The successive growth processes of cell division and cell elongation in maize leaves are characterized by differential DNA methylation that is correlated with differential expression.
Abstract
DNA methylation is an important and widespread epigenetic modification in plant genomes, mediated by DNA methyltransferases (DMTs). DNA methylation is known to play a role in genome protection, regulation of gene expression, and splicing and was previously associated with major developmental reprogramming in plants, such as vernalization and transition to flowering. Here, we show that DNA methylation also controls the growth processes of cell division and cell expansion within a growing organ. The maize (Zea mays) leaf offers a great tool to study growth processes, as the cells progressively move through the spatial gradient encompassing the division zone, transition zone, elongation zone, and mature zone. Opposite to de novo DMTs, the maintenance DMTs were transcriptionally regulated throughout the growth zone of the maize leaf, concomitant with differential CCGG methylation levels in the four zones. Surprisingly, the majority of differentially methylated sequences mapped on or close to gene bodies and not to repeat-rich loci. Moreover, especially the 5′ and 3′ regions of genes, which show overall low methylation levels, underwent differential methylation in a developmental context. Genes involved in processes such as chromatin remodeling, cell cycle progression, and growth regulation, were differentially methylated. The presence of differential methylation located upstream of the gene anticorrelated with transcript expression, while gene body differential methylation was unrelated to the expression level. These data indicate that DNA methylation is correlated with the decision to exit mitotic cell division and to enter cell expansion, which adds a new epigenetic level to the regulation of growth processes.
DNA methylation is the covalent modification of nucleotides in DNA by the addition of a methyl group. In the nuclear genome of higher eukaryotes, 5-methylcytosine is the most important DNA modification (Goll and Bestor, 2005). It is a phenomenon of ancient origin predating plant-animal diversification. However, some differences exist between plant and animal methylome patterning and function, and DNA methylation has been found to be evolutionarily lost in a few species (Feng et al., 2010; Zemach et al., 2010). Eukaryotic DNA methylation is established by DNA methyltransferase (DMT) enzymes that transfer a methyl group from S-adenosyl Met to the fifth carbon of cytosine. These enzymes can largely be subdivided in maintenance and de novo DMTs, depending on whether the recognition site is already methylated or not. Maintenance DMTs conserve the methylation status of symmetrical (palindromic) sites after DNA replication, by recognizing the hemimethylated locus and methylating the newly synthesized strand. In plants, there are two types of maintenance DMTs: DNA METHYLTRANSFERASE (MET) and CHROMOMETHYLASE (CMT). The former methylates CG sites during DNA replication, whereas the latter methylates CHG (H = A, C, or T) sites located in chromatin in which histone 3 is dimethylated on Lys-9 (Goll and Bestor, 2005). De novo DMTs methylate previously unmethylated DNA and are DOMAINS REARRANGED METHYLTRANSFERASEs (DRMs) in plants, since the motifs in the catalytic methyltransferase domain are reshuffled through a circular permutation. DRMs are mainly known for methylating asymmetric CHH sites but are able to de novo methylate cytosines in any sequence context and are guided by 24-nucleotide short interfering RNAs in a process called RNA-directed DNA methylation. These short interfering RNAs are generated through the action of plant-specific RNA polymerases and the RNA interference machinery (Law and Jacobsen, 2010). Also DRM-like (DRML) proteins, catalytically mutated DRM paralogs, play an important role in de novo methylation (Saze et al., 2012). Lastly, DNMT2-type methyltransferases have been classified as DMTs because of the apparent presence of a DNA methylase domain, but they are in fact most likely RNA methyltransferase enzymes (Goll et al., 2006). Plant DNA methylation is removed passively through DNA replication or actively by removal and replacement of the methylated C by a DNA glycosylase and the base excision repair mechanism (Law and Jacobsen, 2010).
Plant genomes bear methylation in CG, CHG, and CHH contexts, as opposed to only CG methylation in animals, with the notable exception of embryonic stem cells and neurons (Lister et al., 2009, 2013). Moreover, the highest levels of DNA methylation have been found in plants, with up to 50% in some species. Together with histone modifications and chromatin remodeling, DNA methylation determines the epigenetic state of the genome, not only at the global level, affecting large chromosomal domains or even entire chromosomes, but also at very specific sites, such as individual genes (Suzuki and Bird, 2008). Plant DNA methylation is known to play an important role in genomic imprinting and genomic protection from transposable elements (TEs) and other repetitive DNA sequences. Furthermore, it regulates the expression of multiple genes (Jaenisch and Bird, 2003) and has been implied to play a role in gene splicing (Regulski et al., 2013). The influence of DNA methylation on development is especially evident in the case of mutations in DMT genes, which are embryo lethal in mice and lead to developmental abnormalities in plants (Goll and Bestor, 2005). The role of DNA methylation in mammalian development has been thoroughly studied in the context of embryonic development, differentiation of pluripotent stem cells, and aberrances associated with cancer progression (Smith and Meissner, 2013). In plants, DNA methylation plays a role in endosperm and embryo development (Choi et al., 2002), vernalization, hybrid vigor (Groszmann et al., 2011), and fruit ripening (Zhong et al., 2013). Tissue-specific differences in plant methylomes have been known for some time, such as the specific methylation changes during maize (Zea mays) leaf development that help steer the tissue-specific expression of certain genes (Tolley et al., 2012). However, the identification of differentially methylated genes that are developmentally and tissue-specifically regulated in plants remains largely unexplored.
Maize is a monocotyledonous grass with a bulky genome consisting largely of repetitive elements in which DNA methylation ranges from 86% to 65%, 74% to 50%, and 5.4% to 5% of CG, CHG, and CHH sequence contexts, respectively (Gent et al., 2013; Regulski et al., 2013). Much research on DNA methylation has been carried out in maize, but mostly in the framework of paramutation, imprinting, and transposon regulation (Regulski et al., 2013). Growing maize leaves is an interesting system in which to study plant organ growth, as at a given time point during their development, all processes of growth are represented in a single leaf. At the base of the leaf, cells are dividing (division zone [DZ]), while at the more distally located transition zone (TZ), cells start to expand and differentiate (expansion zone [EZ]). When cells stop expanding and reach their mature size, they become part of the mature zone (MZ; Nelissen et al., 2013). Previous work has shown that the size of the DZ largely determines final leaf size (Rymen et al., 2007; Nelissen et al., 2012). Here, we use the growth zone of the maize leaf to study the dynamics of DNA methylation during leaf growth by methylation-sensitive amplified polymorphism (MSAP) analysis on dividing, transitioning, elongating, and mature cells. An equal portion of hypermethylation and hypomethylation occurred throughout the growth zone, of which the majority of differential methylation was observed between the DZ and TZ. Surprisingly, only a minority of the differentially methylated sites was associated with TEs, and most differentially methylated sites mapped away from the centromeric regions. Almost 85% of these differentially methylated sites that changed over the developmental gradient mapped in or close to coding sequences. Our data indicate that the majority of the genic differentially methylated sites in the growing maize leaf are at the beginning and end of the coding sequences, and immediately upstream and downstream, opposite to what was seen for steady-state methylated gene bodies (Lister et al., 2008). An inverse correlation between DNA methylation and expression level was observed in genes with differentially methylated sites in the promoter and 5′ regions of the coding sequence. This correlation was not found for genes that are differentially methylated in the central and 3′ regions of the gene body. Many genes that were found to be differentially methylated in the growing maize leaf have functions in chromatin modification, gene regulation, and development.
RESULTS
Maize Maintenance Methyltransferases Are Highly Expressed in Dividing Tissue
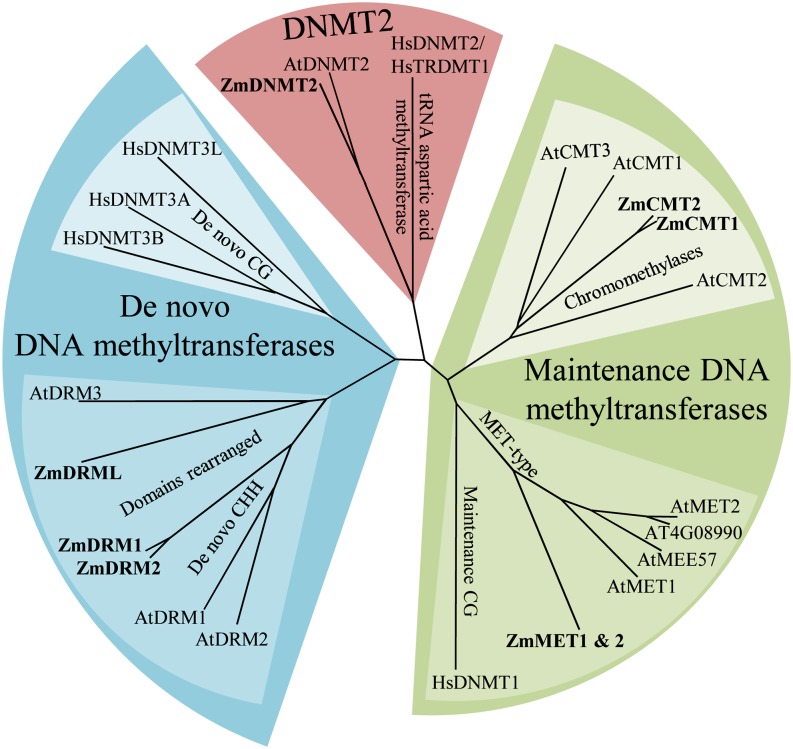
Since DNA methylation patterns are established by DMTs, we set out to profile the expression of all maize DMTs across the developing leaf. First, the encoding genes needed to be denominated and the gene product correctly classified. As in other plant species, four types of DMTs are encoded by the maize genome: MET, CMT, DRM, and DNMT2. However, the nomenclature for the maize orthologs of every type has been assigned in a less straightforward manner. One representative of MET, CMT, and DRM was originally denominated ZMET1, ZMET2, and ZMET3, respectively (Cao et al., 2000). Furthermore, a total of seven genes encoding maize DMT domains were identified by the Chromatin Database Consortium and called DMT101 to DMT107 (Gendler et al., 2008). In order to eliminate further confusion concerning maize DMT nomenclature, we renamed them according to protein functionality and orthology (Fig. 1; Supplemental Table S1). The maize cv B73 genome (release 5b.60) in fact encodes eight proteins with DMT domains. Two genes, GRMZM2G334041 and GRMZM2G333916, encode MET-type DMTs and thus can be denominated as ZmMET1 and ZmMET2, respectively. They are inversely oriented on chromosome 7, 12.3 kb from each other, and are 99.5% identical in the coding sequence. The CMT-type maintenance DMTs are also represented by two genes in maize: GRMZM2G025592 and GRMZM2G005310, which we call ZmCMT1 and ZmCMT2, respectively. As is the case in Arabidopsis (Arabidopsis thaliana), there also are three DRM-type de novo DMT-encoding genes in the maize genome. ZmDRM1 (GRMZM2G092497) and ZmDRM2 (GRMZM2G137366) encode a functional DRM domain and are, like the CMTs, land plant specific and quite different from the animal de novo methyltransferase enzymes, DNMT3A and DNMT3B (Cao et al., 2000). ZmDRML (GRMZM2G065599) encodes a DRM-like protein, similar to AtDRM3, in which the functional motifs of the methyltransferase domain have been mutated. Lastly, GRMZM2G157589 encodes ZmDNMT2, homologous to a protein originally called DNMT2 in animals. However, the animal protein was found to methylate a cytosine at position 38 in Asp tRNA. Therefore, its name was changed to tRNA ASPARTIC ACID METHYLTRANSFERASE1, the first RNA cytosine methyltransferase to be identified (Goll et al., 2006).
Figure 1.
Phylogeny of the DMT domain enzymes of maize (Zm), Arabidopsis (At), and humans (Hs). Three types of methyltransferase enzymes can be distinguished: maintenance (green), de novo (blue), and DNMT2-like (red). The maize proteins are indicated in boldface. Maintenance methyltransferase enzymes fall into two categories: the DMTs that methylate the CG motif (dark green) and are conserved between animals (DNMT1) and plants (MET), and the plant-specific DMTs that methylate the CHG motif (CMTs; light green). ZmMET1 and ZmMET2 are 99.4% identical and, therefore, are indicated as one branch. Animal and plant de novo methyltransferases differ in the arrangement of the methylase domains, causing a difference in target motifs: CG in animals (DNMT3; light blue) and CHH in plants (DRM; dark blue). The DNMT2 lineage encodes a DMT domain, but they are most likely RNA methyltransferase enzymes, as animal DNMT2 was found to methylate Asp tRNA, and was renamed tRNA ASPARTIC ACID METHYLTRANSFERASE1. [See online article for color version of this figure.]
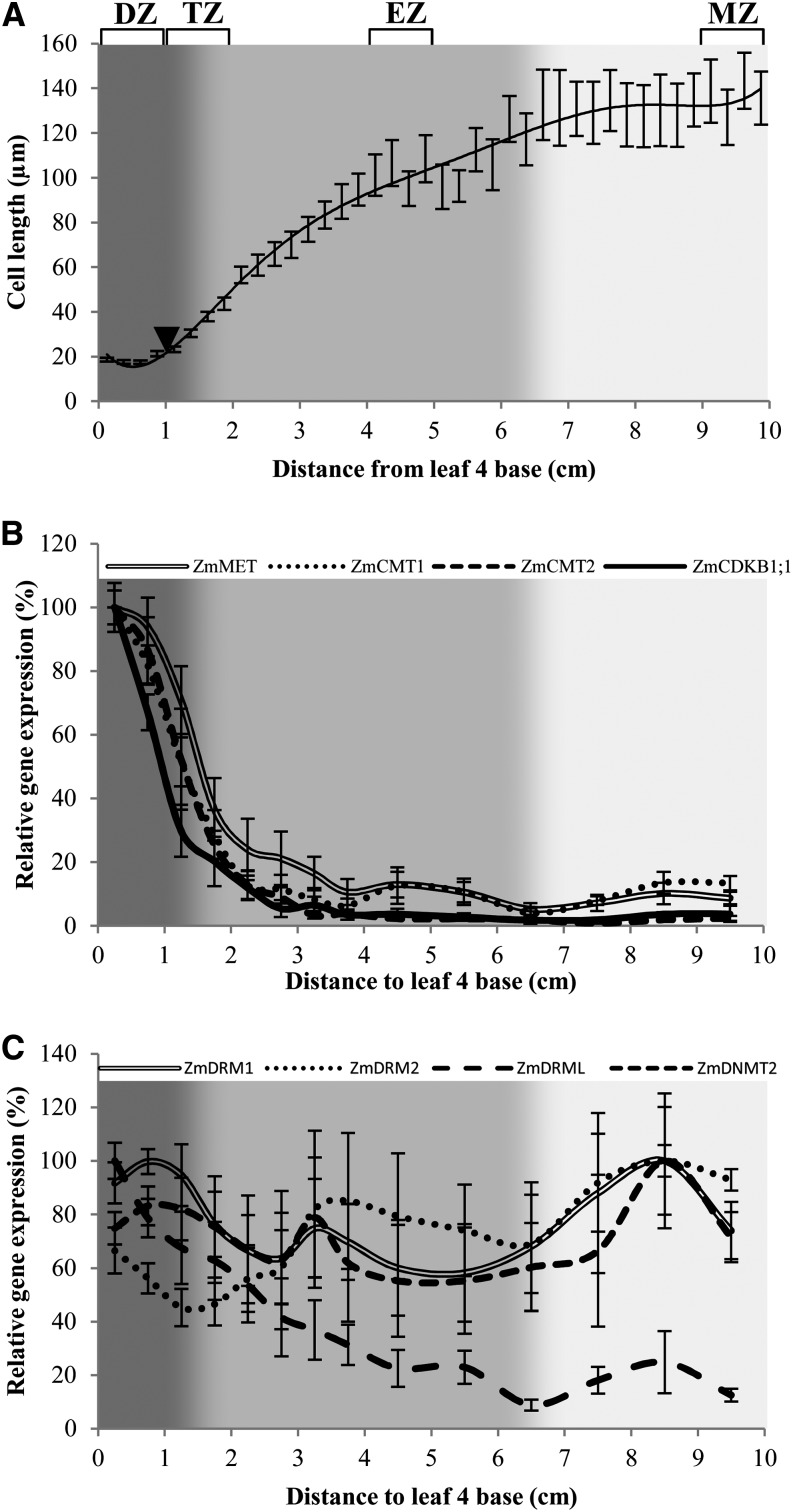
Samples for MSAP and expression analysis were prepared from growing maize leaves. Therefore, growth of the fourth leaf of maize cv B73 was measured over time. The first days after leaf emergence from the sheath are characterized by steady-state growth, during which leaf length increases linearly, since the speed of leaf growth (leaf elongation rate) is constant. After 5 d, the leaf elongation rate diminishes as the leaf approaches its final length. During the steady-state growth phase, the different zones of the fourth leaf are present, and representative samples for the DZ, TZ, EZ, and MZ were dissected according to the microscopic analysis that determined the positions of the zones based on the average cell length profile and the position of the DZ (Fig. 2A). The most basal part (the first 1 cm) of the maize leaf represents proliferating tissue (the DZ), as cells are continuously dividing and maintain a constant average cell length (18.30 ± 0.15 µm). The last dividing cell of the cv B73 leaf lies in the second 1 cm, at 1.23 ± 0.04 cm (n = 4) from the base of the leaf. At this stage, cells also start elongating, so the second 1 cm of the maize leaf (the TZ) consists of both dividing and elongating tissue. In the following centimeters (the EZ), cell length increases rapidly. Closer to the tip of the leaf, cells stop elongating and attain their mature cell length (the MZ).
Figure 2.
Cellular profile and DMT expression along the maize leaf. A, Cellular behavior throughout the first 10 cm of maize leaf 4, 2 d after its emergence, with the first 1 cm consisting of small, dividing cells (DZ), followed by a transition (TZ) toward elongating tissue (EZ), and finally matured cells (MZ). The samples for MSAP representing these tissues are indicated on top (braces), as is the end of the DZ (arrowhead). DMT expression was checked across the whole maize leaf. B and C, Maize maintenance methyltransferase (ZmMET, ZmCMT1, and ZmCMT2) expression (B) is highest in the dividing cells at the base of the maize leaf. The expression sharply decreases until reaching minimal levels 3 to 4 cm from the leaf base. A very analogous expression pattern is found for CDKB1;1, the expression of which is highly correlated with cell division activity (Rymen et al., 2007; Nelissen et al., 2012). The de novo methyltransferases (C) ZmDRM1 and ZmDRM2 are expressed steadily across the zones, as is ZmDNMT2. ZmDRML, on the other hand, has an expression profile similar to the maintenance DMTs.
For expression analysis, a fine-sampling method was applied, harvesting consecutive samples across the first 10 cm of the maize leaf. Combined with a kinematic analysis, this allows us to correlate the expression levels obtained by quantitative PCR to the cellular processes (Nelissen et al., 2012, 2013). The expression of the maintenance DMT transcripts ZmMET (P = 1.3e-13), ZmCMT1 (P = 2.2e-16), and ZmCMT2 (P = 6.5e-14) follows more or less the same profile as that of ZmCDKB1;1 (P = 6.7e-8; Fig. 2B), encoding a CYCLIN-DEPENDENT KINASE and used here as a proxy for cell division (Rymen et al., 2007; Nelissen et al., 2012). The expression of ZmMET1 and ZmMET2 cannot be distinguished because of their high sequence similarity. This expression pattern is in agreement with their role in methylation maintenance, as high levels of them are necessary to replicate methylation patterns on newly synthesized DNA strands (Goll and Bestor, 2005). On the other hand, the expression of ZmDNMT2 (P = 0.251) and the de novo methyltransferases ZmDRM1 (P = 0.216) and ZmDRM2 (P = 0.055) remains constant across the different zones (Fig. 2C). Fluctuations in transcript abundance are observed, especially in the younger tissue, but these expression changes are not significant at the 5% level. Notably, the expression of ZmDRML is broadly similar to that of the maintenance DMTs, with high expression in the dividing tissue and lower expression in the more mature zones (P = 6.7e-9; Fig. 2C). This reduction, however, occurs more gradually compared with the expression patterns of ZmMET, ZmCMT1, and ZmCMT2, which reach their minimal levels immediately after the TZ.
Most Differential DNA Methylation Takes Place in Young Developing Tissue
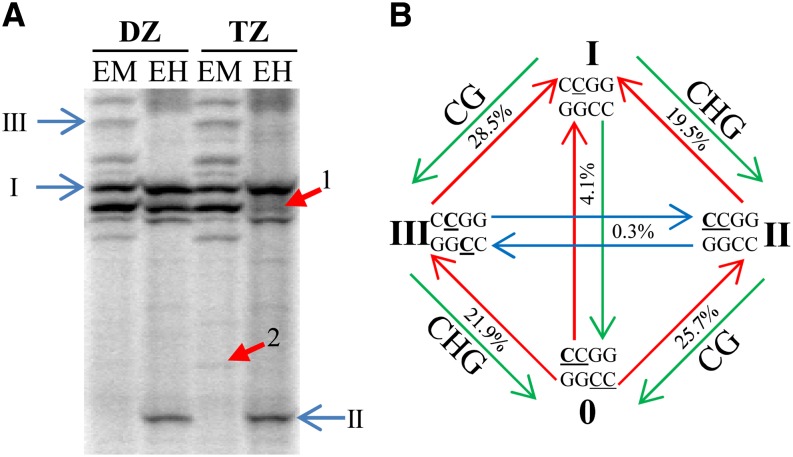
Next, we investigated DNA methylation within the developing maize leaf and the differences between the four zones (DZ, TZ, EZ, and MZ). Therefore, genomic DNA of these maize leaf zones was extracted from maize seedlings during steady-state leaf growth. MSAP analysis was carried out, applying 64 primer combinations, which amplify approximately 9,168 fragments smaller than 800 bp (Rombauts et al., 2003), covering 1.56% of all CCGG sites in the maize genome. The methylation state of these CCGG sites for every zone can be deduced after methylation-sensitive restriction cleavage with HpaII and MspI (Supplemental Table S2). A total of 28,714 patterns of restriction fragments on the gel were scored, the majority of which (95.1%) did not exhibit differences over the four zones. These monomorphic bands represent loci with a stable methylation state across the leaf and are subdivided into three methylation states according to the sensitivity of the locus to HpaII/MspI restriction (McClelland et al., 1994). Almost half (48.1% ± 1.4%) of the loci were found to be stably unmethylated, whereas, regarding stable methylation in the maize leaf, full CG methylation was the dominant form, represented by 36.7% ± 1.19% of all investigated CCGG sites. Hemi-CHG (where H = C) methylation, henceforth referred to as CHG methylation, represented 15.2% ± 0.3%. Stable methylation of both cytosines in the CCGG tetramer inhibits the restriction of both enzymes and cannot be visualized on an MSAP gel, as it yields no bands. However, differential methylation of this locus can be identified, since changing its methylation state into one of the three other methylation states can be visualized (Fig. 3A).
Figure 3.
Example and explanation of the MSAP gel-banding pattern. A, MSAP gel comparing pools of DZ and TZ samples. Each sample is restricted by EcoRI + MspI (EM) and EcoRI + HpaII (EH). Both stably (I, II, and III) and differentially (1, I→III; 2, 0→III) methylated sites are represented, for which the explanation is provided in B. B, I, Nonmethylated CCGG; II, CHG methylation, where H = C; III, CG methylation; 0, heavy CCGG methylation. For additional information, see Supplemental Table S2. Briefly, CCGG means either methylation of both cytosines (CCGG) or only the outer cytosine (CCGG). Differential methylation is indicated by arrows: hypomethylation (red), hypermethylation (green), and both hypomethylation and hypermethylation (blue). A change from 0 to any other MSAP pattern from one zone to the next means hypomethylation. Similarly, a change from I to any other MSAP pattern signifies hypermethylation. Only a change from II to III and III to II can be interpreted as both hypomethylation and hypermethylation of the locus. The overall occurrence of each transition (sum of both hypomethylation and hypermethylation percentages) is represented. The sequence context affected, being either CG or CHG, is indicated at the edges. [See online article for color version of this figure.]
For the remaining methylated bands, we did observe changes in the MSAP pattern, representing differential methylation, either between the two biological repeats or between the zones (1.9% and 3.0% of all observed bands, respectively). The latter represents reproducible changes in DNA methylation across the developing maize leaf, which is the interest of our research. A total of 217 reproducibly differentially methylated bands were identified and cut from the gel for amplification. Of these, 81% exhibited a change in methylation pattern between at least two zones. The remaining 19% were marked by a gradual change in intensity of the MSAP pattern. This could be an increase (17 bands) or decrease (20 bands) in intensity or rather a gradual transition from one MASP pattern in the DZ to another in the MZ (four bands). These gradual methylation changes are most likely caused by unequal changes in the methylation state of the different cell types that make up the tissues of the harvested samples (Xiong et al., 1999; Peraza-Echeverria et al., 2001; Cervera et al., 2002). For our objectives, the most interesting loci were the CCGG sites that abruptly changed their methylation from one zone to the next. Of these, 39% exhibited a change in methylation between the DZ and TZ, 37% between the TZ and EZ, and 24% between the EZ and MZ. Most loci thus undergo a change in the young, growing tissue, either at the end of the DZ or toward the beginning of the EZ.
These data show that many sequences were differentially methylated at the transition from cell division to cell expansion, a transition that was previously shown to play a crucial role in determining leaf size (Nelissen et al., 2012). To identify more sequences that are differentially methylated between the DZ and TZ, we decided to zoom in on the transition between division and elongation, using additional primer combinations on pools of DZ and TZ samples, now covering a total of 6.25% of all CCGG sites (±62,778 restriction fragments smaller than 800 bp; Rombauts et al., 2003; Fig. 3A). A total of 208 additional bands representing differentially methylated loci between the DZ and TZ were cut for sequencing. These compiled data over the two screens indicate that simple methylation changes are most common (95.6%) between two zones (Fig. 3B; methylation changes occurring at the edges). Among these, methylation changes affecting CG sites (54.2%) are more abundant than those occurring at CHG sites (41.4%). Changes between mCG and mCHG, on the other hand, are extremely rare (blue arrows). More specifically, a replacement of CmCG by mCCG occurred only once in our data set, and the inverse was not found. These represent the occurrence of both a hypomethylation and a hypermethylation event at the same CCGG site. Also, changes between heavy and low methylation are quite rare (4.1%). This means that hypomethylation (1%) or hypermethylation (3.1%) of both Cs at the same CCGG site between two zones is not a common form of differential methylation in the maize leaf. The relative abundance between the different zones, of all 12 possible methylation changes that can be identified using MSAP, is summarized in Supplemental Table S3.
Most Differentially Methylated Single-Locus Sequences Map in or Close to Genes and Away from the Centromeres
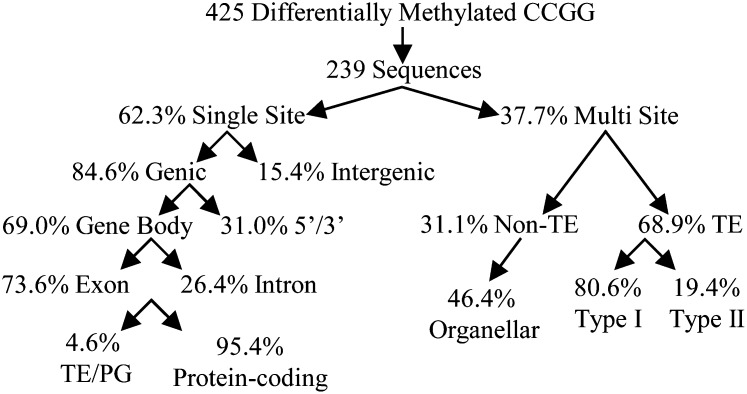
Successfully amplified bands were sequenced and BLAST searched against the maize cv B73 genome (version 5b.60). Sequences that map to multiple locations have a high probability of being a TE and thus were BLASTed to the maize TE database (www.maizetedb.org; Fig. 4). The latter multilocus sites represent 37.7% of the sequences in our data set, and since these sequences mapped to two or more loci in the genome, the exact site of differential methylation could not be identified. A minority of the multilocus sequences mapped to a limited number of sites in the maize genome, some of which also had a copy on an organellar genome. The majority of the multilocus sequences were highly repetitive TEs for which the location could not be identified. These sequences could be classified according to the transposon type (Supplemental Table S4). Type II (DNA) transposons were mostly found in the hypermethylated fraction (nine of 26) and less in the more abundant hypomethylated class (three of 35). Overall, type I (retro)transposons are the most abundant in our data, as they are also most abundant in the maize genome (Baucom et al., 2009). However, the distribution of retroelement families within the data set does differ significantly (P < 0.001) from the natural distribution. This is mainly because the most prevalent maize TE family, Huck, is underrepresented (Supplemental Table S5 [adapted from Baucom et al., 2009]).
Figure 4.
Identification of differentially methylated sequences. Differentially methylated bands were amplified and sequenced. Successful sequences were BLASTed against the maize cv B73 genome (version 5b.60). Sequences mapping to a single location (single site) either map in or in close proximity to a gene (genic) or not (intergenic). Genic sequences map either to a transcript-coding region (gene body) or upstream/downstream from it (5′/3′). Most gene body methylation was found in exons of protein-coding genes. Sequences mapping to multiple sites in the maize genome were either TEs, mapping to numerous sites in the maize genome, or mapped to a limited number of sites in the genome (non-TE). Most TEs are type I (retro)transposons, and about half of the oligomapping sequences had at least one copy on the plastid of the mitochondrial genome (organellar).
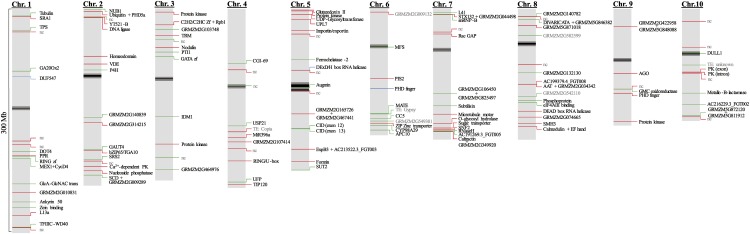
Of all sequences obtained from differentially methylated bands, more than 60% could be mapped to a single location in the maize genome (Fig. 4). Most of these mapped on or in proximity to (up to 5 kb) coding sequences. If the differentially methylated CCGG sequence lies within the transcript-coding region, this is referred to as gene body methylation and was almost always found to encode a protein. In eight instances, a differentially methylated gene body was identified by more than one sequence. Also for the sites that mapped to a locus in proximity of one or more genes, in most cases at least one of these genes was protein coding. All unique loci were mapped on the 10 maize chromosomes, showing an equal distribution over the chromosomes and the chromosome arms (Fig. 5; Supplemental Fig. S1A). However, on average, the sites of differentially methylated sequences were located away from the centromeres and the pericentromeric regions (Supplemental Fig. S1B), which are generally gene poor.
Figure 5.
Mapping of differentially methylated single-locus sequences. All single-locus differentially methylated sequences that mapped to the 10 maize chromosomes are represented. Hypermethylated sequences are indicated in green, hypomethylated sequences are indicated in red, and sequences that undergo both hypomethylation and hypermethylation are indicated in blue. Hits that are not in the vicinity of coding regions are indicated as noncoding (nc) in gray. Pseudogenes and TEs are also indicated in gray. Genes that have a known function, homology, or discernible domain are indicated as such, and the other genes are indicated by their gene code. If the differentially methylated locus lies in the vicinity of two protein-coding genes, both are mentioned. Centromeres and pericentromeric regions are indicated in black and dark gray, respectively.
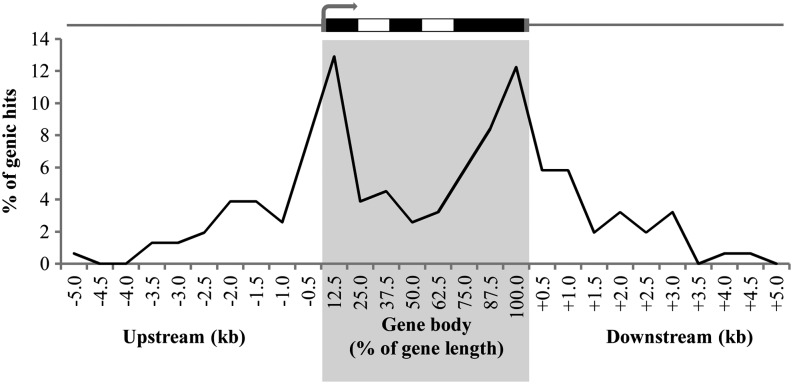
Next, we studied the distribution of differential methylation that mapped to a single location with respect to the gene body (Fig. 4). Of the genic hits, two-thirds map in the gene body and one-third map upstream or downstream (up to 5 kb). The majority of the differentially methylated CCGG sites that map to a gene lie within an exon. However, when examining the site of differential methylation within exons and introns, the distribution of methylation is not significantly different from a random distribution (P > 0.25). When mapping the sites of differential methylation in and close to coding sequences (Fig. 6), we found that the sites of differential DNA methylation are not distributed equally throughout the gene body (P < 0.001). The number of differentially methylated sites is highest around the gene extremities. More specifically, the number of differentially methylated sites is higher than expected in the first 10% (P < 0.02) and the last 20% (P < 0.001) of the gene body. Also, the highest amount of differentially methylated sites in the upstream and downstream sequences was found in the first 0.5 kb before and 1.0 kb after the coding sequences. Similar results were obtained when mapping the hits with respect to the start and stop codons (Supplemental Fig. S2A) or when mapping hits without scaling the gene body around the gene start and stop sites (Supplemental Fig. S2B). Several genes were identified by multiple differentially methylated sequences, and in two cases, two different CCGG sites within the same gene were found to be differentially methylated. The first, a CTC-interacting domain-encoding gene (GRMZM5G829738) orthologous to human ATAXIN2, is hypermethylated in two exons between the DZ and TZ. The second gene (GRMZM2G139157) encodes a protein kinase with a ubiquitin-conjugating domain PK/UbiC, which is differentially methylated at two consecutive CCGG sites in an exon and an intron between the EZ and MZ.
Figure 6.
Locations of genic hits with respect to the gene body. The positions of all genic hits were plotted up to 5 kb upstream and downstream of the gene. To adjust for different gene lengths and the presence of introns, the genic position is expressed as a percentage of the gene body length, with 0% and 100% representing the start and end of the transcript, respectively. A canonical gene model is represented above the graph, with exons, introns, and untranslated regions indicated in black, white, and dark gray, respectively.
Many Differentially Methylated Genes Are Involved in Gene Regulation, Transcription, and Development
The MSAP data set was mined for functional enrichment using PLAZA (http://bioinformatics.psb.ugent.be/plaza; Van Bel et al., 2012). Electron transport was enriched when considering all differentially methylated genes (P = 0.01), whereas zinc ion binding (P = 0.021) was enriched in the TZ-EZ transition, and ATP-dependent helicase (P = 0.025), chlorophyll binding (P = 0.042), and light-harvesting complex (P = 0.027) activities were enriched in the EZ-MZ transition. The DZ-TZ data set is enriched for binding (P = 7.86E-4) and catalytic activity (P = 0.0017), and the genes that were hypomethylated between the DZ and TZ were enriched for ATP binding (P = 0.007) and helicase activity (P = 0.046). To determine the function of these genes, we identified the function of each of the gene products through the presence of protein domains and orthology with Arabidopsis genes. Information about the 95 sequences found to be hypermethylated and hypomethylated between the DZ and TZ is summarized in Supplemental Table S6, A and B, respectively. The 54 remaining sequences were found to be differentially methylated between the TZ and EZ, the EZ and MZ, and multiple zones and are represented in Supplemental Table S7, A to C, respectively.
At least seven genes with various functions in chromatin remodeling were found to be differentially methylated or in the vicinity of a differentially methylated locus. A histone acetyltransferase (GRMZM2G371912) orthologous to Arabidopsis INCREASED DNA METHYLATION1 (IDM1; Qian et al., 2012) and three plant homeodomain (PHD)-encoding genes were undergoing exonic differential methylation. PHDs are found in chromatin modifiers and transcriptional regulators, where they are often responsible for the binding of methylated histones (Sanchez and Zhou, 2011). The telomere-binding SINGLE MYB HISTONE3 (SMH3 [GRMZM2G023667]; Marian et al., 2003), a gene highly orthologous to the ARGONAUTE (AGO)-encoding gene (AC189879.3_FGT003) involved in RNA-induced silencing (Hutvagner and Simard, 2008), and a Sucrose Nonfermentable2 (SNF2)-encoding gene (GRMZM2G313553) were hypomethylated. The SNF2 domain-encoding proteins are helicase-related ATPases that drive chromatin-remodeling complexes (Ryan and Owen-Hughes, 2011). Other DNA-interacting domains are found in two zinc finger proteins, a DNA ligase and a DNA helicase. Also, several genic hits are involved in transcription and transcriptional regulation: an RNA polymerase clamp, a TATA-binding interacting protein, four transcription factors, two RNA helicases, RNaseH, and two splicing factors. Several cytoskeleton-related proteins were identified, such as a tubulin (GRMZM2G407869), kinesin (GRMZM2G338928), exosin (GRMZM2G172602), augmin-like (GRMZM2G041878), and two actin-organizing proteins (GRMZM2G142779 and GRMZM2G113174). Moreover, α-tubulin-encoding genes, of which one was present in this data set, have been shown to be differentially methylated in different maize tissues (Lund et al., 1995). In addition, several classes of growth-related genes were represented in the data set: a β-expansin (EXPB3), GALACTURONOSYLTRANSFERASE4 (cell wall synthesis), PHOSPHATIDYLINOSITOL SYNTHASE2 (cell membrane synthesis), two genes involved in vesicle-associated transport (ANKYRIN REPEAT PROTEIN50 and SYNTAXIN132), a CYCLIN D4 (CYCD4), and an ANAPHASE-PROMOTING COMPLEX10 (APC10)-like gene (Eloy et al., 2011) indicate regulation of transcripts involved in cell division and expansion processes.
Interestingly, at least five genes were found that are known to be involved in development. The MYB-like transcription factor DIVARICATA (GRMZM2G079458) determines dorsoventral asymmetry, promoting ventral identity (Galego and Almeida, 2002). This gene got promoter hypermethylated between the DZ and TZ. GRMZM2G076257 is an ortholog of Arabidopsis DEFECTIVELY ORGANIZED TRIBUTARIES4, a pentatricopeptide repeat protein that regulates vasculature development (Petricka et al., 2008). Also, this gene was hypermethylated between the DZ and TZ in its gene body. Trehalose-6-phosphate (T6P) synthase (GRMZM2G099860) is hypermethylated between DZ and TZ. T6P synthase is involved in the biosynthesis of T6P and trehalose, both having important functions in plant growth and development. Trehalose is an osmoprotectant that influences maize inflorescence architecture, whereas T6P is an important signaling molecule involved in embryo development, vegetative growth, and leaf senescence (O’Hara et al., 2013). Also, the presence of a GIBBERELLIN 20-OXIDASE2 ortholog (GRMZM2G099467), which is hypermethylated in the TZ, was notable, as our previous experiments revealed a pivotal role for GAs in maize leaf development, specifically in the TZ (Nelissen et al., 2012). A hypomethylated CCGG site between the EZ and MZ maps onto the microRNA (MiR) ZmMiR396a stem-loop precursor. In Arabidopsis, MiR396 regulates leaf growth through the regulation of GROWTH REGULATING FACTORs (GRFs; Zhang et al., 2009). More specifically, seven out of nine AtGRFs are targeted by MiR396 (Debernardi et al., 2012). Similarly, all but two out of 18 maize GRFs carry the MiR396 target motif. These two genes, ZmGRF4 and ZmGRF10 (Zhang et al., 2008), are not down-regulated in the elongating tissue (Supplemental Fig. S3). Other abundant protein domains are associated with protein turnover, regulation of protein function, sugar metabolism, and transport.
Only Differential Methylation in Upstream Genic Regions Is Correlated with Gene Expression
We then addressed the question of whether the expression of differentially methylated genes was correlated with the methylation state. A total of 43 genes were selected for expression analysis using quantitative reverse transcription PCR. The expression for genes that undergo differential methylation between the DZ and TZ was investigated over the first 4 cm, whereas the expression of genes that undergo methylation changes in the TZ-EZ or the EZ-MZ was analyzed over the full first 10 cm of the developing maize leaf.
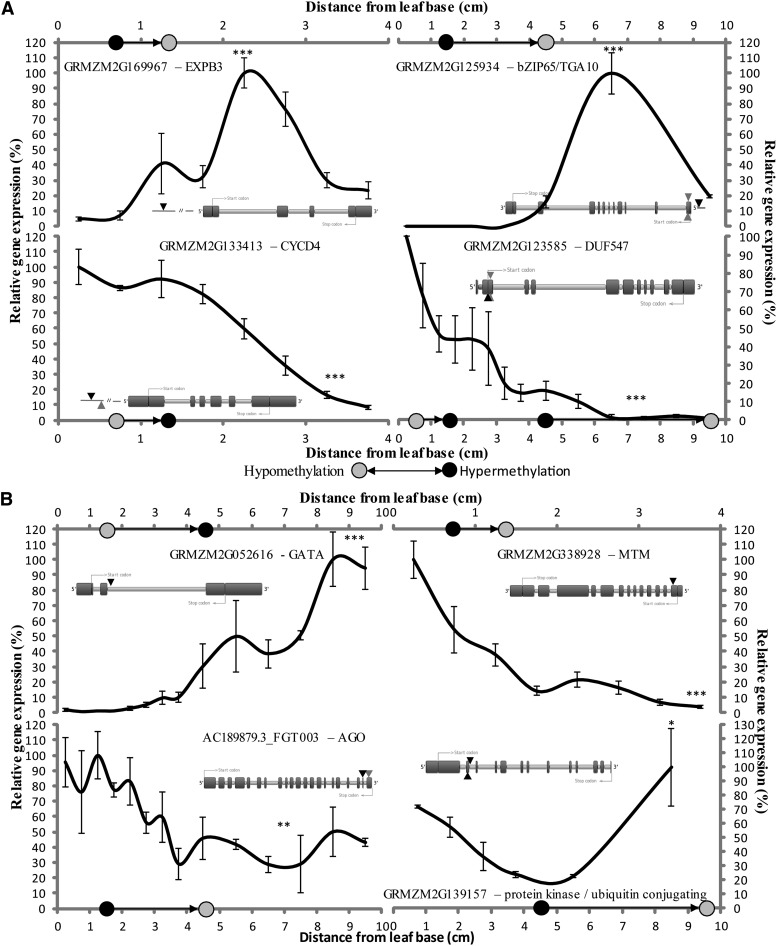
For 10 out of 11 genes undergoing differential methylation in the promoter and 5′ regions of the gene body (up to the start codon), an inverse correlation between DNA methylation and expression was found, eight of which were significant at the 5% level (Supplemental Fig. S4). All four genes undergoing hypermethylation upstream of the gene between the DZ and TZ (Supplemental Fig. S4A) were up-regulated in the following centimeters. EXPB3 (GRMZM2G169967), for example, was 20-fold up-regulated (P = 6.5e-05) in the second 1 cm and down-regulated afterward (Fig. 7A). Similarly, two genes undergoing promoter hypomethylation between the TZ and EZ were up-regulated in the more mature zones (Supplemental Fig. S4C). The most extreme case (GRMZM2G125934) encodes a basic Leucine Zipper (bZIP) protein orthologous to Arabidopsis bZIP65/TGACG MOTIF-BINDING PROTEIN10 (TGA10). This gene was virtually not expressed in dividing and early-elongating tissue but was highly up-regulated in late-elongating tissue, after hypomethylation, and subsequently down-regulated in mature tissue (P = 8.7e-08). Four out of five genes undergoing 5′ hypermethylation between the DZ and TZ were subsequently down-regulated (Supplemental Fig. S4, B and D). For three of these genes, this change in expression was significant. One gene (GRMZM2G123585), encoding a Leu zipper and a Domain of Unknown Function (DUF547), underwent differential methylation at a site 5 bp downstream of the start codon. This site was not only hypermethylated between the DZ and TZ but also hypomethylated between the EZ and MZ. The expression was drastically reduced between the DZ and MZ: almost 7-fold between the DZ and EZ, and dropping toward 1% of its original expression level in the MZ (P = 1.3e-08). DIVARICATA (GRMZM2G031441) was down-regulated after having its promoter hypermethylated between the DZ and TZ (P = 0.072; Supplemental Fig. S4B).
Figure 7.
Expression of genes undergoing differential methylation in the 5′ region (A) and in the remainder of the gene body (B). A, Relative expression of genes undergoing differential methylation in the promoter or 5′ region of the gene body. The CCGG sites of EXPB3, TGA10, and CYCD4 are found in the promoter, whereas for DUF547 it is located immediately downstream of the start codon (ATGGCCGG). B, Relative expression of genes undergoing differential methylation in the remainder of the gene body. The CCGG sites of MTM (for microtubule motor family protein, or kinesin) and AGO are found in an exon and that of GATA is found in an intron. The PK/UbiC-encoding gene has two differentially methylated CCGG sites, found in an exon and the following intron. Zones between which methylation changes take place (DZ-TZ, TZ-EZ, and EZ-MZ) are indicated by black arrows. The location of differential methylation with respect to the gene is represented by a black arrowhead on the gene model. If other possible differential methylation CCGG sites are present, they are indicated by gray arrowheads. *P = 0.01–0.05, **P = 0.001–0.01, ***P ≤ 0.001.
Thirteen genes for which differential methylation was found in the remainder of the gene body were profiled (Supplemental Fig. S5), and only for three of these was the expression anticorrelated with their methylation status. The expression of five of these genes actually showed a significant positive correlation with DNA methylation. For example, an ATP-binding microtubule motor family protein-encoding gene (GRMZM2G338928) was 26-fold down-regulated when a site in its first exon became hypomethylated (Fig. 7B). Oppositely, hypermethylation was sometimes also associated with an increase in gene expression. Hypermethylation of a site in the second intron of a GATA zinc finger transcription factor-encoding gene (GRMZM2G052616) between the TZ and EZ was associated with its up-regulation in the more mature zones. For several of the genes with a positive correlation between methylation and expression, multiple CCGG sites were identified in the MSAP analysis as differentially methylated. The AGO-like domain-encoding gene (AC189879.3_FGT003) was down-regulated (3.4-fold; P = 0.0016) after hypomethylation in its next-to-last exon, but showed a second CCGG site in its last exon. Next, for one of the few genes for which methylation negatively correlated with its expression, we found two hypomethylated sites. The gene encoding both a protein kinase and ubiquitin-conjugating domain (GRMZM2G139157) had both its second exon and adjacent intron hypomethylated between the EZ and MZ. Expression of the gene was up-regulated 4.4-fold in the mature tissue (P = 0.02). This might be the evidence of a large-scale hypomethylation event across the gene body, causing up-regulation. Lastly, we analyzed the expression of genes for which differential methylation was found downstream of the stop codon (Supplemental Fig. S6A) or for which a methylation change was found to be gradual (Supplemental Fig. S6B). Also in these cases, no correlation between methylation and expression was found.
By compiling all expression data (Fig. 8), it became evident that only 5′ methylation correlates negatively with expression. Of the 28 transcriptionally profiled genes, the expression of 14 transcripts anticorrelated with expression, 10 of which were found in the 5′ region of a gene. Eight genes showed a positive correlation between gene expression and methylation, all but one found in the gene body. Six genes were not found to be differentially expressed, only one of which showed differential methylation in the 5′ region.
Figure 8.
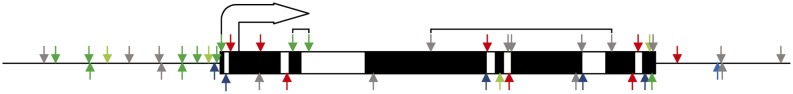
Summary of quantitative PCR expression data in relation to differential methylation. Correlation between differential methylation and expression is indicated as colored arrows. A green arrow indicates that a methylation change at this site led to up-regulation or down-regulation of a profiled gene, after it was hypomethylated or hypermethylated, respectively. A light green arrow means the differential expression was not found to be significant at the 5% level. A red arrow indicates an opposing effect: hypomethylation leading to down-regulation or hypermethylation leading to up-regulation. A blue arrow means that the gene was not found to be differentially expressed, and a gray arrow indicates a gene for which no expression in the sampled zones of the maize leaf could be found. Arrows that are linked represent methylation changes within the same gene. The gene is represented, from start codon to stop codon, as a rectangle, with exons in black and introns in white. The site of the start codon is indicated as an arrow.
DISCUSSION
The Methylation State of the Developing Maize Leaf
Here, we present the analysis of DNA methylation over the different growth zones of the developing maize leaf. More than 50% of the investigated CCGG sites were found to be stably methylated. Because the use of restriction enzymes does not allow us to distinguish some forms of methylation, MSAP results tend to yield an underestimation of genomic DNA methylation (Xiong et al., 1999). Indeed, recent whole-genome bisulfite sequencing of the maize genome in unfertilized ears revealed that of all Cs in CG and CHG contexts, 86% and 74%, respectively, are methylated (Gent et al., 2013). A second whole-genome methylome profiling effort of coleoptile tissue found lower, albeit still substantial, values: 65%, 50%, and 5% (Regulski et al., 2013). Generally, we found that full CG methylation was more represented than hemi-CHG methylation in the leaf, which is in agreement with previous MSAP experiments carried out on leaves of other grass species, such as sorghum (Sorghum bicolor; Zhang et al., 2011) and rice (Oryza sativa; Wang et al., 2011a). Moreover, all plant species for which whole-methylome profiles have been established, predominantly have methylation in a CG context (Lister et al., 2008; Zemach et al., 2010; Gent et al., 2013). Furthermore, we showed that, although the majority of genomic sequences remained stably (un)methylated in the leaf cells, a small number (3%) of the loci underwent methylation changes as the cells changed from a dividing state into an elongating state or as they differentiated into mature leaf cells.
Moreover, almost 40% of the methylation changes were found between cells that are fully dividing and those that are transitioning into elongating cells. Both the cell cycle machinery and cytosine methylation are strongly conserved systems in eukaryotes. However, DNA methylation has been evolutionarily lost in cell cycle model species, such as Saccharomyces cerevisiae and Caenorhabditis elegans (Goll and Bestor, 2005). This suggests that DNA methylation itself does not play a role in cell cycle regulation in these lower eukaryotes. In higher eukaryotes, on the other hand, cytosine methylation and DNA methylation are strongly interconnected, and DNA methylation might impose an additional level of regulation. The progress from dividing to elongating tissue also implies that gradually fewer cells are dividing and undergoing maintenance methylation. This means that the shift between dividing and elongating cells is also a shift between a continuous interchange in hemimethylation and full methylation in the DZ and a more stable methylation state in the EZ (Goll and Bestor, 2005). However, this shift is also associated with important developmental changes determining growth. The cell cycle is characterized by a tightly orchestrated regulation of specific genes, and an exit from the cell cycle leads to the expression of different classes of genes altogether (Inzé and De Veylder, 2006). Moreover, previous experiments from our laboratory have indicated that the TZ, which contains both dividing and elongating tissue, largely determines the speed of leaf growth and final leaf length in maize (Nelissen et al., 2012). Also, Arabidopsis leaf growth is characterized by a robust spatiotemporal regulation of cell division and cell elongation (Andriankaja et al., 2012). Therefore, we focused further screening for methylation changes on the DZ and TZ.
It is also between these two zones that maize maintenance methyltransferase expression was drastically reduced. Yet, although hypomethylation seemed to be slightly more abundant than hypermethylation between the DZ and TZ, overall, hypomethylation and hypermethylation were found to occur in more or less equal amounts. Most likely, the maintenance-type enzymes are required in high number during cell division, during which a great amount of cytosine methylation needs to occur after every round of DNA replication (Goll and Bestor, 2005). At later stages of cellular development, only basal levels of these enzymes are necessary to reinforce cytosine methylation by de novo DMTs, which are stably expressed, and other chromatin modifications established by histone-modifying enzymes (Vaillant and Paszkowski, 2007). Interestingly, the functional DMT genes (MET, CMT, and DRM) are all present in duplicate in the maize genome. Moreover, they all share high homology. This is especially true for the MET genes, which are 99.4% identical at the protein level. Most likely, these genes were quite recently duplicated to compensate for the expansion of the maize genome and its large proportion of repetitive (transposable) elements (Baucom et al., 2009).
Not all methylation changes identifiable with the MSAP technique occurred with the same frequency. Most strikingly, changes between hemi mCCG and full CmCG, indicative of a demethylation and a remethylation event at the same CCGG site, were found to be almost nonexistent. Similar results were found in MSAP experiments conducted in Arabidopsis (Cervera et al., 2002), pepper (Capsicum annuum; Portis et al., 2004), and maize (Lu et al., 2008). Also, the occurrence of two methylation changes between two zones was quite uncommon. Going from one zone to the next, almost all CCGG sites would thus undergo simple one-step hypomethylation or hypermethylation. Most of these differential methylation events occurred at CG sites, in which most DNA methylation takes place (Law and Jacobsen, 2010). It is important to note that not all MSAP pattern changes can be deduced in a straightforward manner. If one or more CCGG sites are present within the amplified MSAP band, the differential methylation pattern might not be caused by a change in the methylation state of the outer CCGG but by an opposing change in (one of) the inner CCGG site(s). Indeed, for 42 of the 149 single-locus differentially methylated sites, one or more CCGG sites were found inside the amplified MSAP sequence (Supplemental Tables S6 and S7).
Differential Methylation Targets 5′ and 3′ Edges of Genic Regions, But Only 5′ Correlates with Expression
Genic loci that underwent differential methylation showed enrichment at the 5′ and 3′ edges of the gene body. This pattern of differential methylation is the exact opposite of the stable methylation patterns that have been identified in several tissues of higher plant species such as Arabidopsis, rice, and poplar (Populus spp.; Feng et al., 2010). In these species, the steady-state gene body methylation is generally high in the central part of genes, with 5′ and 3′ terminal regions being devoid of DNA methylation, which implies an important function in gene regulation. This pattern has been proposed to play a role in transcriptional regulation, with moderately expressed genes being highly methylated in the gene body. An alternative role might lie in exon definition, since the majority of gene body methylation is found in exons (Saze and Kakutani, 2011), or the inhibition of transcriptional initiation from spurious promoters (Lauria and Rossi, 2011). Indeed, almost three-quarters of the differentially methylated sites identified in this experiment were found in an exon. However, in our study, a high prevalence of methylation changes was found at the start and stop of the gene. This might be an indication of versatile gene regulation, whereas the central body methylation is steadily maintained depending on the overall expression level of the gene. There are different ways in which gene body DNA methylation can influence RNA polymerase action and general transcript formation. First, DNA methylation could shield spurious transcript start sites within the gene in order to avoid the formation of truncated gene products, which could be deleterious to cellular function (Lauria and Rossi, 2011). Second, DNA methylation might simply identify which sequences within the transcript are exons and which are introns, as splicing generally coincides with transcription. Indeed, more and more evidence is accumulating that epigenetic modifications, such as DNA methylation, are influencing gene splicing also in maize (Regulski et al., 2013). In this case, differential methylation between tissues could coincide with differential splicing, giving rise to different transcripts of the same gene. However, we did not find compelling evidence that differential methylation in the gene bodies influences differential splicing of the transcripts. Further research will be needed to investigate this phenomenon.
The upstream, or 5′, sequences encompassing the promoter as well as the downstream, or 3′, sequences can contain gene regulatory elements and are subjected to DNA methylation (Suzuki and Bird, 2008; Yu et al., 2013). In this study, we found a correlation between gene expression and DNA methylation of the 5′ portion of the gene. Differential methylation of both the promoter and the portion before and around the start codon has an adverse effect on gene expression. This is reminiscent of CpG island methylation in animals, which is associated with gene promoters but often extends into the 5′ untranslated region and even the first intron (Suzuki and Bird, 2008). No correlation could be found between differential methylation of the central and 3′ part of the gene body or sequences downstream of the gene. In animals, though, several studies of the methylome during the development of both somatic and cancer cells have indicated a correlation between intragenic methylation and gene expression (Kulis et al., 2013). No correlation could be observed between gene expression and differential methylation of the 3′ part of the gene body or sequences downstream of the gene. However, previous steady-state methylome data indicated that gene body methylation might not have a direct influence on the transcription of the gene per se but is rather correlated with the polymerase function in a parabolic fashion (Zemach et al., 2010). This means that moderately expressed genes are most likely heavily methylated, whereas highly and lowly expressed genes tend to bear little gene body methylation.
As the MSAP technique only yields information about the methylation state of one CCGG site, it does not give information about the methylation status of surrounding cytosines. Therefore, we cannot show the causality of the changes in methylation status at the 5′ part of the gene to the difference in expression level, but merely show an increased correlation. In addition, there might also be other factors, such as histone modifications and binding of transcription factors, which are influenced by changes in methylation and, hence, have an effect on their expression (Jaenisch and Bird, 2003). Nevertheless, this study demonstrates a strong correlation between methylation and developmental transitions, providing a basis for further, more detailed studies. For example, bisulfite sequencing would yield valuable information on the neighboring sequences and the overall methylation status of genes.
The Majority of Multiple-Locus Sequences Are (Retro-)Transposons
Roughly 25% of the maize genome consists of non-TE DNA and only 6% of protein-coding genes (Schnable et al., 2009). Maize TEs can be categorized into two classes: RNA (retro- or type I) transposons and DNA (type II) transposons. The former class is by far the largest, occupying 76% of the genome, whereas DNA transposons make up 8.6% of the genome (Schnable et al., 2009). However, more than 70% of the obtained sequences did not exhibit TE-like features. It is unlikely that this is the consequence of a bias caused by the primers used, since we used all 64 possible HpaII/MspI-specific primers in combination with five EcoRI-specific primers, covering a wide range of restriction sites in the genome. A problematic amplification of repetitive sequences could in part explain the bias. However, even if all failed amplifications were TEs, non-TE sequences would still make up more than 40% of the data set. Moreover, at least 34.7% of the hits in our data set were mapped to protein-coding genes, and only 9.6% were not in the vicinity (5 kb in either direction) of a gene. Judging from these data, we could see that genic sequences were more prone to differential methylation than repetitive DNA. Similar results were obtained from MSAP experiments in sorghum (Zhang et al., 2011), rice (Wang et al., 2011a), and maize (Lauria et al., 2004).
Less than 38% of the acquired sequences could not be mapped to a single locus in the maize genome, and most of those turned out to be TEs. When combining transposons from both the single-locus and multiple-locus data sets, only 24.1% of the MSAP hits were found to be TEs. From these, 66.3% were type I TEs, 23.7% were type II TEs, and 10% were TEs of unknown origin (i.e. annotated as being a TE in the maize genome but not when BLASTed to miazetedb.org). There seem to be more type II TEs than expected in the maize genome. This can be explained by the fact that these TEs are generally of lower copy number and silenced less robustly than the very abundant RNA retrotransposons. Several members of the type II TE class are known for their mutagenic abilities. For example, CACTA TEs (11 of 19 type II TEs in our data set) are known to be mobilized in mutants with a compromised DNA methylation machinery in Arabidopsis (Kato et al., 2004). In maize, a process called developmental relaxation of TE silencing causes a burst of TE transcriptional activity in the shoot apical meristem, after which silencing needs to be reestablished (Martínez and Slotkin, 2012). Indeed, 75% of type II TEs are found in the hypermethylated fraction of our data set. This could be evidence of a progressive silencing of DNA TEs in older zones in order to prevent mutation due to transposition.
The long terminal repeat (LTR) retrotransposon order occupies by far the most space in the maize genome (74.6%). This is almost entirely caused by the presence of two superfamilies, Gypsy (46.4%) and Copia (23.7%; Baucom et al., 2009). Also in our data set, all but one of the retrotransposons found are members of the LTR order of retrotransposons. However, within this order, the prevalence of members of the different families differs from the abundance in the maize genome (Supplemental Table S5). Most LTR-type retroelements identified in this study fall within the 20 most prevalent TE families of the maize genome. However, the most prevalent maize TE, Huck, is found only once in our data set. Moreover, the average GC content of Huck elements is 60%, which is much higher than the average GC content of the maize repetitive fraction (48%) and the average genomic GC content (47%; Meyers et al., 2001). The Giepum TE, on the other hand, is more than eight times less present in the maize genome but eight times more present in our data set. The reason for this bias remains to be investigated.
The remaining multihit sequences mapped to two or more loci of the genome encoding the same sequence. Some of these sequences have at least one copy in a plastid genome. Combined with the four single-locus genes of organellar origin (all found in the EZ-MZ data set), a total of 17 organelle-related genes were found using the MSAP approach. Since organellar genomes are of bacterial origin, they exhibit mainly methylation of adenines and not of cytosines (Vanyushin and Ashapkin, 2011). Integration of organellar sequences in the nuclear genome is very common in many plant species, especially those that are outbreeding and have large genomes (Ayliffe et al., 1998). Seeing that over 99% of plastid and 95% of mitochondrial genomic sequences have at least one copy in the maize nuclear genome (Kumar and Bendich, 2011), we most likely are visualizing DNA methylation in the nuclear copies.
Differential Methylation along the Developing Maize Leaf Potentially Affects Many Processes
An interesting finding is the fact that some of the genes that are differentially methylated are themselves involved in epigenetic processes, either directly (IDM1, SMH3, AGO, SNF2, and PHD genes) or indirectly. For example, GRMZM2G311883 is a gene homologous to Arabidopsis MODIFIER OF SNC1-1 14 (MOS14), which is known to form a link between splicing and RNA-dependent DNA methylation (Zhang et al., 2013). Also, genes involved in cell cycle regulation, such as CYCD4, APC10, TIP120-encoding (Wang et al., 2011b), FORMIN8-like (Xue et al., 2011), and AUGMIN6-like (Hotta et al., 2012), are represented in the data set, as are developmental processes affecting the vasculature (Petricka et al., 2008), meristem maintenance (Liu et al., 2009), and cell shape and plant architecture (O’Hara et al., 2013). Several genes that are differentially methylated in the older zones play a known role in development. For example, the family of CORN CYSTATIN (CC) genes is known to play a role in development, and more specifically, CC5, which was found to be hypermethylated, is induced under drought stress (Massonneau et al., 2005). Also, MiR396a influences development by targeting the destruction of the GRF genes (Debernardi et al., 2012). Another interesting gene is PHOSPHATIDYLINOSITOL SYNTHASE2 (PIS2), which underwent two hypomethylation events in the older zones. Recently, it was found that overexpression of ZmPIS in maize enhanced abscisic acid-mediated drought stress tolerance (Liu et al., 2013).
Therefore, it is tempting to speculate that DNA methylation regulates the aforementioned processes in the developmental framework of the growing maize leaf. However, our data show that differential methylation affecting the 5′ region of the gene is correlated with gene expression, whereas this is not the case for the remainder of the gene. This is especially striking in the case of the MiR396a-encoding gene, which is specifically up-regulated in the TZ and again down-regulated in the EZ. Hypomethylation of the CCGG site within the gene body, however, occurs between the EZ and MZ. Exactly how these methylations affect gene function remains unknown. They might be involved in processes other than gene expression as such or counteracted by other epigenetic marks, such as histone modification.
CONCLUSION
It is already known that maize leaf growth is strongly regulated by fine-tuning the development of the different zones in the maize leaf. Especially the size of the DZ is an important factor affecting growth, and this is regulated through differential gene expression and precise hormone deposition (Rymen et al., 2007; Nelissen et al., 2012). Here, we show that the consecutive developmental zones of the growing maize leaf are correlated to differential DNA methylation. Especially when occurring upstream of the gene start and the 5′ end of the gene, differential DNA methylation is associated with expression changes of genes that need to be expressed in a specific developmental context.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type maize (Zea mays ‘B73’) plants were grown in soil (Saniflor; NV Van Israel) in a growth room under controlled conditions: photoperiod (16/8 h light/dark), temperature (25.5°C), relative humidity (50%), and light intensity (170 mmol m−2 s−1 photosynthetically active radiation at the plant level). The growth room was illuminated by a combination of high-pressure sodium vapor lamps (RNP-T/LR/400W/S/230/E40; Radium) and metal halide lamps with quartz burners (HRI-BT/400W/D230/E40; Radium).
Growth, Cellular Analysis, and Sampling
Plants were measured daily to determine leaf elongation rates of leaf 4 (n = 20). For experimental manipulations, leaf 4 was harvested 2 d after appearance (12 d after sowing; Vegetative or V stage 2), at which point the ligule is only a few millimeters from the base, during steady-state leaf growth. For cellular analysis, the first 10 cm of the leaf was cut into 1-cm pieces. These samples were cleared overnight with ethanol:acetic acid (3:1). Epidermal cell length profiles of cell files along the proximal-distal axis were established with a differential interference contrast microscope (AxioImager; Zeiss) and image-analysis software (AxioVision; Zeiss) on samples fixed using lactic acid (n = 3). The size of the DZ was determined as the distance between the leaf base and the most distally located mitotic figure in 4′,6-diamino-phenylindole-stained leaves with a fluorescence microscope (n = 4). For MSAP analysis, the first, second, fourth, and tenth 1 cm were harvested as representatives of the DZ, TZ, EZ, and MZ, respectively. For expression analysis, the most basal 4 cm of the maize leaf was harvested in 5-mm pieces, whereas the following 6 cm were harvested in 1-cm pieces. Samples for MSAP and expression analysis were harvested and flash frozen in liquid N2.
Nucleotide Isolation
Total DNA for MSAP analysis was isolated form individual samples with the cetyl-trimethyl-ammonium bromide method. This DNA was quantified using the quantIT dsDNA High-sensitivity Assay Kit (Invitrogen) and LUMIstar Galaxy (BMG Labtech) according to each manufacturer’s instructions. Total RNA for expression analysis was isolated from pooled samples (five plants) with the guanidinium thiocyanate-phenol-chloroform extraction method using TRI-reagent (Sigma-Aldrich). First-strand complementary DNA was synthesized from 1 µg of total RNA with the iScript kit (Bio-Rad Laboratories) according to the manufacturer’s instructions.
MSAP Analysis of DNA Methylation
The maize methylome was screened using the MSAP protocol (Xiong et al., 1999). MspI and HpaII are used as frequent isoschizomeric restriction enzymes with differential sensitivity toward CCGG methylation, each in combination with EcoRI as a cytosine methylation-insensitive restriction enzyme (Supplemental Table S8). Briefly, 390 ng of genomic DNA underwent restriction-ligation using EcoRI-HpaII/MspI and EcoRI (E)-specific and HpaII/MspI (HM)-specific adapters (Supplemental Table S9), at 37°C for 4 h, and diluted 10-fold. The restriction-ligation mix was the template for preamplification PCR, using primers with one selective nucleotide, and diluted 20-fold. This dilution was the template for selective amplification, using primers with three selective nucleotides, in which the EcoRI primers were labeled with [γ-33P]ATP. For the first screen, 65 selective primers were applied in all 64 possible combinations (E+AAC × HM+NNN) to two biological replicates of DZ, TZ, EZ, and MZ. For the second screen, 68 selective primers (E+AAA/G/T and E+ACA × HM+NNN) were used in all 256 possible combinations on pools of DZ and TZ samples. Hence, for DZ-TZ, 320 primer combinations were tested. Samples (3 µL) were loaded onto a 4.5% denaturing polyacrylamide gel, and electrophoresis was performed at constant power of 100 W. Labeled bands were blotted onto Whatman 3MM blotting paper and visualized on Amersham Hyperfilm ECM (GE Healthcare).
Identification of Differentially Methylated Loci
Polymorphic banding patterns, representing differential methylation, were identified by eye. One representative band was cut from the blotting paper using a razor blade, resuspended in deionized water (1.5 μS cm−1), reamplified with Pwo SuperYield DNA Polymerase (Roche), and sequenced using the E-specific primer. Successfully sequenced loci were BLASTed to the maize genome (www.maizesequence.com, currently replaced by ensembl.gramene.org/Zea_mays), and the differentially methylated CCGG site was identified as the CCGG site at the 3′ end of the sequence or the closest CCGG site. When more than one genomic site was identified, the sequence was BLASTed to the maize TE database (maizetedb.org) in order to determine if the underlying sequence was transposon derived and which type it was. If only one locus was identified and not found to overlap with a gene, we scanned 5 kb upstream and downstream for the presence of one. The largest transcript was chosen to represent each gene, and the position of each single locus CCGG site was calculated with respect to both the transcription and translational start and stop sites of this transcript.
Transcriptional Analysis
Maize CDKB1;1 was identified earlier as a marker for mitotic activity (Rymen et al., 2007). All remaining primers (Supplemental Table S10) were designed with the Beacon Designer 4.0 software and the default settings (Premier Biosoft International). The transcripts were quantified using quantitative reverse transcription PCR with the Lightcycler 480 (Roche Applied Science) and SYBR Green I Master Kit (Roche Applied Science). PCR was done in triple technical replicates according to the manufacturer’s guidelines. Melting curves were generated to check primer specificity, and relative quantification was carried out according to Ramakers et al., 2003 with 18S rRNA (5′-ACCTTACCAGCCCTTGACATATG-3′ and 5′-GACTTGACCAAACATCTCACGAC-3′) as a housekeeping gene. Three biological replicates were averaged for analysis.
Statistical Analysis
The distribution of CCGG sites within the gene body and the occurrence of transposons in the data set were statistically analyzed using Pearson’s χ2 test. Transcript profiles were statistically evaluated based on an ANOVA for the factor position in R (www.r-project.org). P values for distances between DMLs on the maize chromosomes were calculated using a two-sample Student’s t test assuming unequal variance in Excel. All error bars depict se.
Sequence data from this article can be found in the Supplemental Table S11.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of differentially methylated single-locus sequences across the chromosomal arms and between different chromosomal sites.
Supplemental Figure S2. Locations of differentially methylated sites in or close to coding sequences.
Supplemental Figure S3. Expression of the maize MiR396a- and GRF-encoding genes.
Supplemental Figure S4. Expression of genes undergoing differential methylation in the promoter and 5′ regions of the gene body.
Supplemental Figure S5. Expression of genes undergoing differential methylation in the central part of the gene body.
Supplemental Figure S6. Relative expression of genes undergoing differential methylation downstream of the coding sequence and gradual methylation change in the gene body.
Supplemental Table S1. DMT-encoding genes and alternative names.
Supplemental Table S2. MSAP patterns and possible DNA methylation states.
Supplemental Table S3. Relative abundance of differential methylation patterns.
Supplemental Table S4. Differentially methylated TEs in the multilocus data set.
Supplemental Table S5. Abundance of the 20 most prevalent maize TEs.
Supplemental Table S6. Single-locus differentially methylated sequences identified between the DZ and TZ.
Supplemental Table S7. Single-locus differentially methylated sequences identified between the TZ and EZ, EZ and MZ, and multiple zones.
Supplemental Table S8. DNA methylation sensitivity of MSAP restriction enzymes.
Supplemental Table S9. Primers used for MSAP analysis.
Supplemental Table S10. Primers used for quantitative reverse transcription PCR analysis.
Supplemental Table S11. Sequences generated from differential MSAP bands.
Acknowledgments
We thank the Systems Biology of Yield group for the stimulating scientific environment and fruitful discussions, Dr. Annick Bleys for help in preparing the manuscript, and Wilson Ardiles Diaz for help in sequencing the MSAP bands.
Glossary
- DMT
DNA methyltransferase
- DZ
division zone
- TZ
transition zone
- EZ
expansion zone
- MZ
mature zone
- MSAP
methylation-sensitive amplified polymorphism
- TE
transposable element
- PHD
plant homeodomain
- T6P
trehalose-6-phosphate
- LTR
long terminal repeat
Footnotes
This work was supported by the Agency for Innovation by Science and Technology in Flanders (“Strategisch Basisonderzoek” grant no. SB093354), by the Interuniversity Attraction Poles Programme (grant no. IUAP P7/29, “Study of Maize-Arabidopsis-Root-Shoot” growth), initiated by the Belgian State, Science Policy Office, and by Ghent University (“Bijzonder Onderzoeksfonds Methusalem” grant no. BOF08/01M00408 and Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” grant no. 01MRB510W).
Present address: School of Pure and Applied Sciences, Kisii University, P.O BOX 408-40200 Kisii, Kenya.
These authors contributed equally to the article.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Scott NS, Timmis JN. (1998) Analysis of plastid DNA-like sequences within the nuclear genomes of higher plants. Mol Biol Evol 15: 738–745 [DOI] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL. (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, Jacobsen SE. (2000) Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci USA 97: 4979–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera MT, Ruiz-García L, Martínez-Zapater JM. (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol Genet Genomics 268: 543–552 [DOI] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF. (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, Hemerly AS, Beemster GTS, Inzé D, Ferreira PCG. (2011) The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J 68: 351–363 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galego L, Almeida J. (2002) Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev 16: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler K, Paulsen T, Napoli C. (2008) ChromDB: the chromatin database. Nucleic Acids Res 36: D298–D302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK. (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albert N, Fujimoto R, Helliwell CA, Dennis ES, Peacock WJ. (2011) Epigenetics in plants: vernalisation and hybrid vigour. Biochim Biophys Acta 1809: 427–437 [DOI] [PubMed] [Google Scholar]
- Hotta T, Kong ZS, Ho CMK, Zeng CJT, Horio T, Fong S, Vuong T, Lee YRJ, Liu B. (2012) Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell 24: 1494–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet (Suppl) 33: 245–254 [DOI] [PubMed] [Google Scholar]
- Kato M, Takashima K, Kakutani T. (2004) Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 168: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Queirós AC, Beekman R, Martín-Subero JI. (2013) Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta 1829: 1161–1174 [DOI] [PubMed] [Google Scholar]
- Kumar RA, Bendich AJ. (2011) Distinguishing authentic mitochondrial and plastid DNAs from similar DNA sequences in the nucleus using the polymerase chain reaction. Curr Genet 57: 287–295 [DOI] [PubMed] [Google Scholar]
- Lauria M, Rossi V. (2011) Epigenetic control of gene regulation in plants. Biochim Biophys Acta 1809: 369–378 [DOI] [PubMed] [Google Scholar]
- Lauria M, Rupe M, Guo M, Kranz E, Pirona R, Viotti A, Lund G. (2004) Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Howell S. (2009) Overexpression of an Arabidopsis gene encoding a subtilase (AtSBT5.4) produces a clavata-like phenotype. Planta 230: 687–697 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhai S, Zhao Y, Sun B, Liu C, Yang A, Zhang J. (2013) Overexpression of the phosphatidylinositol synthase gene (ZmPIS) conferring drought stress tolerance by altering membrane lipid composition and increasing ABA synthesis in maize. Plant Cell Environ 36: 1037–1055 [DOI] [PubMed] [Google Scholar]
- Lu Y, Rong T, Cao M. (2008) Analysis of DNA methylation in different maize tissues. J Genet Genomics 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Lund G, Messing J, Viotti A. (1995) Endosperm-specific demethylation and activation of specific alleles of α-tubulin genes of Zea mays L. Mol Gen Genet 246: 716–722 [DOI] [PubMed] [Google Scholar]
- Marian CO, Bordoli SJ, Goltz M, Santarella RA, Jackson LP, Danilevskaya O, Beckstette M, Meeley R, Bass HW. (2003) The maize Single myb histone 1 gene, Smh1, belongs to a novel gene family and encodes a protein that binds telomere DNA repeats in vitro. Plant Physiol 133: 1336–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez G, Slotkin RK. (2012) Developmental relaxation of transposable element silencing in plants: functional or byproduct? Curr Opin Plant Biol 15: 496–502 [DOI] [PubMed] [Google Scholar]
- Massonneau A, Condamine P, Wisniewski JP, Zivy M, Rogowsky PM. (2005) Maize cystatins respond to developmental cues, cold stress and drought. Biochim Biophys Acta 1729: 186–199 [DOI] [PubMed] [Google Scholar]
- McClelland M, Nelson M, Raschke E. (1994) Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res 22: 3640–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tingey SV, Morgante M. (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11: 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Coppens F, Dhondt S, Fiorani F, Beemster GTS. (2013) Kinematic analysis of cell division in leaves of mono- and dicotyledonous species: a basis for understanding growth and developing refined molecular sampling strategies. Methods Mol Biol 959: 247–264 [DOI] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, Kamiya Y, Inzé D, Beemster GTS. (2012) A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol 22: 1183–1187 [DOI] [PubMed] [Google Scholar]
- O’Hara LE, Paul MJ, Wingler A. (2013) How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol Plant 6: 261–274 [DOI] [PubMed] [Google Scholar]
- Peraza-Echeverria S, Herrera-Valencia VA, Kay A. (2001) Detection of DNA methylation changes in micropropagated banana plants using methylation-sensitive amplification polymorphism (MSAP). Plant Sci 161: 359–367 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Clay NK, Nelson TM. (2008) Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J 56: 251–263 [DOI] [PubMed] [Google Scholar]
- Portis E, Acquadro A, Comino C, Lanteri S. (2004) Analysis of DNA methylation during germination of pepper (Capsicum annuum L.) seeds using methylation-sensitive amplification polymorphism (MSAP). Plant Sci 166: 169–178 [Google Scholar]
- Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. (2012) A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MTA, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S, Van De Peer Y, Rouzé P. (2003) AFLPinSilico, simulating AFLP fingerprints. Bioinformatics 19: 776–777 [DOI] [PubMed] [Google Scholar]
- Ryan DP, Owen-Hughes T. (2011) Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol 15: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Fiorani F, Kartal F, Vandepoele K, Inzé D, Beemster GTS. (2007) Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol 143: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. (2011) The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T. (2011) Differentiation of epigenetic modifications between transposons and genes. Curr Opin Plant Biol 14: 81–87 [DOI] [PubMed] [Google Scholar]
- Saze H, Tsugane K, Kanno T, Nishimura T. (2012) DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol 53: 766–784 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220 [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Tolley BJ, Woodfield H, Wanchana S, Bruskiewich R, Hibberd JM. (2012) Light-regulated and cell-specific methylation of the maize PEPC promoter. J Exp Bot 63: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Paszkowski J. (2007) Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol 10: 528–533 [DOI] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K. (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin BF, Ashapkin VV. (2011) DNA methylation in higher plants: past, present and future. Biochim Biophys Acta 1809: 360–368 [DOI] [PubMed] [Google Scholar]
- Wang WS, Pan YJ, Zhao XQ, Dwivedi D, Zhu LH, Ali J, Fu BY, Li ZK. (2011a) Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J Exp Bot 62: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, He FF, Ma XX, Mao CZ, Hodgman C, Lu CG, Wu P. (2011b) OsCAND1 is required for crown root emergence in rice. Mol Plant 4: 289–299 [DOI] [PubMed] [Google Scholar]
- Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q. (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet 261: 439–446 [DOI] [PubMed] [Google Scholar]
- Xue XH, Guo CQ, Du F, Lu QL, Zhang CM, Ren HY. (2011) AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol Plant 4: 264–278 [DOI] [PubMed] [Google Scholar]
- Yu DH, Ware C, Waterland RA, Zhang J, Chen MH, Gadkari M, Kunde-Ramamoorthy G, Nosavanh LM, Shen L. (2013) Developmentally programmed 3′ CpG island methylation confers tissue- and cell-type-specific transcriptional activation. Mol Cell Biol 33: 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, et al. (2013) The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J 32: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DF, Li B, Jia GQ, Zhang TF, Dai JR, Li JS, Wang SC. (2008) Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). Plant Sci 175: 809–817 [Google Scholar]
- Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D. (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xu C, von Wettstein D, Liu B. (2011) Tissue-specific differences in cytosine methylation and their association with differential gene expression in sorghum. Plant Physiol 156: 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]