypeptide patterns exhibited by captured proteins by either zipcode sequence were essentially identical (Crofts et al., 2010). Direct binding studies showed, however, that RBP-A and RBP-I interacted much weaker with the 3′ UTR zipcode than the 5′ CDS (Fig. 3). This preferential binding is somewhat surprising in that both zipcode sequences are required for restricted localization of prolamine RNAs to the PB-ER (Hamada et al., 2003b). Although these RBPs may have different affinities to these zipcode sequences, a more likely possibility is that stable binding of RBP-A and RBP-I to the 3′ UTR zipcode requires other factors to stabilize the interaction. For example, the RNA binding protein, She2p, is essential to induce binding of She3p to ASH1 RNA that encodes a transcriptional repressor and, in turn, to recruit the Myo4p myosin motor protein (Böhl et al., 2000; Long et al., 2000). As RBP-A and RBP-I exist in multiprotein complexes, the absence of one or more proteins may influence the binding efficiency of the protein complex.
Multiple RBP Complexes Bind to the Prolamine mRNA Zipcodes
Our Co-IP studies showed that the five RBPs assemble onto the prolamine zipcode as three multiprotein complexes. Two of these complexes contain a common RBP-J and RBP-K but are distinguishable by containing a third protein, either RBP-A or RBP-I (Figs. 4 and 5). The direct protein interactions between RBP-J and RBP-A or between RBP-J and RBP-I were verified by yeast two-hybrid studies (Fig. 6). In addition, a third complex containing RBP-Q exists and colocalizes with the RBP-A containing multiprotein complex in a RNA-dependent manner (Figs. 4 and 5).
The results from BiFC tests corroborated and extended information on the interaction of these five RBPs. RBP-K and RBP-J can interact with each other as well as to RBP-A or RBP-I. As suggested by the Co-IP, yeast two hybrid, and BiFC results (Figs. 5–7), RBP-A, RBP-I, and RBP-Q do not directly interact with each other. When bound to the zipcode sequences, RBP-Q is not recognized by its antibody, indicating that its antigenic sites are inaccessible, likely because of its interaction with other proteins. Although an antibody-reactive form of RBP-Q is readily detected, this antibody-reactive RBP-Q is not bound to the zipcode sequence.
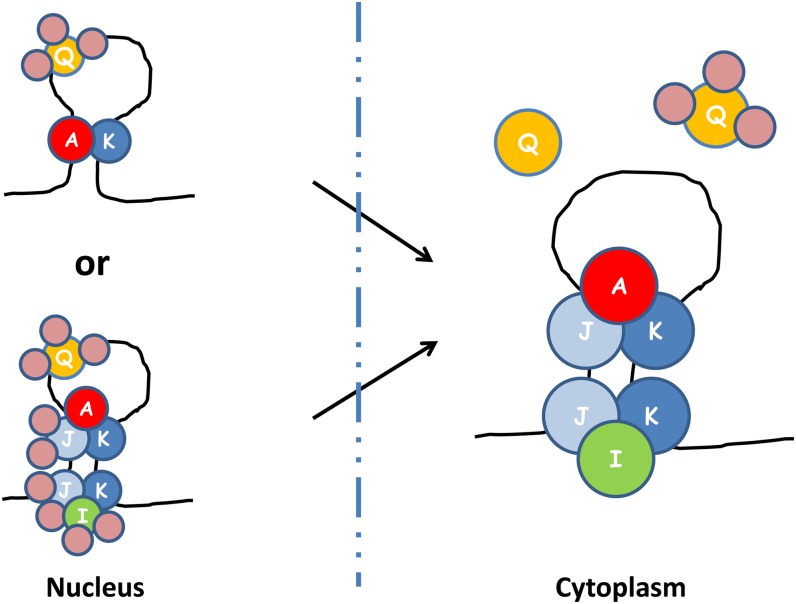
Based on the available information, a model is proposed as shown in Figure 8. The model is based on the relative distributions of the five hnRNPs within the nucleus and cytoplasm, the Co-IP results in the presence and absence of RNase, and the protein interactive results from yeast two hybrid and BiFC. One conclusion based on the available results is the presence of two ternary complexes, A-J-K and I-J-K, which are bound to the same zipcode as independent multiprotein complexes. Both I-J-K and A-J-K multiprotein complexes are present when the zipcode is intact, but only one of the multiprotein complexes is detected with antibodies to either RBP-A or RBP-I when the zipcode is degraded by RNase. As RBP-I is nearly excluded from the nucleus (Fig. 2C), this zipcode assembly containing I-J-K and A-J-K multiprotein complexes likely exists in the cytoplasm.
Figure 8.
A proposed model of prolamine zipcode mRNA-RBP protein assembly complexes based on results obtained by IP, yeast two-hybrid analysis, and BiFC. Note that at least three different RBP complexes bind to the prolamine zipcodes to form two ribonucleoprotein assembly complexes. RBP-A, RBP-J, and RBP-K and RBP-I, RBP-J, and RBP-K are assembled in the cytoplasm as two separate multiprotein complexes on the prolamine zipcode. When associated with RBP-A and RBP-K, RBP-Q is not accessible to its antibody, suggesting that it is bound by other proteins that comprise a third multiprotein family. As BiFC results indicate that heterodimer formation occurs in the nucleus (Figure 7), it is likely that multiprotein complexes A-J-K and I-J-K are formed in the nucleus, where they interact with the prolamine zipcode. If so, RBP-I and RBP-J are bound by other proteins so that they are not accessible to their respective antibodies. A simpler ribonucleoprotein assembly complex in the nucleus would consist of two multiprotein complexes, one containing RBP-Q and a second containing RBP-A and RBP-K. Note that the various multiprotein complexes were arbitrarily drawn on the zipcode stem-loop structure. The actual binding specificity of the various multiprotein complexes to the stem-loop structure remains to be resolved.
The Co-IP studies showed that RBP-Q is associated with RBP-A and RBP-K only when antibodies to the latter two RBPs are used to generate the immunoprecipitate. The inability to capture RBP-Q in association with the other two RBPs using anti-RBP-Q indicates that RBP-Q is sequestered from reacting to its polyclonal antibody, likely due to its assembly in a third multiprotein complex. This indicates the existence of a second zipcode assembly containing at least two multiprotein complexes, one consisting of RBP-Q and a second consisting of RBP-A and RBP-K. As BiFC results showed that all detected RBP interactions that form heterodimers occur in the nucleus, we propose that assembly of ternary A-J-K and I-J-K complexes on the prolamine zipcode takes place in this organelle. As antibodies to RBP-I and RBP-J do not coimmunoprecipitate RBP-Q, these RBPs, like RBP-Q, are also likely to be sequestered from their respective antibody. Moreover, of the five hnRNPs, RBP-Q is the only one that has a nuclear localization signal, suggesting its initial assembly on the prolamine zipcode occurs in the nucleus. As RBP-I is mainly distributed to the cytoplasm, addition of I-J-K complex may facilitate rapid export from the nucleus to the cytoplasm. Alternatively, a simpler ribonucleoprotein assembly complex in the nucleus would consist of two multiprotein complexes, one containing RBP-Q and a second containing RBP-A and RBP-K.
Export from the nucleus to the cytoplasm likely involves extensive remodeling of the ribonucleoprotein assembly. The RBP-Q complex is removed, whereupon it is now assessable to its antibody. The composition of the A-J-K and I-J-K multiprotein complexes are also refashioned where RBP-J and RBP-I are made available to react to their respective antibody.
An obvious question raised here is why there are different multiprotein complexes that bind to the zipcode RNA. As demonstrated in Xenopus laevis (Kress et al., 2004), the localization of the maternal Vg1 RNA to the vegetal pole of the oocyte is initiated in the nucleus where two RBPs, hnRNP I and Vg1RBP/vera interact with each other and with Vg1 RNA. Once exported, the ribonucleoprotein complex is remodeled where the hnRNP I-Vg1RBP/vera complex is released, allowing additional RBPs to interact with Vg1 RNA. The localization of prolamine RNAs shares similar features in assembling the ribonucleoprotein in the nucleus with subsequent remodeling in the cytoplasm.
The presence of multiple protein complexes further exemplifies the complexity of the RNA localization process. The localization element (IV/V RNA) of bicoid mRNA in Drosophila spp. is recognized by at least two different RNA binding complexes. One complex was composed of five different RBPs, which collectively exhibit strong binding specificity (Arn et al., 2003). Although the two complexes could act redundantly, they are likely responsible for different events of the RNA localization process, including linking the mRNA to the transport machinery or anchoring the mRNA at its destination site (Arn et al., 2003). Hence, the presence of two proteins complexes containing a core of RBP-K and RBP-J are likely responsible for one of these steps in RNA localization.
The five RBPs studied here are all hnRNPs, proteins normally involved in RNA splicing and maturation. Prolamine genes, however, lack introns, and therefore, the assembly of specific hnRNPs with the newly transcribed RNA is likely essential for its export from the nucleus and for subsequent events of RNA localization in the cytoplasm. Such a role is supported by the distribution of RBP-A in developing endosperm cells, as viewed by indirect fluorescence microscopy (Crofts et al., 2010). Consistent with the subcellular localization studies depicted in Figure 2C, RBP-A is found not only enriched in the nucleus, but is also closely associated with cytoplasmic microtubules and with the cortical ER (Crofts et al., 2010). The latter location possibly infers a role of this RBP in anchoring of the RNAs at the cortical ER. Likewise, these five RBPs are likely to be involved in the transport and localization of other RNA species. Results from genetic studies suggest that the glutelin and prolamine regulated transport pathways and default transport pathway share common protein components, with one or more unique proteins specifying a specific pathway (Crofts et al., 2005; Doroshenk et al., 2012).
In addition to the five hnRNPs studied here, 10 additional RBPs were specifically captured by the prolamine zipcode sequences. Of these were two additional hnRNPs, several other types of RRM containing RBPs, and five non-RRM RBPs. One or more of these proteins may be responsible for precluding RBP-Q and possibly RBP-K from being recognized by its antibody, while others may be involved in direct binding to the prolamine zipcode sequences. Proteins in addition to these 15 identified may also be involved as well. Liquid chromatography tandem mass spectrometry of IPs generated with RBP-K antibodies identified translation elongation factors and various nucleotide binding proteins (data not shown). The role of these proteins is unclear but may be related to activating translation at the destination site. Finally, RNAs move along actin filaments, a process powered by myosin. The players involved in associating the RNA with myosin have yet to be identified but likely involve one or more of the other 10 RBPs captured by the prolamine zipcode sequences (Crofts et al., 2010). The results obtained here will provide the foundation to pursue studies on identifying the different protein factors that are responsible for specifying each of the steps involved in RNA transport and localization.
MATERIALS AND METHODS
Crude Protein Extraction and Subcellular Fractionation for Rbp Protein Distribution
Wild-type rice (Oryza sativa ‘Kitaake’) seeds were grown in a controlled-environment walk-in chamber using a diurnal cycle at 26°C for 11 h under lights, followed by 13 h at 22°C. Deglumed grains at 3, 5, 7, 10, 14, 21, and 28 DAF were homogenized with 1× SDS-PAGE sample buffer containing 4 m urea at a final concentration of 10 mg mL–1 and then incubated with vigorous shaking at room temperature for 30 min. The extract was briefly centrifuged at 100g for 5 min to remove large starch grains, and then the supernatant fraction was centrifuged at 12,000g for 10 min at 4°C. Thirty microliters of the soluble extract were separated on 10% (v/v) SDS-PAGE gel, while the pellet fraction was extracted with 1× SDS solution and centrifuged, and the resulting pellet extract containing prolamine polypeptides was separated on 12% (v/v) acrylamide gel.
Nuclear and postnuclear fractions were prepared from middeveloping rice seeds per the manufacturer’s instruction using the CelLytic PN Isolation/Extraction Kit (Sigma-Aldrich). Five grams of developing 10- to 13-DAF seeds were ground in 15 mL of nuclei isolation buffer (NIB) at 4°C. The suspension was passed through a 100-mesh filter and then centrifuged at 1,260g for 10 min. The supernatant fluid was collected and concentrated to 5 mL using a Centricon YM-30 and designated the cytoplasmic fraction. The pellet, consisting of intact nuclei and chromatin, was resuspended in 5 mL of NIB containing 0.3% (v/v) Triton X-100 together with 1× protease inhibitor cocktail (Sigma-Aldrich). A portion of the lysate (800 µL) was layered on top of an 800-µL layer of 2.3 m Suc in a 2-mL microcentrifuge tube, which was then subjected to centrifugation at 12,000g for 10 min. The nuclei pellet was washed twice with NIB. Nuclear proteins were extracted by resuspending the pellet in 800 µL of nuclear protein extraction buffer followed by shaking for 30 min. A nuclear protein extract was obtained by centrifugation at 12,000g for 10 min and collecting the supernatant fluid. Forty microliters of cytosolic and nuclear proteins were separated on 10% (v/v) SDS-PAGE gels.
Co-IP Test and RNase Treatment
Antibodies raised against RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q were prepared and affinity purified as previously described (Crofts et al., 2010). Antibodies were cross linked to protein A as described in the manufacturer’s instruction for Co-IP (Pierce Biotechnology).
Seed extracts used for Co-IP studies were prepared by grinding 1 g of deglumed, middeveloping rice seeds (11–15 DAF) in 3 mL of IP buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, pH 8.0, 0.5% [v/v] Nonidet P-40, 1 mm dithiothreitol, 5 unit mL–1 SUPERase In [Ambion], and 1× Protease inhibitor cocktail [Roche]). The homogenate was subjected to centrifugation at 100g for 5 min, followed by a second centrifugation of the 100g supernatant at 12,000g for 10 min at 4°C. The soluble protein extract was incubated with cross-linked RBP antibody-protein A-conjugated agarose beads (Invitrogen) overnight with gentle rotation at 4°C. The antibody beads were washed with 1 mL of cold IP buffer five times. Binding protein was eluted by addition of 100 µL of low pH buffer contained in Crosslink Co-IP Kit (Pierce Biotechnology). Twenty microliters of elute were separated on 10% (v/v) acrylamide gel. For RNase treatment, a mixture containing 10 ng of RNase A and 1 unit of RNase T was added to the IP mixture, incubated for 5 min at room temperature, and then washed and processed as described for the untreated samples.
Immunoblotting
Proteins separated on acrylamide gels were electroblotted onto nitrocellulose membrane (Pall Corporation). The blotted membrane was incubated overnight at 4°C with primary polyclonal antibodies (1:1,000) in 4% (w/v) nonfat milk powder dissolved in 1× Tris-buffered saline (TBS) buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl). After two washes with fresh 1× TBS, the membrane was incubated with goat anti-rabbit horseradish peroxidase conjugated secondary antibody (1:1,000; Pierce Biotechnology) for 1 h at room temperature with gentle shaking. Finally, the membrane was washed two times with 1× TBS/0.1% (v/v) Tween 20 to remove excess antibodies. The antibody-antigen complex was detected by chemiluminescence using the SuperSignalWest Pico Chemiluminescent Substrate (Pierce Biotechnology). The chemiluminescent signals were detected using a FujiFilm LAS-3000 image analyzer.
Biotinylated Zipcode RNA Binding Assay
The biotinylated prolamine RNA fragments were used as previously described (Crofts et al., 2010). Namely, three biotinylated RNA fragments of 5′ CDS zipcode (GAGUUUGAUGUUUUUAGGUCAAAGUUAUAGGCAAUA), 3′ UTR zipcode (UGUAAUGUGUUUUAACAGUAUAGUGGUUCG), and a control RNA (GUUCAGGCCAUAGCGUACGAGCUACAACUCCAGCAA) were synthesized by Integrated DNA Technologies. One hundred picomoles of biotinylated RNA was bound to 75 µg of streptavidin-conjugated magnetic beads (Roche Applied Science) by incubating at 4°C for 30 min. After washing, the biotinylated RNA-streptavidin-conjugated magnetic beads were incubated with 1 mL of seed extract. The seed extract was prepared as described for IP sample preparation. The mixture of protein-biotinylated RNA beads was incubated for 2 h at 4°C with gentle rotation. The magnetic beads were then washed five times with 1 mL of fresh IP buffer and the proteins eluted by treatment with 100 µL of 1× SDS sample buffer. SDS-PAGE and immunoblotting were performed as described above.
Yeast Two-Hybrid Assay
The Matchmaker Library Construction and Screening Kit was used for generation of yeast (Saccharomyces cerevisiae) gene constructs and yeast two-hybrid screening on synthetic dextrose plates containing X-α-gal with deficiency of Trp and Leu (Clontech). For preparation of yeast cells harboring RBPs for two-hybrid analysis, CDS for the various RBPs, prepared from reverse-transcribed RNAs isolated from midstage developing seeds, were amplified and inserted into pGBKT7 and pGADT7 for bait and prey gene constructs, respectively. Primer sets used for PCR amplification are shown in Supplemental Table S1. pGBKT7 and pGADT7 plasmids with RBP coding sequences were transformed into yeast strains Y187 and AH109, respectively, by electroporation using a MicroPulser (Bio-Rad). Control plasmids without inserts were also transformed into each yeast strain as negative controls. Mating and screening the yeast hybridizations were performed according to the manufacturer’s instructions.
Preparation of BiFC Gene Constructs and Transformation into Tomato Protoplast
The pSAT1 plasmids from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org) were used to generate N- and C-terminal fusions to EYFP fragments. Coding sequences of RBP-A, RBP-I, RBP-J, RBP-K, and RBP-Q were amplified with gene-specific primer sets (listed in Supplemental Table S1) harboring restriction sequence against multiple cloning sites on pSAT1 plasmid. The PCR products were cloned in pCR2.1 (Life Technologies), and open reading frames were confirmed by Sanger sequencing (Eurofin MWG Operon). The RBP sequences were then cloned into pSAT1 plasmid containing either nEYFP or cEYFP fragments at N terminus of RBP. Gene constructs are illustrated in Figure 7A.
Protoplasts were isolated from tomato (Solanum lycopersicum) suspension cells (Kim et al., 2012). DNA transformation was accomplished by adding 20 μg of each nEYFP and cEYFP plasmid to 250 µL of protoplasts (2 × 106 cells per mL) followed by 40% (w/v) polyethylene glycol solution (40% polyethylene glycol 4000, 0.4 m mannitol, 0.1 m Ca(NO3)2, pH 8–9) to 20% [w/v] final concentration. After 20 min at room temperature, transformed protoplasts were allow to recover in K3 solution at 25°C for 16 h (Schütze et al., 2009). EYFP fluorescence was observed with a Zeiss LSM510 Meta Laser Scanning Microscope or Leica TCS SP5 confocal system using an excitation wavelength range of 490 to 515 nm and emission intensity of 520 to 560 nm. Control studies using tomato protoplasts cotransformed with EYFP fragment empty vectors are shown in Supplemental Fig. S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Liquid chromatography-tandem mass spectrometry analysis of the 45-kD region identifies four peptides of RBP-I.
Supplemental Figure S2. Tomato protoplast cotransformed with EYFP fragment empty vectors.
Supplemental Table S1. Primer list for yeast two hybridization and BiFC.
Acknowledgments
We thank Drs. Kelly A. Doroshenk, Mio Satoh-Cruz, and Changlin Wang for help with material and sample preparation and Akita Prefectural University for providing access to equipment.
Glossary
- ER
endoplasmic reticulum
- PB-ER
ER membranes that delimit the prolamine intracisternal granules
- UTR
untranslated region
- CDS
coding regions
- hnRNP
heterogeneous nuclear ribonucleoprotein
- RRM
RNA recognition motif
- DAF
d after flowering
- Co-IP
coimmunoprecipitation
- RNase
ribonuclease
- BiFC
biomolecular fluorescence complementation
- NIB
nuclei isolation buffer
- TBS
Tris-buffered saline
- IP
immunoprecipitation
Footnotes
This work was supported by National Science Foundation grants (nos. DBI–0605016 and IOS–1021699), National Science Foundation Intergovernmental Personnel Act Funds, Project 0590, Agricultural Research Center, College of Agricultural, Human, and Natural Resource Sciences, Washington State University, and the Japanese Society for the Promotion of Science in the form of a Grant-in-Aid for Young Scientists (to A.J.C.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Arn EA, Cha BJ, Theurkauf WE, Macdonald PM. (2003) Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell 4: 41–51 [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. (2000) She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 19: 5514–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Crofts N, Whitelegge JP, Okita TW. (2010) Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 231: 1261–1276 [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Satoh M, Ogawa M, Kumamaru T, Satoh H. (2005) The role of mRNA and protein sorting in seed storage protein synthesis, transport, and deposition. Biochem Cell Biol 83: 728–737 [DOI] [PubMed] [Google Scholar]
- Doroshenk KA, Crofts AJ, Morris RT, Wyrick JJ, Okita TW. (2012) RiceRBP: a resource for experimentally identified RNA binding proteins in Oryza sativa. Front Plant Sci 3: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. (2004) Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963 [DOI] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Choi SB, Wang C, Singh S, Kawai N, Franceschi VR, Okita TW. (2003a) The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW. (2003b) Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Moinuddin SG, Atwell KM, Costa MA, Davin LB, Lewis NG. (2012) Opposite stereoselectivities of dirigent proteins in Arabidopsis and schizandra species. J Biol Chem 287: 33957–33972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Yoon YJ, Mowry KL. (2004) Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol 165: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Francis-Lang H, Flament A, Norvell A, Schüpbach T, Ish-Horowicz D. (1999) Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell 98: 171–180 [DOI] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lee LY, Wu FH, Hsu CT, Shen SC, Yeh HY, Liao DC, Fang MJ, Liu NT, Yen YC, Dokládal L, et al. (2012) Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell 24: 1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Mowry KL. (2007) Ribonucleoprotein remodeling during RNA localization. Differentiation 75: 507–518 [DOI] [PubMed] [Google Scholar]
- Li X, Franceschi VR, Okita TW. (1993) Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 72: 869–879 [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. (2000) She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AS, Moran-Jones K, Shan J, Munro TP, Snee MJ, Hoek KS, Smith R. (2002) Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem 277: 18010–18020 [DOI] [PubMed] [Google Scholar]
- Marchand V, Gaspar I, Ephrussi A. (2012) An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Curr Opin Cell Biol 24: 202–210 [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. (2009) mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518 [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Patel MV, Rose A, Tiedge H. (2011) Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J Cell Biol 194: 441–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. (2011) Translation-independent localization of mRNA in E. coli. Science 331: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Ohad N, Shichrur K, Yalovsky S. (2007) The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol 145: 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM. (2007) How does an mRNA find its way? Intracellular localisation of transcripts. Semin Cell Dev Biol 18: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C. (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- St Johnston D. (2005) Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 6: 363–375 [DOI] [PubMed] [Google Scholar]
- Stewart M. (2007) Ratcheting mRNA out of the nucleus. Mol Cell 25: 327–330 [DOI] [PubMed] [Google Scholar]
- Washida H, Kaneko S, Crofts N, Sugino A, Wang C, Okita TW. (2009a) Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol 50: 1710–1714 [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Doroshenk KA, Satoh-Cruz M, Nagamine A, Katsube-Tanaka T, Ogawa M, Kumamaru T, Satoh H, Okita TW. (2012) RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J 70: 471–479 [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Kaneko S, Crofts N, Sakulsingharoj C, Kim D, Choi SB, Hamada S, Ogawa M, Wang C, et al. (2009b) Identification of cis-localization elements of the maize 10-kDa δ-zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J 60: 146–155 [DOI] [PubMed] [Google Scholar]
- Yano T, López de Quinto S, Matsui Y, Shevchenko A, Shevchenko A, Ephrussi A. (2004) Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell 6: 637–648 [DOI] [PubMed] [Google Scholar]