Anther indehiscence at high temperatures is coordinately regulated by sugar and auxin.
Abstract
Male reproduction in flowering plants is highly sensitive to high temperature (HT). To investigate molecular mechanisms of the response of cotton (Gossypium hirsutum) anthers to HT, a relatively complete comparative transcriptome analysis was performed during anther development of cotton lines 84021 and H05 under normal temperature and HT conditions. In total, 4,599 differentially expressed genes were screened; the differentially expressed genes were mainly related to epigenetic modifications, carbohydrate metabolism, and plant hormone signaling. Detailed studies showed that the deficiency in S-ADENOSYL-l-HOMOCYSTEINE HYDROLASE1 and the inhibition of methyltransferases contributed to genome-wide hypomethylation in H05, and the increased expression of histone constitution genes contributed to DNA stability in 84021. Furthermore, HT induced the expression of CASEIN KINASEI (GhCKI) in H05, coupled with the suppression of starch synthase activity, decreases in glucose level during anther development, and increases in indole-3-acetic acid (IAA) level in late-stage anthers. The same changes also were observed in Arabidopsis (Arabidopsis thaliana) GhCKI overexpression lines. These results suggest that GhCKI, sugar, and auxin may be key regulators of the anther response to HT stress. Moreover, PHYTOCHROME-INTERACTING FACTOR genes (PIFs), which are involved in linking sugar and auxin and are regulated by sugar, might positively regulate IAA biosynthesis in the cotton anther response to HT. Additionally, exogenous IAA application revealed that high background IAA may be a disadvantage for late-stage cotton anthers during HT stress. Overall, the linking of HT, sugar, PIFs, and IAA, together with our previously reported data on GhCKI, may provide dynamic coordination of plant anther responses to HT stress.
Temperature is an important physical parameter affecting all living organisms on Earth, and global warming has major implications for plant development, growth, reproduction, and yield (Ahuja et al., 2010). Increasing temperatures may induce the instability of crop yields, mainly because the reproductive phase is vulnerable to the effects of increasing global temperature. In particular, male reproductive organs in higher plants are known to be more sensitive to temperature increases than female organs (Peet et al., 1998). Decreased crop yields have mainly occurred because high temperature (HT) increases the probability of male sterility (Hedhly et al., 2009; Zinn et al., 2010).
In flowering plants, male reproductive development includes a series of complicated events (Goldberg et al., 1993). During this process, the young microspore stage has been reported to be the most sensitive to temperature changes. In the tapetum, the innermost cell layer of the anther is known to be the most sensitive cell layer to environmental stress during the young microspore stage (Suzuki et al., 2001). Additionally, some researchers have reported that the flowering stage and the young microspore stage of rice were most sensitive to HT stress, showing poor anther dehiscence during flowering and disrupted tapetum functions during young microspore development (Sato et al., 2002; Endo et al., 2009). Cotton (Gossypium hirsutum) produces a characteristic indeterminate inflorescence, and flower bud development is asynchronous. The anther stage can be very different among buds according to bud length (Ma et al., 2012). In general, the buds of cotton begin to appear at 40 to 45 d after seedling emergence, and the cotton plant reaches a peak in budding and flowering in late July; interestingly, in China, HT occurs most frequently in July. Thus, cotton is seriously subjected to yield decreases of approximately 110 kg ha−1 for each 1°C increase in ambient temperature (Singh et al., 2007), due to HT causing cotton pollen abortion, anther indehiscence, and boll shedding. However, few studies have focused on the molecular mechanisms of HT-induced reproductive sterility in cotton, although other crops, such as rice (Oryza sativa; Jagadish et al., 2007), wheat (Triticum aestivum; Saini et al., 1984), and barley (Hordeum vulgare; Oshino et al., 2007), have been investigated.
In recent decades, accumulating transcriptomic and proteomic studies on plant species have identified stress-response pathways that function during anther development (Frank et al., 2009; Jagadish et al., 2010). Several genes that play roles in the anther development of angiosperms have been cloned and studied using various molecular techniques (Zhang et al., 2011). Meanwhile, environmental stresses were regarded as important factors affecting crop reproduction, among which heat stress was focused on due to the increasing global temperature (Mittler et al., 2012). Plants usually utilize valuable resources to modify their metabolism and protect themselves against HT damage through an intricate mechanism involving perception, transduction, and exchange of signals. Plants putatively possess four sensors, inward calcium flux, histones, unfolded proteins in the endoplasmic reticulum, and unfolded proteins in the cytosol, that trigger different signal transduction events (e.g. sugar signaling and plant hormone signaling) to establish a new steady-state balance of metabolic processes by altering the composition of certain transcripts, proteins, metabolites, and lipids (McClung and Davis, 2010; Suzuki et al., 2012). In our previous study, HT induced CASEIN KINASEI (GhCKI) expression earlier in an HT-sensitive cotton line, and overexpression of GhCKI regulated anther abortion by inhibiting the activities of starch synthases, disrupting the homeostasis of Glc, abscisic acid (ABA), and reactive oxygen species and causing male sterility (Min et al., 2013). In the last 2 years, many reports have demonstrated that PHYTOCHROME-INTERACTING FACTOR genes (PIFs) were required for hypocotyl elongation at HT by regulating auxin biosynthesis (Franklin et al., 2011). Additionally, soluble sugar also regulates auxin biosynthesis through PIF proteins (Sairanen et al., 2012). Thus, there is an intimate and complex regulation network involving sugar signaling and plant hormone signaling under HT. However, few genes and transcriptomes have been studied in cotton to improve our understanding of how male sterility occurs and how the regulatory mechanism functions during anther development. Additionally, the means by which regulatory networks manage male sterility after HT has not yet been studied.
To better understand how cotton addresses HT stress during the male reproductive stage, we used RNA sequencing (RNA-seq) to produce relatively complete transcriptome data during cotton anther development. Based on the analysis of complex regulatory networks and differentially expressed genes, we found that genes related to epigenetic modifications, starch and Suc synthesis and metabolism, and auxin signaling showed differences in expression. Detailed studies on gene expression, enzyme activity, and the contents of various substances revealed significant differences between HT-tolerant and HT-sensitive cotton anthers.
RESULTS
Kinetics of Cotton Anther Development under HT Stress
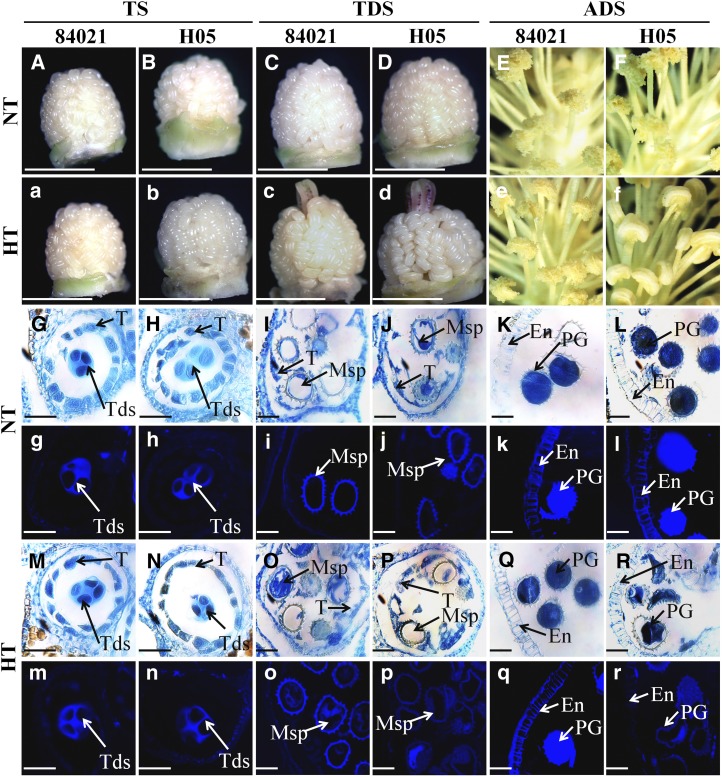
To explore the mechanism of anther abortion under HT stress, two cotton lines, 84021 (HT tolerant) and H05 (HT sensitive), were employed in this study (Supplemental Fig. S1). Obvious HT damage was verified by the appearance of visual symptoms on H05, such as indehiscent anthers, whereas the anthers of 84021 were split normally under HT conditions. No difference was observed between the lines under normal temperature (NT) conditions (Fig. 1, A–F and a–f). To monitor the response of anther tissues to HT, changes in the histology of 84021 and H05 anthers were determined at the tetrad stage (TS; 6- to 7-mm bud), tapetal degradation stage (TDS; 9- to 14-mm bud), and anther dehiscence stage (ADS; more than 24-mm bud) after the plants were exposed to HT for 7 d. Anthers at the same developmental stages exposed to NT were adopted as controls. Little change was observed in the anther development of 84021 after 1 week of treatment with HT compared with that observed with NT based on histological analysis (Fig. 1, G, I, K, M, O, and Q) and aniline blue staining (Fig. 1, g, i, k, m, o, and q). Similarly, the meiotic behavior of pollen mother cells was normal, and the tetrads formed after exposure to HT for 7 d in H05 were similar to those formed under NT (Fig. 1, H, N, h, and n). However, the development of microspores was seriously affected by HT in H05 (Fig. 1, J, P, j, and p), and HT caused defects in the secondary thickening of the anther endothecium wall (Fig. 1, L, R, l, and r). As a result, the anthers of H05 were aborted due to indehiscence and partial male sterility. The different responses between 84021 and H05 anthers were utilized to study changes in the transcriptome after exposure to HT by RNA-seq.
Figure 1.
Schematic representation and histological observation of different cotton anther developmental stages under NT and HT conditions. Similar tissues were used for RNA-seq analysis. A to F and a to f, Anthers from 84021 (A, C, and E) and H05 (B, D, and F) at TS (A and B), TDS (C and D), and ADS (E and F) under NT conditions (controls) and anthers from 84021 (a, c, and e) and H05 (b, d, and f) at TS (a and b), TDS (c and d), and ADS (e and f) under HT conditions. No significant differences between 84021 and H05 buds were detected at TS under the two conditions. Buds at TDS show exposed stigmas in both lines, whereas the H05 plant shows indehiscent anthers and the 84021 plant shows normally split anthers under HT conditions at ADS. G to R, Cross sections of anthers from 84021 (G, I, K, M, O, and Q) and H05 (H, J, L, N, P, and R) at TS (G, H, M, and N), TDS (I, J, O, and P), and ADS (K, L, Q, and R) under NT (G–L) and HT (M–R) conditions. No significant differences were observed in the formation of tetrads after exposure to HT for 7 d in 84021 and H05 anthers. However, microspores and pollen grains were abnormal in H05 anthers at TDS and ADS. g to r, Cross sections of 84021 and H05 anthers were stained with aniline blue to detect callose and secondary wall thickening, as shown by bright blue fluorescence. Anthers of 84021 (g, i, k, m, o, and q) and H05 (h, j, l, n, p, and r) are shown at TS (g, h, m, and n), TDS (i, j, o, and p), and ADS (k, l, q, and r) under NT (g–l) and HT (m–r) conditions. Microspores and pollen grains in H05 anthers at TDS and ADS showed weak blue fluorescence. There was no obvious secondary wall thickening in the anther endothecium wall in H05 after HT. Msp, Microspore; PG, pollen grain; T, tapetum; Tds, tetrads; En, endothecium. Bars = 5 mm (A–D and a–d) and 50 μm (G–R and g–r).
Comparison of HT-Induced Cotton Anther Transcript Profiles between 84021 and H05
Cotton is a polyploid species with large genome size (approximately 2.5 Gb); the draft genome of the wild diploid cotton Gossypium raimondii had not been sequenced and assembled before August 2012 (Wang et al., 2012). Thus, to gain insight into the molecular mechanisms of anther abortion under HT in cotton, RNA-seq was adopted in this study in 2009 to analyze differentially expressed genes (DEGs) during anther development to compare transcriptional alterations under NT and HT conditions. A total of 147,028,153 clean reads from 12 samples with an average length of 49 bp were generated. Due to genome sequence references being unavailable, we mapped the reads to two reference databases (one was cotton contigs containing 65,386 sequences, assembled from multiple genes from different databases, and the other was National Center for Biotechnology Information cotton unigenes). Finally, for each sample, approximately 60% of the reads were not mapped to the cotton contigs, and 37.9% of the mapped reads included 34.8% matched unique reference sequence. There were on average 4,645,018 unambiguous mapped clean reads per sample. For each gene, the transcripts were calculated and subsequently normalized to reads per kilobase per million reads (RPKM; Mortazavi et al., 2008), which represents gene expression levels. The overview of the read information is shown in Supplemental Table S1. No significant differences existed in the number of reads among the 12 samples.
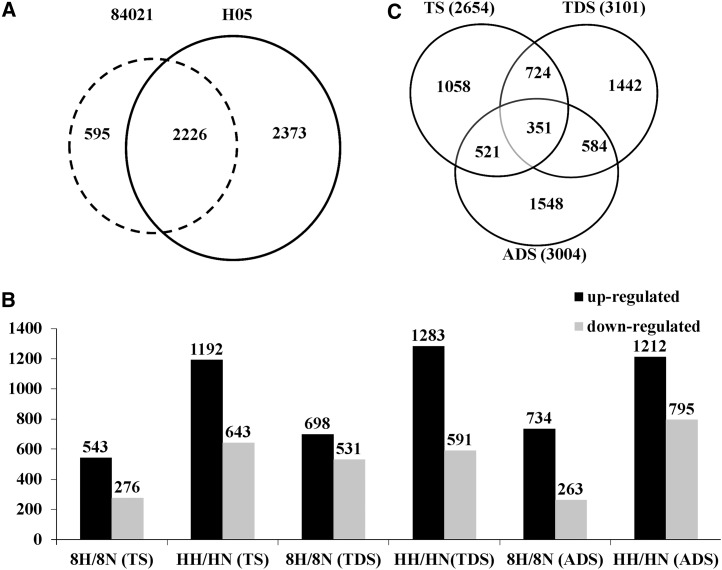
To further investigate HT-induced male sterility in the HT-sensitive cotton line H05, a total of 20,258 genes were collected from cotton anthers that responded to HT. Among these, 5,194 DEGs were filtered with a cutoff of RPKM ≥ 50 (P ≤ 0.001) and an absolute value of log2 ratio ≥ 1 based on a false discovery rate < 0.001 (Supplemental Table S2). These results showed that the number of differentially expressed HT response genes in H05 was significantly greater than that in 84021. In total, 4,599 DEGs were isolated from H05; however, only 2,821 DEGs were identified in 84021, and 2,226 DEGs were identified in both lines, which indicated that there was a significant difference in HT-dependent gene regulation between the two cotton lines (Fig. 2A). To determine the important genes responsible for HT tolerance, we divided the 5,194 DEGs into 25 clusters based on their expression patterns by Genesis, which is based on the K-means method (Supplemental Fig. S2), and found that 595 DEGs showed the same expression pattern between 84021 and H05; the remaining 4,599 DEGs were up- or down-regulated at different anther development stages in the two lines under the two temperature conditions (Fig. 2B; Supplemental Table S3). More genes were up-regulated during HT stress in both lines, and more genes were found to be responsive to HT stress in H05 at all tested stages of anther development. Spatial analysis was also performed on the DEGs to uncover the degree of overlap between the three different anther development stages of 84021 and H05 under HT and NT. There were 2,654, 3,101, 3,004 DEGs during TS, TDS, and ADS, respectively (Fig. 2C). Among these, 819, 1,229, and 997 DEGs of 84021 and 1,835, 1,872, and 2,007 DEGs of H05 were present in the three anther developmental stages. These data indicated that, regardless of the action of HT on 84021 or H05, there were significant numbers of DEGs present in TDS, which is the most sensitive stage of anther development, consistent with previous reports (Suzuki et al., 2001).
Figure 2.
Analysis of DEGs based on RNA-seq data. A, Distribution of 5,194 DEGs. Dashed and solid circles refer to 84021 and H05, respectively. B, Number of differentially expressed genes that were up- or down-regulated at different stages during anther development. 8N and 8H refer to 84021 under NT and HT conditions, respectively; HN and HH refer to H05 under NT and HT conditions, respectively; C, Venn diagram showing the DEGs in each of the three different anther developmental stages of the two cotton lines. The overlapping regions correspond to the number of DEGs present at more than one anther developmental stage.
To dissect the potential functions of 4,599 DEGs (Supplemental Table S3) that were responsive to HT, we searched the nonredundant protein sequence database in GenBank (National Center for Biotechnology Information) using Blast2GO (Götz et al., 2008). Using this approach, 4,205 genes returned BLAST results. Through mapping and annotation, 3,601 genes could be classified into one or more ontologies: 3,065 DEGs participated in biological processes, 2,764 DEGs had molecular functions, and 3,185 DEGs had cellular components. Furthermore, the functional category distribution frequency was calculated based on levels 2 and 3 (Supplemental Fig. S3). At the biological process level, the 3,065 DEGs could be assigned to 23 biological processes, including cellular processes (2,634 DEGs), metabolic processes (2,431 DEGs), responses to stimuli (1,632 DEGs), development, regulation, localization, signaling, reproduction, death, and other processes. Among these biological processes, cellular and metabolic processes each accounted for more than two-thirds of all annotated DEGs. These results implied that cell growth and metabolic activities were strongly affected after HT.
To explore the biological pathways that are important for anther development in response to HT, DEGs were annotated to the reference pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) using KeggArray software (Supplemental Fig. S4A). In total, 2,098 unigenes were assigned to 121 KEGG pathways. When sorting the pathway search results by the number of hits, metabolic pathways (396 members) and biosynthesis of secondary metabolites (225 members) were the most highly represented groups in typical pathways. Genes participating in protein processing in endoplasmic reticulum (57 members), ribosome (53 members), and plant hormone signal transduction (48 members) were enriched. In particular, pathways involved in carbohydrate and energy metabolism, including glycolysis/gluconeogenesis (36 members), starch and Suc metabolism (33 members), pyruvate metabolism (29 members), citrate cycle/tricarboxylic acid cycle (19 members), and oxidative phosphorylation pathways (40 members), were significantly affected during HT. DEGs associated with carbohydrate metabolism, plant hormone synthesis and signal transduction, and endoplasmic reticulum stress, which are hallmarks of anther development under HT, were identified through gene expression profiling (Supplemental Fig. S4B).
Analysis of Genomic DNA Methylation and Histone Modification
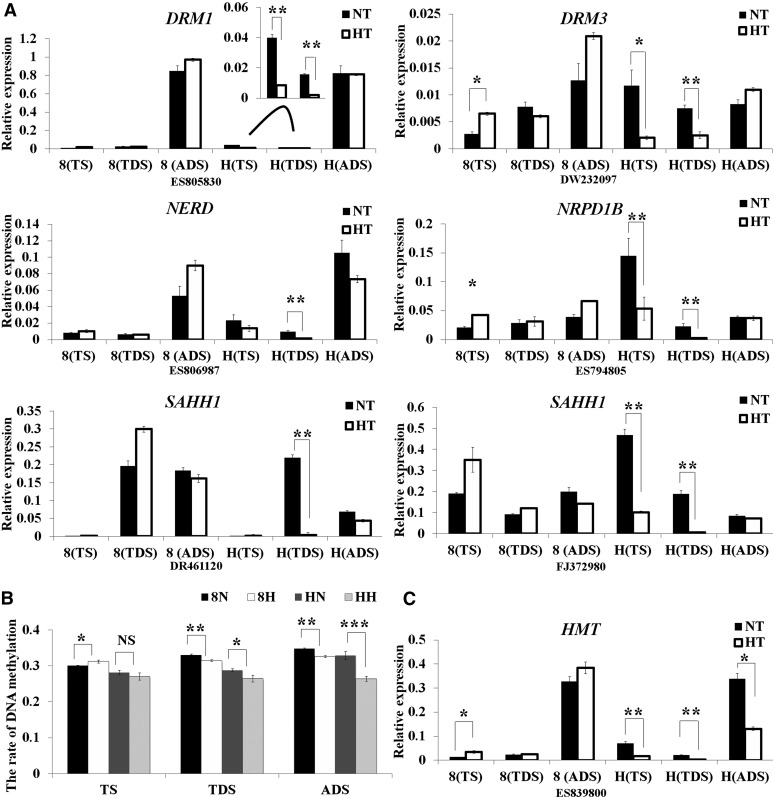
Because global gene expression is mainly regulated by DNA methylation and histone modification (Strahl and Allis, 2000; Martinowich et al., 2003), we focused on characterizing genes involved in DNA methylation and histone modification during the anther development process under HT. The expression of two DNA methyltransferases, DNA CYTOSINE-5-METHYLTRANSFERASE (DRM1) and S-ADENOSYL-l-METHIONINE-DEPENDENT METHYLTRANSFERASE (DRM3), was significantly down-regulated at TS and TDS and showed no significant differences at ADS in H05 by HT. However, no significant differences in the expression levels of either gene were observed in 84021 anthers under HT, except that DRM3 was up-regulated at TS (Fig. 3A). Four genes required for normal DNA methylation, NEEDED FOR RDR2-INDEPENDENT DNA METHYLATION (NERD), NUCLEAR RNA POLYMERASE D1B (NRPD1B), and two S-ADENOSYL-l-HOMOCYSTEINE HYDROLASE1 (SAHH1) genes, were also influenced at TS and TDS anthers and showed no significant differences at ADS anthers in H05 under HT conditions. Compared with DRM1 and DRM3, the four normal DNA methylation genes showed similar expression changes in 84021 and H05 under HT (Fig. 3A). Irrespective of the DNA methyltransferases or genes required for normal DNA methylation, the expression of these genes was suppressed by HT in H05 anthers but not in 84021 anthers. To confirm this result, the genome-wide total DNA methylation level of anthers was measured by HPLC at three anther developmental stages under two conditions. It was found that the total DNA methylation level of H05 was not associated with HT treatment and was always lower than that in 84021. Additionally, HT evidently decreased the degree of DNA methylation in H05 compared with that in 84021 (Fig. 3B), which was in accordance with the results from gene expression analysis.
Figure 3.
Expression profiles of genes in the DNA methylation pathway. A, The expression levels of DNA methylation-related genes identified by RNA-seq were validated by qRT-PCR in 84021 and H05 under two conditions. The black and white columns refer to NT and HT conditions, respectively. 8 and H indicate 84021 and H05, respectively. B, Quantification of the genome-wide total DNA methylation level via HPLC during three anther development stages. The y axis represents the rate of DNA methylation. 8N and 8H refer to 84021 under NT and HT conditions, respectively; HN and HH refer to H05 under NT and HT conditions, respectively. C, The expression of HMT was validated by qRT-PCR in 84021 and H05 under two conditions. Data are presented as means ± se from three biologically independent experiments. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) by Student’s t test. NS, Not significant.
Similar to DNA methylation, several histone constitution- and modification-related genes were found to be differentially expressed in response to HT. A total of 24 histone constitution genes (such as HISTONE H1, HISTONE H2A, HISTONE H2B, HISTONE H3, and HISTONE H4) showed expression peaks at the TS and were subsequently down-regulated during anther development in both lines (Supplemental Fig. S5); the expression of histone constitution genes was higher in 84021 than in H05. Similarly, some genes involved in histone modification were differentially expressed during anther development under HT. HMT, a histone methyltransferase associated with the suppression of gene transcription, had a high expression level at ADS but was found to be down-regulated in H05 upon HT at three anther development stages (Fig. 3C). However, two jumonji C (jmjC) domain-containing genes that function as histone demethylases were also clearly suppressed by HT in H05 (Supplemental Fig. S6). In addition, two HISTONE ACETYLTRANSFERASES (HAT1 and HAT2) and two HISTONE DEACETYLASES (HDA15 and HDA2C) in H05 showed significant differences in expression under HT. The expression of one HISTONE MONOUBIQUITINATION gene, which is involved in the ubiquitylation of histone H2B and the activation of genomic transcription (Liu et al., 2007), was down-regulated by HT in anthers at early developmental stages in H05 (Supplemental Fig. S6). Based on the above, we may deduce that the suppressed expression of DNA methylation-related genes in H05 caused the decreased DNA methylation level; the higher expression of histone constitution genes in 84021 allowed anthers at different developmental stages to maintain DNA stability. The expression of histone modification genes tended to activate gene transcription in both lines, although the degree of change in gene expression in H05 was stronger than that in 84021. These results indicate that more DEGs appeared, and a larger proportion of the DEGs showed increased expression levels in H05 anthers under HT, which is consistent with the results of statistical analysis of the expression patterns. These results also indicated that epigenetic modifications, particularly DNA methylation and histone modification, were involved in anther development under normal conditions and were influenced in both lines but respond differently to HT.
HT Disrupted Cotton Anther Carbohydrate Metabolism Pathways
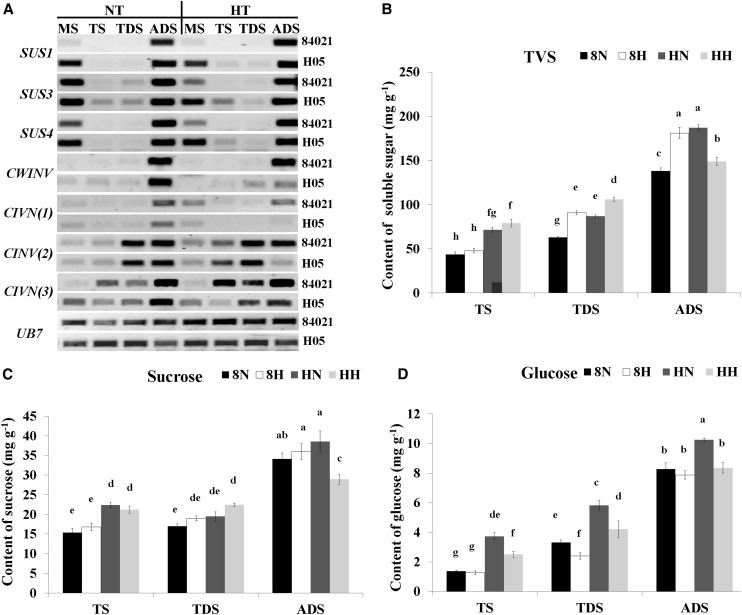
A subset of genes involved in carbohydrate and energy metabolism during cotton anther development under HT was characterized. Among these, 33 genes with homologs that encode major functional enzymes in starch and Suc metabolism pathways in other species were examined during anther development of cotton exposed to HT. The expression profiles of the genes were characterized using RNA-seq analysis and were clustered and displayed in a heat map. The corresponding locations of these enzymes in the starch and Suc metabolism pathways are shown in Supplemental Figure S7. The expression of some genes was also verified by reverse transcription (RT)-PCR (Fig. 4A; Supplemental Fig. S8A), and the results showed significantly higher expression of SUCROSE SYNTHASE1 (SUS1), SUS3, and SUS4 at the meiosis stage in H05 compared with 84021 under NT conditions. There were no significant differences in the expression levels of the three SUS genes between the two lines after HT. However, compared with their levels in 84021, the expression levels of four INVERTASE (INV) genes, which catalyze the hydrolysis of Suc to Glc and Fru, one cell wall invertase (CWINV) and three cytosolic alkaline/neutral invertases (CINV), were significantly reduced by HT in H05 at ADS anthers (Fig. 4A). CWINVs have previously been shown to affect anther sink strength under cold conditions (Oliver et al., 2005). This result suggests that the H05 anther has defects in the conversion of Suc to Glc and Fru under HT, which leads to Suc accumulation and activates the starch synthesis pathway and then affects anther sink strength. To further confirm this result, the contents of total soluble sugar, Suc, and Glc in 12 samples were assessed under NT and HT conditions (Fig. 4, B–D). A detailed analysis showed that the total soluble sugar and Suc contents underwent similar changes in the two cotton lines when subjected to HT at the early anther development periods. At the anthesis stage, the total soluble sugar and Suc contents in anthers increased in 84021; however, significant reductions in the total soluble sugar content and the Suc content were observed in H05 during HT stress (Fig. 4, B and C). The results of Glc content assays showed that the Glc level in the anther increased gradually during 84021 and H05 anther development under NT, and higher background Glc content was observed in H05. When the anthers were subjected to HT stress, the Glc level did not significantly change in the 84021 TS and ADS anthers but was significantly reduced in the TDS anthers. Compared with 84021, the Glc content of H05 was reduced more significantly and much earlier, and significantly reduced Glc content was noted at every anther developmental stage under HT (Fig. 4D). Plant SNF1-RELATED PROTEIN KINASE1 (SnRK1) and HEXOKINASE1 (HXK1) and yeast Saccharomyces cerevisiae (baker's or brewer's yeast) HXK2, YEAST CASEIN KINASE I (YCKI), and cAMP-DEPENDENT PROTEIN KINASE A (cAMP-PKA) are the central metabolic genes involved in sugar sensing and signal transduction pathways in plant and yeast, respectively (Rolland et al., 2006). In our RNA-seq data, the SnRK1 regulatory subunit γ-1 (ES803272) and CKI (JQ713826) were up-regulated in H05 in early developmental stage anthers after HT stress (Supplemental Fig. S8B), which is consistent with our previous study indicating that excessive accumulation of GhCKI led to the reduction of Glc content in anthers (Min et al., 2013). These results indicated that sugar homeostasis, especially the decrease in Glc content in anthers, correlated for anther abortion in H05 under HT.
Figure 4.
Effects of the conversion of Suc and Glc on HT injury in cotton anther development. A, The expression of SUS and INV genes was validated by RT-PCR. SUS genes were SUS1 (FJ713478), SUS3 (DT571793), and SUS4 (U73587). INV genes were CWINV (ES809752), CINV(1) (ES815699), CINV(2) (ES815150), and CINV(3) (ES794929). UB7 gene was UBQUTIN7 (DQ116441). No significant differences in SUS expression and decreased INV expression were observed in H05 under HT. MS, Meiosis stage. B to D, Determination of the partitioning of total soluble sugar (B), Suc (C), and Glc (D) in 12 samples of two cotton lines under NT and HT conditions. 8N and 8H refer to 84021 under NT and HT conditions, respectively; HN and HH refer to H05 under NT and HT conditions, respectively. Data are presented as means ± se from six biologically independent experiments. Values not sharing a common letter are considered statistically significant (shortest significant range; P < 0.05).
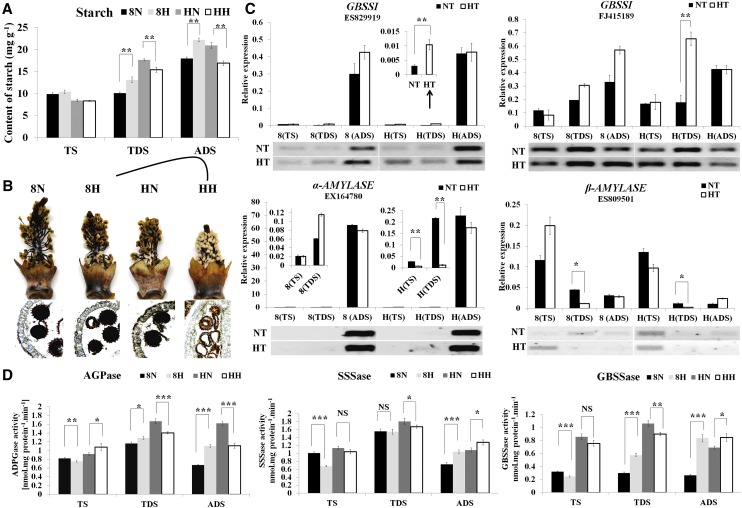
To verify whether H05 was abnormal in starch partitioning, starch content was assessed in 12 samples (Fig. 5A). Before the uninucleate microspore stage, we did not observe significant differences in starch accumulation in either 84021 or H05 during HT compared with the NT condition. At TDS and ADS, strong starch accumulation was observed in the anthers of 84021 under HT. However, significantly reduced starch accumulation was observed in the H05 anthers; it decreased to approximately 87.2% at TDS (17.62 mg g−1 fresh weight for NT and 15.37 mg g−1 fresh weight for HT) and to approximately 19.2% at ADS (20.88 mg g−1 fresh weight for NT and 16.87 mg g−1 fresh weight for HT), consistent with the staining of starch (Fig. 5B). Abnormal starch accumulation affected pollen development, consistent with the KEGG analysis. Generally, the starch accumulation was determined by the interplay between starch biosynthesis and catabolism pathways. To clarify the individual gene expression pattern, the genes involved in starch synthesis and catabolism pathways were analyzed. Expression of the starch synthesis gene GRANULE-BOUND STARCH SYNTHASE (GBSSI) was significantly induced by HT in H05, and the starch catabolism genes α-AMYLASE and β-AMYLASE were significantly reduced in H05 under HT. However, these genes showed no significant differences in 84021 (Fig. 5C). This result indicated that HT suppressed the accumulation of starch in H05, and H05 might acclimate to HT stress by triggering the expression of starch synthesis genes and suppressing the expression of starch catabolism genes.
Figure 5.
Effects of HT on starch synthesis and catabolism in 84021 and H05 anthers under HT and NT conditions. A, Starch content was assessed in 84021 and H05 anthers at three different developmental stages under HT and NT conditions. 8N and 8H refer to 84021 under NT and HT conditions, respectively; HN and HH refer to H05 under NT and HT conditions, respectively. B, Schematic representation and cross observation of starch staining in the anthers at ADS described in A. C, Expression levels of starch synthesis (GBSSI) and catabolism (α-AMYLASE and β-AMYLASE) genes were confirmed by qRT-PCR and RT-PCR in the anthers of 84021 and H05 under two conditions. The x axis indicates the anther developmental stages; 8 and H indicate 84021 and H05, respectively. D, The enzymatic activities of AGPase, SSSase, and GBSSase were assessed. The data in A, C, and D are presented as means ± se from three biologically independent experiments. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) by Student’s t test. NS, Not significant.
To our knowledge, most of the genes encoding the above enzymes belong to multigene families; thus, the starch content is determined not only by the interplay between starch biosynthesis and catabolism pathways but also by the interplay among multigene family members (Hawker and Jenner, 1993). The AGPase is responsible for the conversion of Suc into ADP-Glc; subsequently, starch synthase (GBSS, SOLUBLE STARCH SYNTHASE [SSS] and STARCH BRANCHING ENZYME) acts on the ADP-Glc to generate starch. Thus, the activities of AGPase, SSSase, and GBSSase were investigated (Fig. 5D). The result of the AGPase activity assay was basically consistent with previous data, indicating that H05 TDS and ADS anthers accumulate less starch during HT, indicating that the activities of starch synthase were greatly influenced during anther development under HT. However, increased activities of SSSase and GBSSase were noted in ADS anthers, which might be influenced by the Glc content in H05. Based on our results, the accumulation of Glc and starch in H05 anthers was reduced by HT, and the lack of either Glc or starch in the anther would seriously impair anther development, eventually leading to male sterility.
HT Causes Anther Abortion by Altering Auxin Metabolism and Signaling
Changes in the expression patterns of groups of genes implicated in different phytohormone signaling pathways were identified in anthers under HT (Supplemental Fig. S4B). It was shown that genes involved in auxin metabolism and signaling constituted the largest group, whereas cytokinin, jasmonic acid, and brassinosteroid signaling pathways were also obviously affected by HT, indicating that a complicated cross talk between phytohormones might be involved in the development of cotton anthers during HT (Fig. 6A). By combining the KEGG results with poplar (Populus spp.), Ricinus communis, and Arabidopsis (Arabidopsis thaliana) annotations, genes related to auxin metabolism and signaling could be classified into auxin synthesis, transport, degradation, and response genes. Six/two transcripts related to auxin biosynthesis/indole-3-acetic acid (IAA)-amido synthase were validated by RT-PCR. Auxin biosynthesis genes, such as TRYPTOPHAN BIOSYNTHESIS4, YUCCA2 (YUC2), YUC5, YUC11, NITRILASE2 (NIT2), and NIT4, showed enhanced transcription in H05 compared with 84021 under NT conditions. However, IAA-amido synthases, such as JASMONATE RESISTANT1 and an auxin-responsive GH3 gene family member, showed much lower expression in H05 than in 84021, regardless of the conditions (NT or HT). These results suggest that H05 anthers have higher expression levels of auxin biosynthesis genes and lower expression levels of auxin catabolism genes (Fig. 6B). The expression of auxin transport genes was also validated by RT-PCR (Supplemental Fig. S9). Two auxin influx genes, LIKE AUXIN RESISTANT1 and AUXIN RESISTANT1 (AUX1), were significantly induced by HT in the anthers of 84021 at ADS; however, the expression of the two genes was not significantly different in H05 between the NT and HT conditions. The auxin efflux genes RHAMNOSE BIOSYNTHESIS1 and AUXIN POLAR TRANSPORTER showed reduced expression at TDS in 84021, whereas PIN-FORMED3 showed increased expression at TDS in H05 after exposure to HT. These results indicated that 84021 anthers have greater polar auxin transport ability than H05 under the two conditions. In addition, several AUX/IAA, AUXIN RESPONSE FACTOR and SMALL AUXIN-UP RNA-LIKE genes were differentially expressed, showing sophisticated expression profiles (Supplemental Fig. S9).
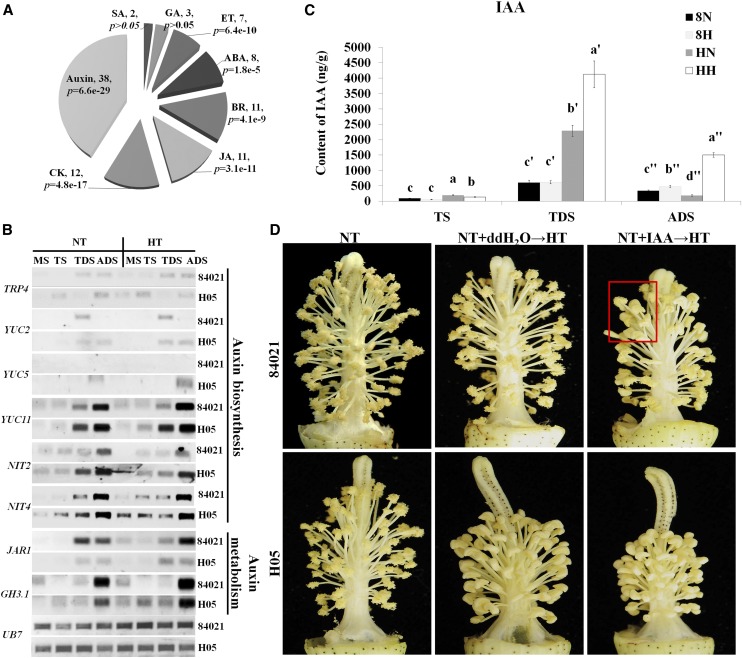
Figure 6.
Effects of the auxin signaling pathway on HT injury of cotton anthers. A, Analysis of the distribution of transcripts in plant hormone signaling pathways. BR, Brassinosteroid; CK, cytokinins; ET, ethylene; GA, GA3; JA, jasmonic acid; SA, salicylic acid. P values were determined by singular enrichment analysis (http://bioinfo.cau.edu.cn/agriGO), and the largest group was related to auxin signaling. B, RT-PCR was used to verify the detailed expression profiles of genes involved in auxin biosynthesis and catabolism. UB7, UBQUTIN7; MS, meiosis stage. C, Detection of endogenous IAA content in 84021 and H05 anthers at TS, TDS, and ADS under HT and NT conditions. H05 showed a higher background IAA concentration at TS and TDS. At all anther developmental stages, H05 showed a more sharply decreased/increased IAA concentration compared with 84021. 8N and 8H refer to 84021 under NT and HT conditions, respectively; HN and HH refer to H05 under NT and HT conditions, respectively. Data are presented as means ± se from six biologically independent experiments. Statistically significant difference analysis was performed at the respective anther developmental stage. Values not sharing a common letter are considered statistically significant (shortest significant range; P < 0.05). D, Representative anther structures are shown for each treatment. NT+ddH2O→HT and NT+IAA→HT indicate that distilled, deionized water and 10−6 m IAA, respectively, were applied four times (at days 7, 5, 3, and 0 before the temperature was increased to the stress point) to flower buds of 84021 and H05, which was followed by sustained HT damage for 7 d. The temperature was then restored to normal for 1 week, and photographs were taken. The red box indicates indehiscent anthers.
To verify the expression pattern of genes involved in auxin metabolism, the concentration of endogenous IAA was measured by HPLC-mass spectrometry. Large differences were found in the concentrations of endogenous IAA between 84021 and H05 at every anther development stage under NT and HT conditions (Fig. 6C). It was very interesting that the content of endogenous IAA was significantly higher in H05 compared with 84021 in TS and TDS anthers under NT. The IAA content decreased at TS under HT in both lines and increased by approximately 40% at TDS only in H05 compared with its level under NT. Moreover, approximately 10- and 4-fold increases in the concentration of IAA were found at ADS in H05 under HT compared with that in H05 under NT and 84021 under HT, respectively. These results indicated that HT may influence the homeostasis of endogenous IAA in anthers, and the higher level of IAA in H05, which was achieved through increasing the expression levels of biosynthesis genes and decreasing the expression of catabolism genes, may contribute to the abnormalities in anther development.
To further understand the role of auxin in anther development under HT, exogenous 10−5 or 10−6 m IAA (and water as a control) was applied to the flower buds of two cotton lines four times on the morning of days 7, 5, 3, and 0 before increasing the temperature to trigger HT injury. The application of 10−5 m IAA caused the loss of flower buds; however, the buds of two cotton lines grew normally with the application of 10−6 m IAA under NT. Therefore, we chose 10−6 m IAA to conduct experiments. After HT damage was allowed to occur for 7 d, the temperature was restored to normal for 1 week, and the phenotypes of the anthers were photographed at anthesis (Fig. 6D). The experiment showed that the application of exogenous IAA before subjecting the plants to HT injury resulted in increased indehiscent anthers in 84021, whereas anther indehiscence could not be suppressed in H05 compared with its level under pure HT treatment (water has been applied to flower buds four times before HT injury). These results indicated that HT exerted a strong effect on the endogenous IAA homeostasis of H05, suggesting that the higher background auxin concentration was caused by higher expression of auxin biosynthesis genes and lower expression of auxin catabolism genes. These changes then caused differential expression of auxin signaling-related genes, which was the primary cause of HT injury and led to abnormalities in anther development.
GhCKI and GhPIFs Might Play Roles in Linking HT, Sugar, and Auxin Signaling Pathways
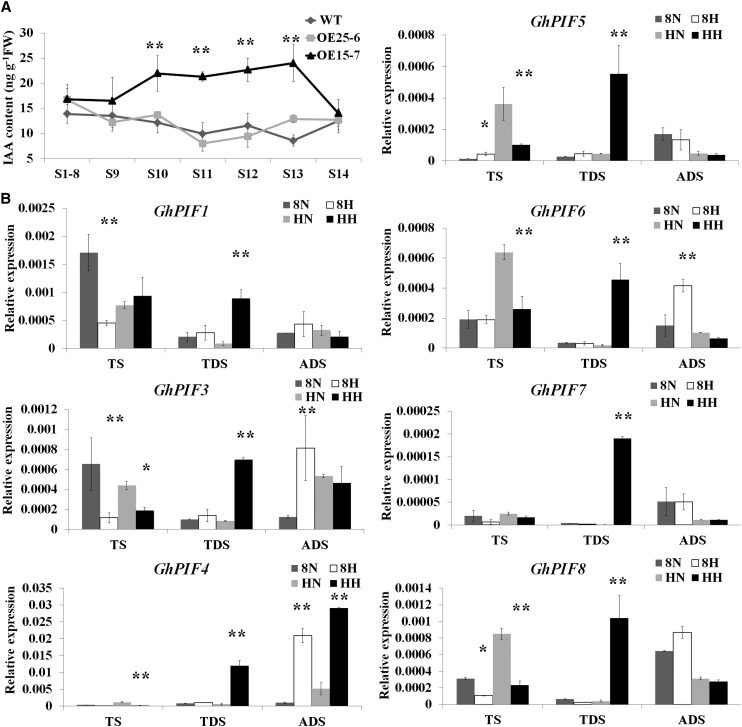
We demonstrated that the sugar and auxin signaling pathway genes were involved in the response of cotton anthers to HT. First, decreased levels of Glc in H05 under HT (Fig. 4D) and in the GhCKI overexpressor line (Min et al., 2013) were detected. Second, the IAA content in H05 anthers was measured under two conditions (Fig. 6C), and the results suggest that the IAA content was reduced in early-stage anthers but increased in late-stage anthers by HT in H05. The probable link between GhCKI and IAA suggests that IAA content should similarly increase in late-stage anthers in the GhCKI overexpression line. We measured the IAA content of the Arabidopsis GhCKI overexpression line in anthers at different developmental stages (Fig. 7A). The results show that increased endogenous IAA levels existed in late-stage anthers, which might be enhanced by the expression of GhCKI, suggesting a link between HT, GhCKI, Glc, and IAA. It is well established that soluble sugars regulate IAA biosynthesis (Sairanen et al., 2012). To further identify the regulator of sugar and IAA, the expression of seven cotton PIFs (GhPIFs) was detected by quantitative reverse transcription (qRT)-PCR (Fig. 7B). These genes are homologous to AtPIFs, which were recently shown to bind to cis-elements in the promoters of multiple genes involved in auxin biosynthesis (Franklin et al., 2011). At TS, the expression of seven GhPIF genes generally was down-regulated by HT in the anthers of both lines, except that the expression of GhPIF1 in H05 was slightly up-regulated and GhPIF5 in 84021 was significantly up-regulated. At ADS, the expression of seven GhPIF genes generally was up-regulated or not significantly different by HT in anthers of both lines. However, HT only significantly induced the expression of GhPIFs in the anthers of H05 at TDS. It is intriguing that the whole change trends in GhPIF expression and IAA content occur in concert, indicating that GhPIFs are intimately involved in the regulation of auxin biosynthesis. Taken together, our data show that HT induced GhCKI expression earlier and more significantly in the anthers of H05, and a high level of GhCKI suppressed the activities of starch synthases and decreased the starch and Glc contents throughout all anther developmental stages. Glc or soluble sugars might regulate auxin biosynthesis via PIF proteins, which might positively regulate auxin biosynthesis under HT in the anthers of the HT-sensitive line H05. Another possibility is that GhCKI directly phosphorylates GhPIF proteins, regulating plant hormone biosynthesis genes and eventually leading to H05 anther abortion (Supplemental Fig. S10). However, this possibility should be further verified in future studies.
Figure 7.
GhCKI and GhPIFs might link HT, sugar, and auxin signaling pathways. A, IAA content was assessed in the buds of Arabidopsis wild type (WT) and two GhCKI overexpression lines with anthers at various developmental stages. The x axis indicates the anther developmental stages. OE25-6, with moderate GhCKI expression, exhibited normal anther dehiscence, similar to the wild type. OE15-7, with high GhCKI expression, was unable to dehisce and release pollen grains (Min et al., 2013). FW, Fresh weight. B, Relative mRNA levels of seven PIF genes in the anthers of 84021 and H05 under the two temperature treatments. 8N and 8H refer to 84021 under NT and HT conditions, respectively; HN and HH refer to H05 under NT and HT conditions, respectively. Data are presented as means ± se from three biologically independent experiments. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01) by Student’s t test.
DISCUSSION
As the global temperature increases, HT damage to crop production becomes a serious factor. HT can affect sexual reproduction, thereby decreasing crop yield. Cotton, as a fiber crop, suffers from HT damage by shedding buds under extreme HT. However, the mechanism of bud shedding under HT is unknown. Based on our previous research, the expression of GhCKI was induced earlier in anthers by HT, which regulated anther abortion and caused bud shedding (Min et al., 2013). To globally identify the genes and pathways participating in cotton anther development under HT, we chose two cotton lines (the HT-tolerant 84021 and the HT-sensitive H05), two temperature regimes (NT, 28°C–35°C/20°C–28°C day/night; HT, 35°C–39°C/29°C–31°C day/night), and three anther development stages (TS, TDS, and ADS), corresponding to crucial stages at which male reproductive organs are sensitive to stresses (Sato et al., 2002; Endo et al., 2009). We then performed RNA-seq analysis and obtained relatively complete transcriptome information that allowed us to fully understand the relationship between anther development and HT.
DNA Methylation and Histone Modification May Regulate Anther Development under HT
Under HT, anthers may alter their developmental process to rapidly adapt to these changes. DNA methylation and histone modification, the two main covalent modifications involved in chromatin structure and remodeling, play important roles in the transition between anther development stages and the acclimation of HT (Jaenisch and Bird, 2003). However, the means by which DNA methylation and histone modification regulate these processes in cotton are not well understood. DNA methylation alters the interactions between proteins and DNA. Histone modification can alter the chromatin state by adding or removing methyl, acetyl, ubiquitin, or phosphate modifications on histone tails (Strahl and Allis, 2000; Martinowich et al., 2003). A decade of study has clarified that these epigenetic modifications are associated with gene expression regulation, providing plant cells with a mechanism for responding to developmental transitions and environmental stress (Jaenisch and Bird, 2003). It has been thought that deficiency in SAHH1 catalyzed the reversible reaction of S-adenosylhomocysteine to homocysteine and adenosine, which was a major contributing factor to genome-wide hypomethylation. Deficiency in the supply of methyl groups for DNA methylation led to the loss of control of global gene expression and metabolic pathways (Ouyang et al., 2012). In this study, the expression level of SAHH1 was significantly reduced in H05 under HT, consistent with the appearance of genome-wide hypomethylation and abundance of DEGs detected in H05 under HT. In addition, S-adenosylhomocysteine is a competitive inhibitor of all methyltransferases, but the expression of most methyltransferases was not significantly altered in the sahh1 mutant (Ouyang et al., 2012). However, the expression of methyltransferase genes, such as DRM1 (Singh et al., 2008) and DRM3 (Henderson et al., 2010), was significantly decreased in the anthers of H05 under HT conditions. Therefore, there were differences between our results and the transcript profiling analysis of the sahh1 mutant. We thus propose that other pathways mediate DNA methylation in cotton anthers in response to HT. It has been thought that heterochromatic small interfering RNAs play key roles in de novo cytosine DNA methylation through a pathway termed RNA-directed DNA methylation (RdDM; Law and Jacobsen, 2010). RNA-DIRECTED DNA METHYLATION1 (RdDM1), ARGONAUTE4, NRPD1b (which is the largest subunit of RNA polymerase Pol V), and KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR1 are required for the biogenesis, loading, functioning, and strengthened functioning of heterochromatic small interfering RNAs in the RdDM pathway, respectively (He et al., 2009; Ye et al., 2012). Intriguingly, through gene expression profiling, we unexpectedly found that most genes involved in the RdDM pathway have decreased expression levels in the anthers of H05 under HT conditions, with the exception of RdDM1. We propose that the RdDM pathway is involved in DNA hypomethylation in H05 under HT conditions.
Histone constitution and modifications regulate gene expression at a level that is superimposed onto DNA methylation (Berger, 2007). Histone constitution genes were generally more highly expressed in 84021 than in H05 under both conditions, providing more stable DNA in 84021 under HT conditions. In addition, two jmjC domain-containing genes involved in histone demethylation had significantly decreased expression in the three stages of anther development in H05 under HT conditions. Thus, we speculate that the suppressed histone demethylation compensated for the DNA hypomethylation in H05 under HT. However, this speculation should be verified in future studies.
Sugar Signaling, Which May Be Regulated by GhCKI, Plays Vital Roles in the Cotton Anther Response to HT
The sugar metabolism pathway plays vital roles in male reproductive organ development and protects against various types of environmental stresses (Frank et al., 2009; Min et al., 2013). To our knowledge, the anther has the highest sink strength in the developing flower, and temperature activates high metabolic activity by consuming large amounts of sugars to support early pollen development (Oliver et al., 2007). Moreover, Glc is the most important form of sugar used for energy in anthers, and starch serves as a marker of pollen maturity and provides an energy reserve for pollen germination. Either the reduction of Glc or the deficit in starch in the anther can dramatically impair pollen development and cause male sterility (Datta et al., 2002; Oliver et al., 2007). Based on expression pattern analysis of starch and Suc metabolism-related genes and the determination of Glc, Suc, and starch contents, we speculated that the reduced Glc and starch influenced male reproductive development under HT. Moreover, previous authors have comprehensively reviewed the three sugar regulatory pathways involved in Glc sensing and signaling in yeast, and HXK2, YCKI, and PKA play vital roles in their respective pathways (Rolland et al., 2006). However, in plants, it appears that SnRKs play a central role in the sugar sensing and signaling pathway (Rolland et al., 2006). Plant SnRKs (especially SnRK1) can complement the yeast sucrose nonfermenting1 (snf1) mutant, which is unable to survive on a nonfermentable carbon source, and activate starch synthesis. Silencing of SnRK1 causes abnormal starch accumulation in pollen grains and male sterility in barley (Zhang et al., 2001). The catalytic subunits of SnRKs, SNF1 KINASE HOMOLOG10 and SNF1 KINASE HOMOLOG11, interact with Snf4 to suppress the snf1 and snf4 mutations in yeast; the snf1 and snf4 mutations were also suppressed by CKI (Kleinow et al., 2000). In our previous studies, the expression of GhCKI was induced earlier in H05 by HT, and overexpression of GhCKI regulated anther abortion by inhibiting the activities of starch synthases, causing increased Glc content in early-stage buds. However, the Glc content of buds of the GhCKI overexpression line gradually decreased as anther development proceeded (Min et al., 2013), consistent with the change in Glc content in H05 under HT compared with that observed under NT conditions. Based on the overexpression of GhCKI in Arabidopsis, the HT-induced earlier expression of GhCKI in cotton H05, and the decreased Glc content in late-stage anthers in both plants, we speculate that a lack of Glc in the anthers during the anther development period caused male sterility. Thus, sugar signaling, and especially the Glc signaling pathway, may be regulated by GhCKI, playing a key role in anther development under HT.
Sugar Signaling May Regulate Auxin Biosynthesis via PIF Proteins during the Response to HT in Cotton Anthers
Sugars play important roles not only as energy sources but also as signaling molecules in the plant defense response. To date, many reports have indicated that sugars may contribute greatly to the homeostasis of plant hormones, such as ABA, jasmonic acid, brassinosteroids, cytokinins, and auxin (Rolland et al., 2006; Sairanen et al., 2012). In addition, HT alters hormone production and signaling (Kotak et al., 2007). Thus, an intimate and complex relationship might exist between sugar signaling and plant hormone signaling under HT. In our previous study, overexpression of GhCKI disturbed the homeostasis of Glc in buds and subsequently triggered the accumulation of ABA. ABA then initiated the antioxidant defense system and disturbed the balance of reactive oxygen species, leading to anther abortion (Min et al., 2013). Meanwhile, the IAA content has been measured in the buds of Arabidopsis lines overexpressing GhCKI (Fig. 7A). The results suggest that GhCKI regulates both the sugar signaling pathway and plant hormone signaling pathways in anthers in response to HT. Furthermore, it is well documented that HT promotes the expression of genes involved in several auxin biosynthesis pathways in certain tissues (Gray et al., 1998). However, Sakata et al. (2010) suggested that the anther tissue-specific reduction of auxin, which leads to the abortion of pollen development, is the primary cause of HT injury; additionally, these authors showed that exogenous application of auxin can reverse plant male sterility caused by HT (Sakata et al., 2010). In this study, the IAA content was decreased in TS but increased in TDS and ADS under HT in H05 (Fig. 6C). Additionally, an increase in indehiscent anthers was observed in 84021 by overapplication of exogenous IAA before HT injury. These results suggest that the auxin concentration plays a very complex role in male fertility during HT.
Previous research has uncovered numerous links between sugar and auxin; for example, exogenous Glc up-regulates auxin biosynthesis (Sairanen et al., 2012). We thus propose that a sugar signaling pathway upstream of the auxin signaling pathway functions in the response of cotton anthers to HT. Like sugar and auxin, the PIF proteins have also been shown to be involved in environmental responses to HT (Leivar and Quail, 2011). PIF1 and PIF4 have been shown to be involved in the temperature regulation of auxin biosynthesis (Sun et al., 2012), and PIF4 has been shown to induce the expression of the auxin biosynthetic genes TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 and CYTOCHROME P450, FAMILY 79, SUBFAMILY B, POLYPEPTIDE2 through binding to their promoter elements (Franklin et al., 2011). Furthermore, Stewart et al. (2011) showed that PIF gene expression was up-regulated by Suc, leading to an increase in auxin levels. In our research, the Glc content of H05 was decreased by HT throughout all anther developmental stages examined, and PIF gene expression was down-regulated at TS but up-regulated in the two later stages of anther development by HT (Fig. 7B). These results suggest that the regulation mechanism between sugar and PIF proteins is likely to be complex because Suc could regulate the expression of PIFs (Stewart et al., 2011), and the phosphorylation status of hexoses may also contribute to the regulation of PIF gene expression (Rolland et al., 2006). Meanwhile, suppressed IAA biosynthesis was observed in the TS anthers under HT, indicating that PIF proteins might act as positive regulators of HT-induced IAA biosynthesis. In support of this explanation, the enhanced expression of PIFs in the TDS and ADS anthers showed a significantly enhanced capacity for the induction of IAA biosynthesis under HT (Fig. 6C). However, other researchers treated a pif1-1/pif3-3/pif4-2/pif5-3 quadruple mutant line and a PIF5 overexpression line with Glc and observed a significantly increased and decreased capacity for Glc induction of IAA biosynthesis in the pif1-1/pif3-3/pif4-2/pif5-3 quadruple mutant and the PIF5 overexpressor, respectively, suggesting that the PIF proteins act as negative regulators of sugar-induced IAA biosynthesis (Sairanen et al., 2012). Although the connections among HT, GhCKI, sugar, PIFs, and IAA during the regulation of the cotton anther response to HT are complex and require further research, it appears that GhCKI and PIF proteins act as master switches during the HT induction of IAA biosynthesis.
CONCLUSION
In this study, we examined the gene expression profiles of cotton anthers in response to HT using an RNA-seq approach. Our results indicate that HT induced higher GhCKI expression in H05, caused Glc deficiency, and regulated PIF gene expression. PIFs positively regulate IAA biosynthesis; thus, sugar and auxin signaling pathways might represent a highly complex and central signaling network. Moreover, we showed that IAA application increased indehiscent anthers in 84021 coupled with an elevated concentration of auxin in TDS, suggesting that a higher background auxin concentration weakens the defense response of the anther against HT. An improved understanding of the transcriptional responses to HT during anther development will help to increase cotton productivity.
MATERIALS AND METHODS
Plant Materials and HT Treatment
Two cotton (Gossypium hirsutum) lines with obvious differences in performance under HT in the field were employed in this study: 84021, which is tolerant to HT, and H05, which is sensitive to HT (Min et al., 2013). The plants were grown in a greenhouse at 28°C to 35°C/20°C to 28°C day/night as a normal condition. During HT treatment, the plants were cultivated at 35°C to 39°C/29°C to 31°C day/night in a greenhouse. As described, bud lengths of 5, 6.5, 9, and 24 mm represented the stages of pollen mother cell transition to meiosis, tetrad, microspore, and anther dehiscence, respectively (Ma et al., 2012). When the plants were treated with HT for 7 d, buds of different lengths (3–5, 6–7, 9–14, and more than 24 mm) were collected under HT and NT. The anthers were excised and immediately frozen in liquid nitrogen; they were then stored at −70°C until use.
Histological Analysis
To determine the responses of 84021 and H05 anthers to HT histologically, the samples were fixed, dehydrated, embedded, sectioned, stained, and examined according to a previous report (Deng et al., 2012). To estimate the deposition of callose and the secondary enhancement of the anther cell wall, the anthers at different developmental stages were stained with a 1% aniline blue solution. To observe where the starch is actually accumulating and in which cells of the anther, cross sections of anthers at the different developmental stages were stained with Lugol solution (Sigma).
Sequencing
Based on a modified guanidine thiocyanate method (Deng et al., 2012), total RNA was isolated from the anthers of 84021 and H05 at three specific stages with or without HT treatment, as shown schematically in Figure 1. Approximately 20 μg of total RNA from each sample was sent to Beijing Genomics Institute, where the mRNA was enriched using oligo(dT) magnetic beads and digested into short fragments; then, the first- and second-strand complementary DNAs were synthesized. After purification and the addition of single A nucleotides, the fragments were ligated to sequencing adaptors and sequenced via Illumina HiSeq 2000. The sequence of the 49 bp representing each transcript was then determined and derived. Gene expression was calculated and subsequently normalized to RPKM (Morrissy et al., 2009).
RT-PCR and qRT-PCR
To confirm the differential expression pattern of the HT-responsive genes selected in the RNA-seq experiments, RT-PCR and qRT-PCR analyses were performed. Gene-specific primers were designed according to the reference unigene sequences (Supplemental Table S4). Total RNA was isolated from the anthers, first-strand complementary DNA was generated using SuperScript III reverse transcriptase (Invitrogen), and qRT-PCR was performed in 15-μL reactions using the method described in our previous study (Min et al., 2013). The cotton UBQUITIN gene was used as a control, and the relative gene expression levels were calculated using the 2−ΔCT method (−ΔCT indicates the minus of [CT,target − CT,UB7], CT indicates the threshold cycle number of the amplified gene) method. The results were obtained from three biologically independent experiments.
Assays of Total Soluble Sugar, Suc, Glc, and Starch Content
Approximately 50 mg (fresh weight) of anther tissue was ground into a homogenate in 1 mL of 80% cold aqueous methanol. The subsequent procedures were performed essentially as described previously (Min et al., 2013). The supernatant was used for sugar assays, and the contents of total soluble sugar, Suc, and Glc were determined as described by Yemm and Willis (1954). The remaining pellet was used to assay starch content using an improved colorimetric method (Buysse and Merckx, 1993).
Starch Synthesis Enzyme Extraction and Activity Measurement
To determine the activities of AGPase, SSSase, and GBSSase, the enzymes were extracted according to the method of Nakamura et al. (1989). Approximately 100 mg of each anther sample was ground to a fine powder in liquid nitrogen. The subsequent procedures were performed essentially as described previously (Min et al., 2013). The supernatant was used for the AGPase and SSSase activity analyses. The remaining pellet was resuspended in 1 mL of extraction buffer, and the GBSSase activity assay was performed. Detailed methods for AGPase, SSSase, and GBSSase activity measurement were described by Nakamura et al. (1989) and Schaffer and Petreikov (1997).
Quantification of Endogenous IAA
For the estimation of endogenous IAA levels, 50 mg (fresh weight) of each sample was homogenized in 1 mL of 80% cold aqueous methanol and subsequently shaken at 4°C overnight. The subsequent extraction procedure was performed according to a previous report (Liu et al., 2012), and the extracts were dissolved in 400 μL of methanol and filtered through a 0.22-μm nylon membrane; they were then stored at −70°C until use. The quantification of endogenous IAA was performed according to a previous report (Liu et al., 2012).
Exogenous IAA Application
To assess the effects of IAA on anther development under HT, the buds of two cotton lines were sprayed with 10−5 or 10−6 m IAA (distilled, deionized water was sprayed as a control). The IAA solutions were sprayed onto all buds of two cotton lines on the morning of days 7, 5, 3, and 0 before increasing the temperature to induce HT injury. The application of 10−5 m IAA caused the loss of flower buds; however, the buds of two cotton lines grew normally with the application of 10−6 m IAA under NT. After four applications of 10−6 m IAA or distilled, deionized water under NT, HT damage was triggered and allowed to occur for 7 d. Then, the temperature was restored to normal for 7 d and the phenotypes of the anthers were photographed at anthesis.
Statistics
The graphical data represent the results of multiple independent experiments (n ≥ 3), and means ± se are shown. The P values of the distribution of transcripts in plant hormone signaling pathways were evaluated through singular enrichment analysis comparing the DEGs in plant hormone signaling pathways in this study with a reference database by AgriGO (http://bioinfo.cau.edu.cn/agriGO). The physiological parameters (sugar and IAA) of 84021 and H05 were compared using a one-way ANOVA and shortest significant range (P < 0.05) post hoc analysis, and values not sharing a common letter were considered statistically significant. The statistical significance of gene expression level, DNA methylation rate, and the enzymatic activities of starch synthase was determined using the two-tailed unpaired Student’s t test, and P < 0.05 was considered statistically significant.
Sequence data from this article can be found in the GenBank/EMBL data libraries (see Supplemental Tables S2 and S4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of the field performance of HT-tolerant 84021 and HT-sensitive H05 buds and anthers under HT.
Supplemental Figure S2. Cluster analysis was performed by Genesis, which is based on the K-means method.
Supplemental Figure S3. Gene Ontology functional classification analysis of 4,599 DEGs.
Supplemental Figure S4. KEGG metabolism pathway analysis of 4,599 DEGs.
Supplemental Figure S5. Expression analysis of genes involved in histone constitution.
Supplemental Figure S6. Seven genes involved in histone modification were differentially expressed during anther development under HT.
Supplemental Figure S7. Overview of the putative starch and Suc metabolism pathway involved in cotton anther development under HT.
Supplemental Figure S8. The expression of particular genes involved in the sugar signaling pathway was verified by RT-PCR.
Supplemental Figure S9. Detailed expression profiles of the genes involved in the auxin signal transduction pathway by RT-PCR.
Supplemental Figure S10. Schematic diagram of the GhCKI and GhPIFs, linking HT, sugar, and auxin signaling.
Supplemental Table S1. Overview of read number by RNA-seq.
Supplemental Table S2. A total of 5,194 DEGs were filtered with RPKM ≥ 50 and the absolute value of log2 ratio ≥ 1.
Supplemental Table S3. A total of 4,599 DEGs showed the different expression patterns between 84021 and H05.
Supplemental Table S4. Oligonucleotides used in this study.
Acknowledgments
We thank Dongqin Li for assistance with liquid chromatography/mass spectrometry.
Glossary
- HT
high temperature
- ABA
abscisic acid
- RNA-seq
RNA sequencing
- NT
normal temperature
- TS
tetrad stage
- TDS
tapetal degradation stage
- ADS
anther dehiscence stage
- DEGs
differentially expressed genes
- RPKM
reads per kilobase per million reads
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- RT
reverse transcription
- qRT
quantitative reverse transcription
- IAA
indole-3-acetic acid
- RdDM
RNA-directed DNA methylation
Footnotes
This work was supported by the National Cotton Agricultural Research System (grant no. CARS–18–09), the National High-Tech 863 Program (grant no. 2011AA10A102), and the Ministry of Agriculture of China (grant no. 2013ZX08005–004).
The online version of this article contains Web-only data.
References
- Ahuja I, de Vos RC, Bones AM, Hall RD. (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15: 664–674 [DOI] [PubMed] [Google Scholar]
- Berger SL. (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Buysse J, Merckx R. (1993) An improved colorimetric method to quantify sugar content of plant tissue. J Exp Bot 44: 1627–1629 [Google Scholar]
- Datta R, Chamusco KC, Chourey PS. (2002) Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol 130: 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X. (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Tsuchiya T, Hamada K, Kawamura S, Yano K, Ohshima M, Higashitani A, Watanabe M, Kawagishi-Kobayashi M. (2009) High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol 50: 1911–1922 [DOI] [PubMed] [Google Scholar]
- Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N. (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60: 3891–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM. (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker J, Jenner C. (1993) High temperature affects the activity of enzymes in the committed pathway of starch synthesis in developing wheat endosperm. Funct Plant Biol 20: 197–209 [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. (2009) An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedhly A, Hormaza JI, Herrero M. (2009) Global warming and sexual plant reproduction. Trends Plant Sci 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Deleris A, Wong W, Zhong X, Chin HG, Horwitz GA, Kelly KA, Pradhan S, Jacobsen SE. (2010) The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 6: e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet (Suppl) 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Craufurd PQ, Wheeler TR. (2007) High temperature stress and spikelet fertility in rice (Oryza sativa L.). J Exp Bot 58: 1627–1635 [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C. (2000) Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J 23: 115–122 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li X, Xiao J, Wang S. (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Koornneef M, Soppe WJ. (2007) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Wei H, Song M, Pang C, Liu J, Wang L, Zhang J, Fan S, Yu S. (2012) Transcriptome profiling analysis reveals that flavonoid and ascorbate-glutathione cycle are important during anther development in upland cotton. PLoS ONE 7: e49244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302: 890–893 [DOI] [PubMed] [Google Scholar]
- McClung CR, Davis SJ. (2010) Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr Biol 20: R1086–R1092 [DOI] [PubMed] [Google Scholar]
- Min L, Zhu L, Tu L, Deng F, Yuan D, Zhang X. (2013) Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J 75: 823–835 [DOI] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P. (2012) How do plants feel the heat? Trends Biochem Sci 37: 118–125 [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA. (2009) Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19: 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yuki K, Park SY, Ohya T. (1989) Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol 30: 833–839 [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R. (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Oliver SN, Van Dongen JT, Alfred SC, Mamun EA, Zhao X, Saini HS, Fernandes SF, Blanchard CL, Sutton BG, Geigenberger P. (2005) Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ 28: 1534–1551 [Google Scholar]
- Oshino T, Abiko M, Saito R, Ichiishi E, Endo M, Kawagishi-Kobayashi M, Higashitani A. (2007) Premature progression of anther early developmental programs accompanied by comprehensive alterations in transcription during high-temperature injury in barley plants. Mol Genet Genomics 278: 31–42 [DOI] [PubMed] [Google Scholar]
- Ouyang B, Fei Z, Joung JG, Kolenovsky A, Koh C, Nowak J, Caplan A, Keller WA, Cui Y, Cutler AJ, et al. (2012) Transcriptome profiling and methyl homeostasis of an Arabidopsis mutant deficient in S-adenosylhomocysteine hydrolase1 (SAHH1). Plant Mol Biol 79: 315–331 [DOI] [PubMed] [Google Scholar]
- Peet M, Sato S, Gardner R. (1998) Comparing heat stress effects on male-fertile and male-sterile tomatoes. Plant Cell Environ 21: 225–231 [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Saini H, Sedgley M, Aspinall D. (1984) Development anatomy in wheat of male sterility induced by heat stress, water deficit or abscisic acid. Funct Plant Biol 11: 243–253 [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K. (2012) Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T, Oshino T, Miura S, Tomabechi M, Tsunaga Y, Higashitani N, Miyazawa Y, Takahashi H, Watanabe M, Higashitani A. (2010) Auxins reverse plant male sterility caused by high temperatures. Proc Natl Acad Sci USA 107: 8569–8574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Peet MM, Thomas JF. (2002) Determining critical pre- and post-anthesis periods and physiological processes in Lycopersicon esculentum Mill. exposed to moderately elevated temperatures. J Exp Bot 53: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Petreikov M. (1997) Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol 113: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Zubko E, Meyer P. (2008) Cooperative activity of DNA methyltransferases for maintenance of symmetrical and non-symmetrical cytosine methylation in Arabidopsis thaliana. Plant J 56: 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Prasad P, Sunita K, Giri S, Reddy KR. (2007) Influence of high temperature and breeding for heat tolerance in cotton: a review. Adv Agron 93: 313–385 [Google Scholar]
- Stewart JL, Maloof JN, Nemhauser JL. (2011) PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS ONE 6: e19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Takeda H, Tsukaguchi T, Egawa Y. (2001) Ultrastructural study on degeneration of tapetum in anther of snap bean (Phaseolus vulgaris L.) under heat stress. Sex Plant Reprod 13: 293–299 [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, et al. (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 46: 859–870 [DOI] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. (1954) The estimation of carbohydrates in plant extracts by anthrone. Biochem J 57: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L. (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG. (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J 28: 431–441 [DOI] [PubMed] [Google Scholar]
- Zinn KE, Tunc-Ozdemir M, Harper JF. (2010) Temperature stress and plant sexual reproduction: uncovering the weakest links. J Exp Bot 61: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]