Transcriptome analysis indicates that ectopic expression of MADS domain transcription factors mimics the induction of somatic embryogenesis, providing a mechanism for its enhancement in culture.
Abstract
Somatic embryogenesis (SE) is a poorly understood process during which competent cells respond to inducing conditions, allowing the development of somatic embryos. It is important for the regeneration of transgenic plants, including for soybean (Glycine max). We report here that constitutive expression of soybean orthologs of the Arabidopsis (Arabidopsis thaliana) MADS box genes AGAMOUS-Like15 (GmAGL15) and GmAGL18 increased embryogenic competence of explants from these transgenic soybean plants. To understand how GmAGL15 promotes SE, expression studies were performed. Particular genes of interest involved in embryogenesis (ABSCISIC ACID-INSENSITIVE3 and FUSCA3) were found to be directly up-regulated by GmAGL15 by using a combination of quantitative reverse transcription-polymerase chain reaction and chromatin immunoprecipitation. To look more broadly at changes in gene expression in response to GmAGL15, we assessed the transcriptome using the Affymetrix Soybean Genome Array. Interestingly, the gene expression profile of 35Spro:GmAGL15 explants (0 d in culture) was found to resemble nontransgenic tissue that had been induced for SE by being placed on induction medium for 3 d, possibly explaining the more rapid SE development observed on 35Spro:GmAGL15 tissue. In particular, transcripts from genes related to the stress response showed increased transcript accumulation in explants from 35Spro:GmAGL15 tissue. These same genes also showed increased transcript accumulation in response to culturing nontransgenic soybean explants on the medium used to induce SE. Overexpression of GmAGL15 may enhance SE by making the tissue more competent to respond to 2,4-dichlorophenoxyacetic acid induction by differential regulation of genes such as those involved in the stress response, resulting in more rapid and prolific SE.
AGAMOUS-Like15 (AGL15; Arabidopsis Genome Initiative identifier At5g13790) is an Arabidopsis (Arabidopsis thaliana) MADS domain transcription factor that accumulates to its highest level in developing embryos (Heck et al., 1995; Rounsley et al., 1995). Although AGL15 expression is not exclusive to embryos, and roles in the control of flowering time have been reported (Adamczyk et al., 2007), the level of accumulation in tissues outside the embryo is generally too low to detect by protein gel blot (Perry et al., 1996). However, immunologically related protein can be detected in developing embryos from a range of species and developmental contexts (Perry et al., 1996, 1999). Constitutive expression of AGL15 promotes development in an embryonic mode in at least two systems in Arabidopsis, and loss of function, in one system as a double mutant with the related MADS box gene AGL18, decreases somatic embryo tissue production (Harding et al., 2003; Thakare et al., 2008).
To extend our knowledge to a crop plant, we isolated a soybean (Glycine max) ortholog of AGL15 (referred to henceforth as GmAGL15; Thakare et al., 2008). We found that biolistic transformation with 35Spro:GmAGL15 constructs significantly increased the number of transformants obtained compared with controls (Thakare et al., 2008). One, but not the only, possible mechanism for this is by enhancing the regeneration of transformed cells via somatic embryogenesis (SE). We have since regenerated transgenic plants and report here on the enhancement of SE by 35Spro:GmAGL15 in soybean. We isolated a related gene that encodes a protein most similar to AGL18 in Arabidopsis and report on increased SE by 35Spro:GmAGL18 as well.
SE is not well understood, particularly what makes some cells competent to form somatic embryos. Understanding the underlying mechanism is important because SE is a more accessible model to understand zygotic embryogenesis that occurs embedded in maternal tissue. SE is also relevant to agriculture, in that genetic engineering generally requires the regeneration of transformed cells into plants. This regeneration may be by organogenesis or SE. The ability for SE varies depending on plant, tissue, age, and even particular genotype of a plant species (Karami and Saidi, 2010; Yang and Zhang, 2010). As a MADS domain transcription factor, GmAGL15 most likely promotes SE by controlling gene expression. The promotion of SE by GmAGL15 provides a unique opportunity to compare this tissue with control tissue to examine how GmAGL15 might induce SE, and we report here on the characterization of the transcriptome in response to increased GmAGL15 accumulation. A number of orthologs of Arabidopsis genes that encode key transcription factors necessary for normal seed development, and sufficient for the expression of embryo-specific programs outside of the seed context, are regulated by GmAGL15. These include orthologs of ABSCISIC ACID-INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and AGL18 that we show are directly regulated by GmAGL15.
To extend our understanding of the mechanism by which GmAGL15 promotes SE, we assessed the transcriptome in response to the expression of GmAGL15 via the 35S promoter. In addition to the regulation of genes encoding key embryo transcription factors, genes involved in the stress response, many of them linked to SE, were found to be up-regulated earlier in 35Spro:GmAGL15 tissue compared with the wild type.
RESULTS
Overexpression of GmAGL15 Increases the Frequency of Soybean SE
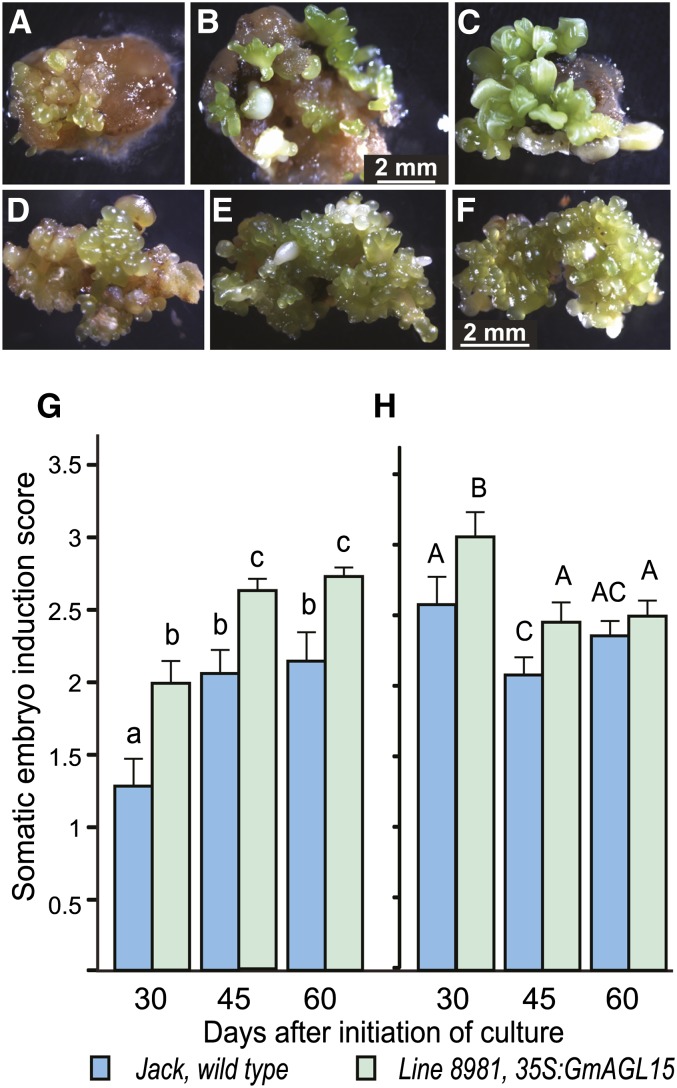
We previously reported that a 35Spro:GmAGL15 transgene significantly increased the number of putative transformants for the relatively embryogenic soybean cv Jack from an average of 14 (for the complementary DNA [cDNA] construct) to 16.6 (for the genomic DNA [gDNA] construct) putative transformants per biolistic transformation compared with eight for the empty vector control (Thakare et al., 2008). GmAGL15 could be facilitating a variety of steps during transformation, with one possible mechanism being the enhancement of regeneration of the transformed cells. We have now regenerated transgenic plants expressing the gene from chromosome 11 (two genes encode putative orthologs of AGL15 in soybean: Glyma11g16105 and Glyma12g17721). To test whether tissue containing the transgene is more competent for SE, the frequencies of SE for the cv Jack wild type and transgenic lines expressing 35Spro:GmAGL15 were compared by placing immature cotyledon explants from 4- to 5-mm embryos onto D40 medium (40 mg L−1 2,4-dichlorophenoxyacetic acid [2,4-D]) that is used to induce SE. As shown in Figure 1, A to C, the development of embryos on explants from two different lines of 35Spro:GmAGL15 was more rapid and robust than the control on D40 medium. To quantitate, somatic embryo induction was scored as described by Meurer et al. (2001) at different days after placement in culture (dac). Individual explants (21–25 per plate) were scored as 0 if no embryos were produced, 1 if one to five embryos were present, 2 if six to 15 embryos were present, and 3 if more than 15 embryos were present. The score for each plate was calculated by summing the score for each explant on the plate and then dividing by the total number of explants. The average was calculated among all plates of a given genotype. Scores are shown in Figure 1G for one of the transgenic lines (line 8981). The embryo induction score of line 8981 was significantly increased compared with nontransgenic cv Jack at all time points assessed. A second transgenic line also produced more embryos than the control at 45 dac (Supplemental Fig. S1).
Figure 1.
Overexpression of GmAGL15 enhances the production and proliferation of somatic embryos in soybean. A to C, Immature cotyledon explants cultured on D40 medium. A, Control (cv Jack). B and C, Two independent 35Spro:GmAGL15 lines. All images are the same magnification, and explants are the same age. D to F, Somatic embryo clusters induced on D40 were moved to D20 medium at approximately 6 weeks for proliferation. D, Control (cv Jack). E and F, Two independent lines expressing 35Spro:GmAGL15. G, Initiation of somatic embryogenesis on D40 medium. H, Proliferation of somatic embryos induced on D40 upon subculture to D20 medium. See text for the scoring systems. Means and se for at least nine plates of explants per genotype per time point are shown. Different letters indicate significant differences at P < 0.05.
After initiation on D40 medium, somatic embryos were subcultured to D20 medium (20 mg L−1 2,4-D) to allow the proliferation of somatic embryos (Meurer et al., 2001). Specifically, we transferred tissue that consisted of clusters of globular structures that were somewhat green in color, because cv Jack had been shown previously under conditions we used to produce green somatic embryos, even though other cultivars may be embryogenic but not green in color (Meurer et al., 2001; Kita et al., 2007). The scoring system used to quantitate proliferation is described in “Materials and Methods.” Overexpression of GmAGL15 also impacts on the proliferation of somatic embryos. As shown in Figure 1, D to F, the transgenic lines produced somatic embryos more robustly than the control upon subculture, with a greater fraction of the proliferating explants retaining green color. With extended periods of time without subsequent subculture to fresh D20 medium, somatic embryo tissue did continue development, but scores for proliferating transgenic tissue remained significantly higher than the wild type at 45 dac after subculture to D20 medium (Fig. 1H). With regular subculture to fresh D20 medium approximately each 30 d, 35Spro:GmAGL15 tissue remained more uniformly proliferative with a green color than the nontransgenic control (Supplemental Fig. S2).
A Gene Related to GmAGL15, Glyma02g33040 (GmAGL18), Also Impacts on SE in Soybean
The MADS box gene most closely related to AGL15 (At5g13790) in Arabidopsis is AGL18 (At3g57390), and these two gene products have redundant functions in the promotion of SE (Thakare et al., 2008) and in the control of flowering time (Adamczyk et al., 2007). A protein BLAST search of the soybean proteome database was performed using Arabidopsis AtAGL18 protein, and the top scoring match was encoded by Glyma02g33040. An alignment of predicted proteins encoded by AtAGL18 and Glyma02g33040 was performed using ClustalW2 (Fig. 2A; 49% identity). A phylogenetic tree was constructed using protein sequences encoded by GmAGL15 (chromosome 11 and 12 genes), AtAGL15, AtAGL18, Glyma02g33040, and two closely related proteins to AGL15 and AGL18 in Arabidopsis, SHORT VEGETATIVE PHASE/At2g22540 and AtAGL24/At4g24540 (Parenicová et al., 2003). As shown in Figure 2B, the protein encoded by Glyma02g33040 was most closely related to that of AtAGL18; thus, we refer to gene Glyma02g33040 as GmAGL18. Based on data available in the Soybean eFP Browser, transcript from this gene accumulates primarily in flowers and seeds, especially during early seed development (Libault et al., 2010; Supplemental Fig. S3).
Figure 2.
Isolation of a putative ortholog of Arabidopsis AGL18 from soybean. A, Alignment of predicted proteins encoded by Arabidopsis (At) AGL18 and an ortholog from soybean (Gm) generated using ClustalW2. B, Phylogenetic tree generated using ClustalW2 and the predicted protein sequences encoded by AGL15, AGL18, and closely related AGL24 and SVP MADS box genes from Arabidopsis and three genes from soybean.
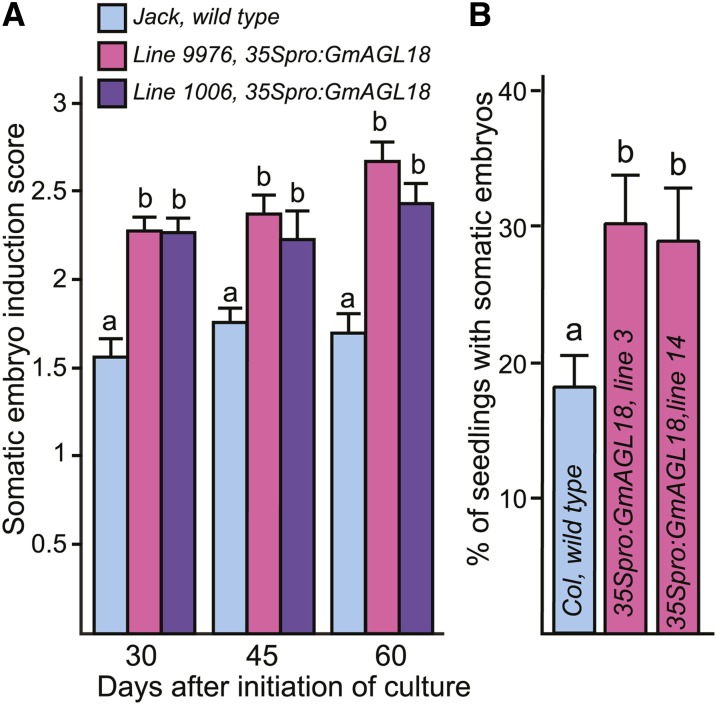
Because AGL18 has redundant functions to AGL15 in Arabidopsis (Adamczyk et al., 2007; Thakare et al., 2008), we investigated whether the overexpression of GmAGL18 from the 35S promoter would enhance SE in soybean. Upon culturing immature cotyledon explants from transgenic lines (35Spro:GmAGL18, line 9976, gDNA; line 1006, cDNA) and cv Jack wild type on D40 medium, somatic embryos developed on explants from the two transgenic lines more extensively than the control (Supplemental Fig. S4). Somatic embryo induction was scored as described above. As shown in Figure 3A, the scores of embryogenesis from transgenic lines were significantly higher than those of controls at three time points on D40 medium.
Figure 3.
Promotion of SE by overexpression of Glyma02g33040 (GmAGL18) in soybean and Arabidopsis. A, Somatic embryo production on immature cotyledon explants plated on D40 medium. Means and se for at least nine plates of explants per genotype per time point are shown. Different letters indicate significant differences at P < 0.05. B, Effect of the overexpression of GmAGL18 on the production of somatic embryos from the shoot apical region of Arabidopsis seedlings when seeds are allowed to complete germination in liquid medium containing 2,4-D. “Wild type” indicates the nontransgenic Arabidopsis control. Means from three biological replicates and se are shown. Different letters indicate significant differences at P < 0.05. [See online article for color version of this figure.]
We transformed Arabidopsis with 35Spro:GmAGL18 to examine the effects of GmAGL18 accumulation on SE in this plant. A shoot apical meristem somatic embryo (SAM SE) system of Mordhorst et al. (1998) was used in which mature seed is allowed to complete germination in liquid medium containing 2,4-D, and seedlings will, at some frequency, form somatic embryos from the shoot apical region (Supplemental Fig. S5). Two different transgenic lines with 35Spro:GmAGL18 were tested, and 28% to 30% of “seedlings” had embryo development at the meristem (Fig. 3B). This was a significant increase compared with the nontransgenic control, which showed 18% SAM SE development.
Genes Important for Embryogenesis Are Direct Targets of GmAGL15
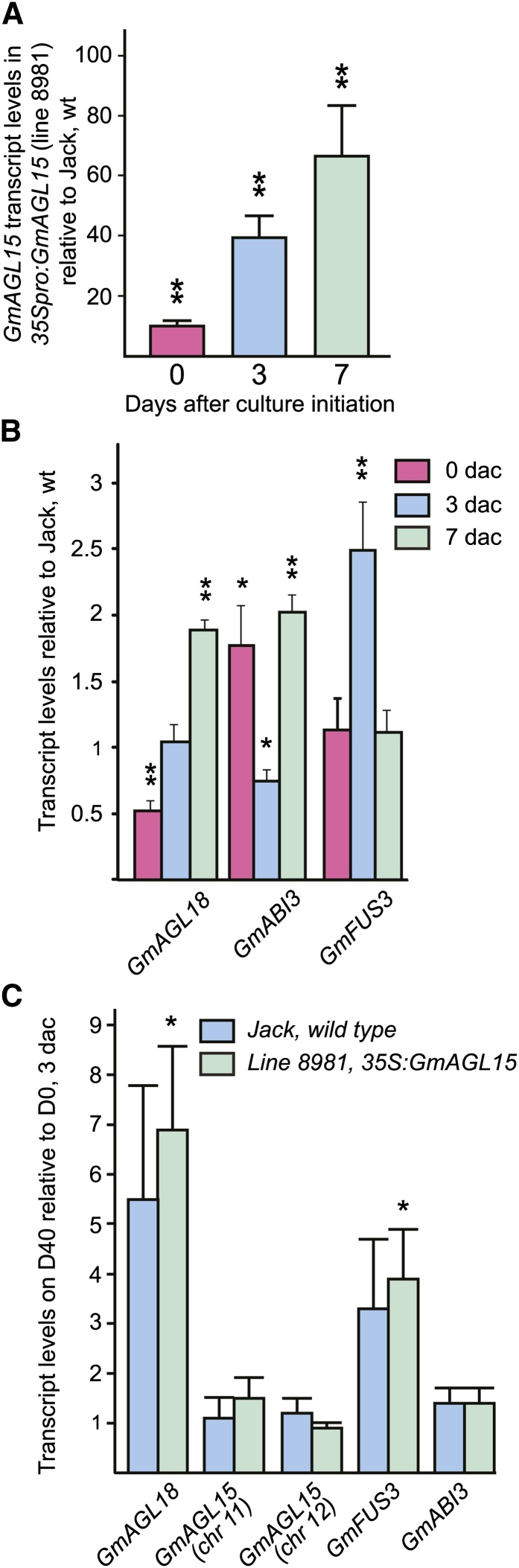
The so-called AFL (for ABI3, FUS3, and LEAFY COTYLEDON2) proteins are B3 domain transcription factors necessary for normal seed development and sufficient, when ectopically expressed, to lead to the ectopic expression of seed-specific genes, and in the case of LEC2, to ectopic embryos (Stone et al., 2001; Tamminen et al., 2001; Gazzarrini et al., 2004). Because GmAGL15 promotes SE in soybean (Fig. 1), we investigated whether increased GmAGL15 accumulation impacts on the expression of AFL orthologs in soybean. Potential soybean orthologs encoding ABI3 and FUS3 were identified by protein BLAST, but no convincing ortholog of LEC2 was found. The Glyma loci identifiers are found in Table I. For ease of understanding, the putative loci are referred to henceforth as Gm followed by the Arabidopsis gene name. To investigate how these genes and GmAGL18 are regulated by GmAGL15, gene expression, as determined by transcript abundance, was assessed using quantitative reverse transcription (qRT)-PCR. As shown in Figure 4A, GmAGL15 was truly overexpressed in cotyledon explants from transgenic line 8981 (35Spro:GmAGL15), and the fold change compared with cv Jack wild type increased with time in culture. At 0 and 7 dac, transcript accumulation from GmABI3 was significantly increased in 35Spro:GmAGL15 compared with the wild type; however, GmAGL18 showed decreased transcript abundance in 35Spro:GmAGL15 at 0 d and increased transcript abundance at 7 d (Fig. 4B). The transcripts of FUS3 significantly increased at 3 dac on D40 medium in transgenic tissues compared with the control; however, ABI3 showed decreased transcript accumulation compared with the wild type at this stage.
Table I. Putative orthologs in Arabidopsis and soybean.
Predicted proteins encoded by select Arabidopsis genes were used to search the proteome using Phytozome version 8.0 and the soybean proteome database version 1.0. The predicted proteins from the top matches were then used to search the Arabidopsis proteome and confirm the strongest match within this organism. For the GST and EP3 genes, the soybean gene was identified as being of interest first and used to search the Arabidopsis proteome. Asterisks indicate selections used for further analysis.
| Arabidopsis Genome Initiative Identifier | Arabidopsis Gene Name (No. of Amino Acids) | Soybean Locus Identifier (No. of Amino Acids) | Identity | Positive | Gap | E Value |
|---|---|---|---|---|---|---|
| % | ||||||
| At3g57390 | AGL18 (256) | Glyma02g33040* (265) | 49 | 62 | 9 | 5e-44 |
| At3g24650 | ABI3 (720) | Glyma08g47240* (717) | 45 | 56 | 11 | e-125 |
| Glyma18g38490 (662) | 44 | 54 | 10 | e-122 | ||
| At3g26790 | FUS3 (313) | Glyma16g05480* (375) | 56 | 69 | 14 | 3e-66 |
| At5g62690 | TUB2 (450) | Glyma03g15020* (449) | 94 | 96 | 0 | 0.0 |
| At3g09270 | GSTU8 (224) | Glyma03g16600* (220) | 56 | 74 | 0 | 2e-68 |
| At3g54420 | AtEP3 (273) | Glyma11g13270* (294) | 77 | 86 | 0 | 2e-92 |
Figure 4.
Transcript accumulation from soybean genes in isolated immature cotyledon explants at 0, 3, and 7 dac on D40 medium. A, GmAGL15 transcript accumulation in 35Spro:GmAGL15 compared with cv Jack wild type. Overexpressors show increased GmAGL15 transcript at 0 dac, and the fold increase compared with the wild type increased over time in culture. B, Transcript accumulation from select soybean orthologs to genes involved in Arabidopsis embryogenesis in 35Spro:GmAGL15 compared with cv Jack wild type. C, Transcript accumulation from cv Jack wild type and from 35Spro:GmAGL15 cultured for 3 d on D40 medium compared with 3 d on D0 medium. Data shown are means and se for three to five biological replicates. Asterisks indicate significant differences at **P < 0.01 and *P < 0.05. [See online article for color version of this figure.]
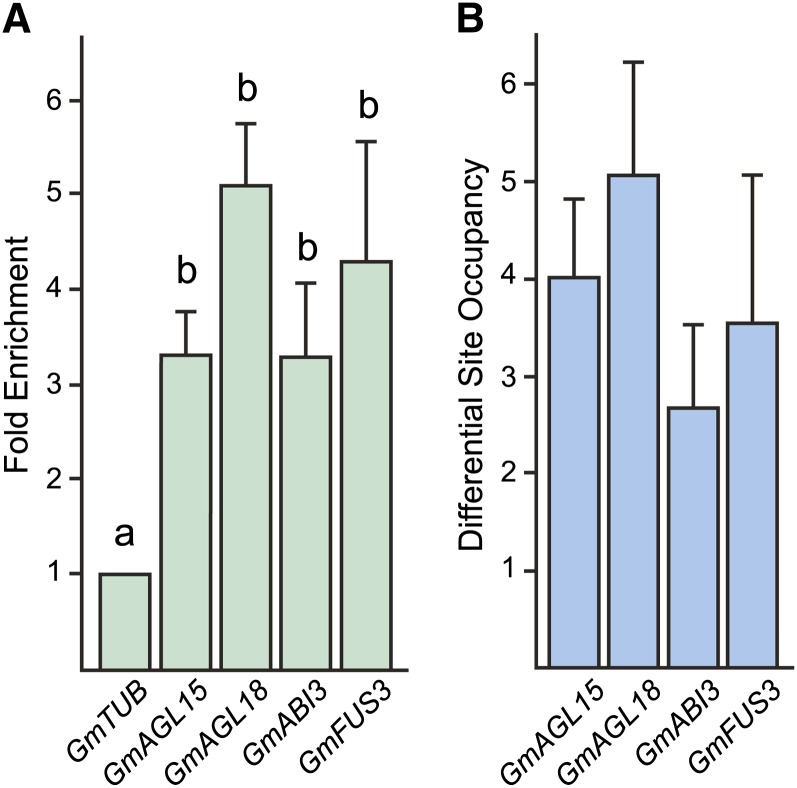
In order to investigate whether these genes may be directly regulated by GmAGL15, we assessed whether GmAGL15 may bind to regulatory regions of these genes in vivo. The regulatory regions were examined for potential binding sites for MADS domain proteins, which are called CArG motifs for C-A/T-rich-G (Norman et al., 1988; Sommer et al., 1990), and oligonucleotides were designed to these regions to perform chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR; Supplemental Table S1). Three independent biological replicates of DNA recovered by ChIP using anti-AGL15-specific antiserum (raised against Brassica napus AGL15; Perry et al., 1996) or preimmune serum as a control were generated, and qPCR was used to quantitate the association of DNA fragments with GmAGL15 in the immune precipitation compared with controls. Fold change measures the amount of amplicon corresponding to the suspected target in the immune precipitation compared with the preimmune control. As shown in Figure 5A, regulatory regions of GmAGL18, GmABI3, and GmFUS3, as well as GmAGL15 itself, were specifically associated with GmAGL15 compared with a nonbound control DNA fragment (GmTUBULIN). Another measure of the specific association is differential site occupancy (DSO), which compares the amount of the target amplicon in the immune precipitation with that of a control amplicon that is not expected to be bound by AGL15 (GmTUB2) in the same immune precipitation. All of the genes were also significantly enriched compared with the nonbound control within the same immune precipitation (Fig. 5B). These results indicate that these genes are directly regulated by GmAGL15 in soybean.
Figure 5.
Verification of the in vivo association of GmAGL15 with select DNA fragments. A, Fold enrichment calculations from qPCR on at least three independent ChIPs. Fold enrichment compares the amplicon of suspected target fragments recovered in immune ChIP and preimmune control ChIP. Results are normalized to the nonbound control fragment from GmTUB2. B, DSO calculations from qPCR on at least three independent ChIPs. Recovery of target fragments by coimmunoprecipitation with anti-AGL15 serum was compared with the recovery of a nonbound control DNA fragment (GmTUB2) in the same immune precipitation. Means and se are shown. Different letters indicate significant differences at P < 0.05. All DSOs for targets are significant. [See online article for color version of this figure.]
Besides regulation by GmAGL15, some of these genes responded to the 2,4-D in the medium used to induce SE. As shown in Figure 4C, while neither GmAGL15 encoding genes on chromosome 11 or 12 showed a significant up-regulation in response to 3 dac on D40 medium compared with 3 dac on D0 medium (the same as D40 but lacking the 2,4-D; D0 is also identical to Murashige and Skoog 0 medium [Liu et al., 1992] but has a pH of 7 and 3% Suc, as does D40, rather than a pH of 5.8 and 2% Suc, as does Murashige and Skoog 0 medium), GmAGL18 and GmFUS3 showed increased transcript after culture in both cv Jack wild type and 35Spro:GmAGL15 tissue, with the increase being significant for the 35S:GmAGL15 tissue. GmABI3 transcript showed no such response to 2,4-D in the medium (Fig. 4C).
Functional Classification of GmAGL15-Responsive Genes
A microarray experiment was conducted to more globally identify genes that may be responsive to GmAGL15 accumulation and play a role in SE in soybean. Cotyledon explants from young embryos (4–5 mm) of cv Jack wild type and transgenic line 8981 (35Spro:GmAGL15, O/E in Supplemental Data Set S1) were collected at 0, 3, and 7 dac on D40 medium and used to isolate mRNA to generate probes for expression arrays. Partek GS was used to determine statistically significant up- and down-regulated genes. Cutoffs used were P < 0.05 and ratios of transcript abundance in line 8981 compared with cv Jack of 1.5 or greater for up-regulation and 0.67 or less for down-regulation. At 0, 3, and 7 dac on D40 medium, some genes were consistently down-regulated in response to GmAGL15 overexpression (Supplemental Table S2) and others were consistently up-regulated compared with the wild type (Supplemental Table S3). The data shown are from biological duplicates at each time point per genotype.
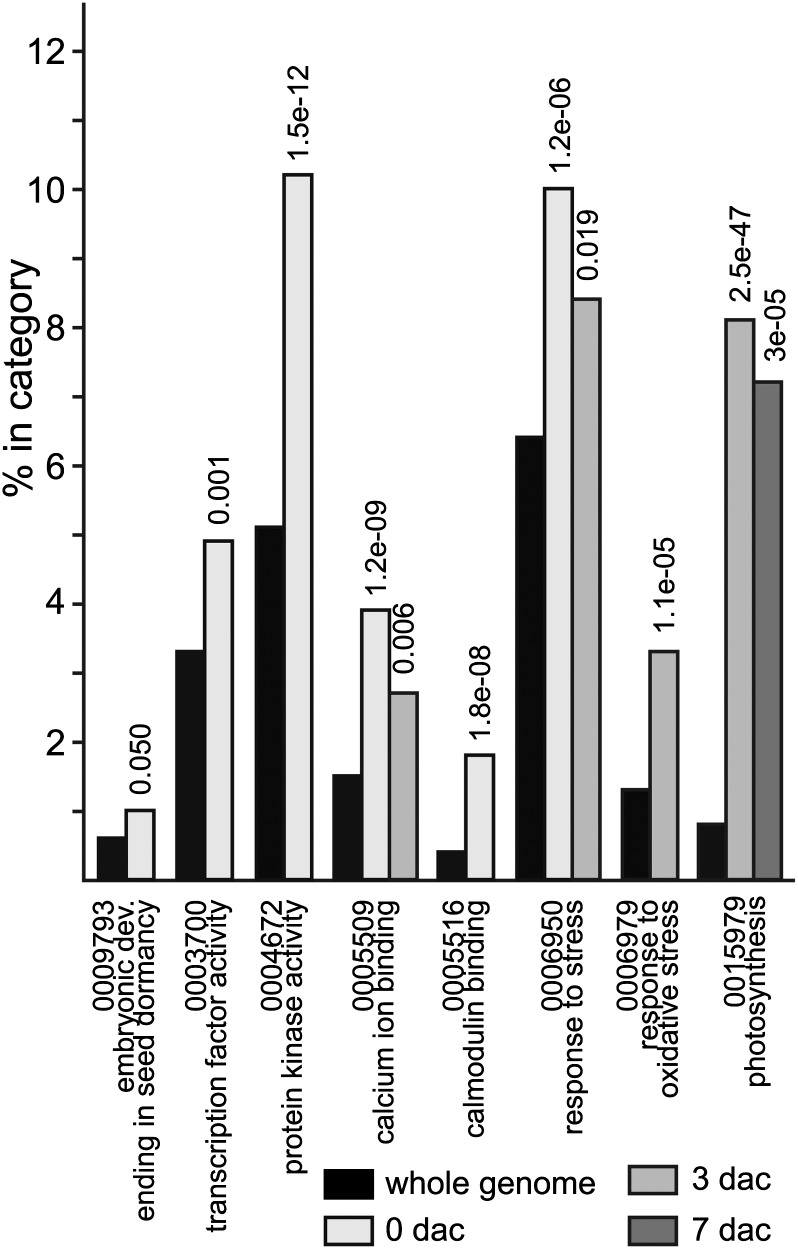
When AgriGO was used to assess categories that were overrepresented with a hypergeometric statistical test and using the Affymetrix soybean genome chip probe identifiers (Du et al., 2010), genes encoding products involved in response to stress were enriched at 0 and 3 dac, with an overrepresentation of genes responsive to salt and osmotic stress at 0 dac and to oxidative stress at 3 dac. Stress-responsive genes were also overrepresented in the 0-dac down-regulated lists, in particular response to oxidative stress, although genes encoding products involved in response to hydrogen peroxide were 0.6% of the 0-dac up-regulated list compared with 0.3% for the whole genome (P = 0.024). Genes encoding transcription factors were overrepresented only in the 0-dac up-regulated list. Genes encoding products involved in photosynthesis were enriched in the 0-dac GmAGL15-repressed list, but at 3 and 7 dac, genes in this category were enriched in the GmAGL15-expressed list. Genes encoding products involved in calcium ion and calmodulin binding were also up-regulated at the early time points. Select enriched Gene Ontology (GO) categories are shown in Figure 6.
Figure 6.
Functional categorization of GmAGL15-responsive genes using AgriGO. Select GO categories are shown for which there is a significant overrepresentation in lists of genes that show increased transcript accumulation in 35Spro:GmAGL15 compared with cv Jack wild type in the immature cotyledon explants either upon excision (0 dac) or after 3 and 7 d on D40 medium compared with the percentage in the category in the whole genome. P values are shown above the bars.
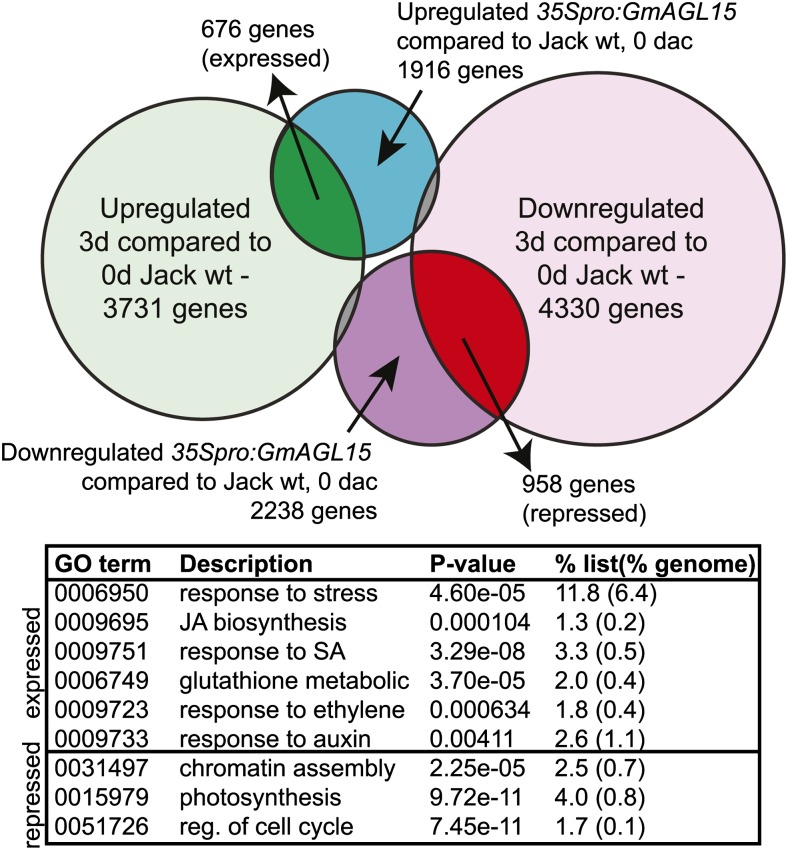
Surprisingly, the number of genes with differential transcript accumulation in 35Spro:GmAGL15 tissue compared with the wild type was largest at 0 dac and progressively smaller at 3 and 7 dac on D40 (Supplemental Data Set S1). This may be in part because 0-dac 35Spro:GmAGL15 tissue resembled, to some extent in terms of gene expression, cv Jack that had been cultured for 3 d. Of the list significantly (P < 0.05) up-regulated by GmAGL15 (1.5 or greater), over one-third (35%) were also significantly up-regulated in cv Jack at 3 dac compared with 0 dac (P < 0.05 and fold change of 1.5 or greater; Fig. 7). When those on both lists were analyzed with AgriGO with a hypergeometric statistical test, a number of GO terms in Biological Processes were overrepresented, some of which are shown in Figure 7. Of the repressed list in response to increased GmAGL15 at 0 dac, 43% were significantly down-regulated by culturing cv Jack on D40 for 3 d (Fig. 7). Only 5% to 6% showed the opposite pattern.
Figure 7.
Overlap between genes responsive to 35Spro:GmAGL15 at 0 dac compared with cv Jack wild type (wt) and genes responsive to induction of SE by culturing cv Jack wild type on D40 medium for 3 d. Considerable overlap exists for genes with increased transcript abundance in response to 35Spro:GmAGL15 and in cv Jack wild type in response to culturing on D40 induction medium. Considerable overlap also exists for genes with reduced transcript abundance in both comparisons. The 676 genes that overlap as “expressed” and 958 genes that overlap as “repressed” were analyzed for overrepresented GO terms using the probe identifiers and AgriGO, and select categories from Biological Processes are shown. [See online article for color version of this figure.]
To perform pathway analysis, PathExpress was used (http://bioinfoserver.rsbs.anu.edu.au/utils/PathExpress/; Goffard and Weiller, 2007; Goffard et al., 2009). Although particular pathways were identified as being statistically overrepresented in the set of genes responsive at each time point when compared with the whole genome, some of the same pathways were present on lists for both up- and down-regulated genes, either at the same or different time points (Supplemental Table S4).
Genes Related to SE Are Precociously Up-Regulated in 35Spro:GmAGL15 Explants
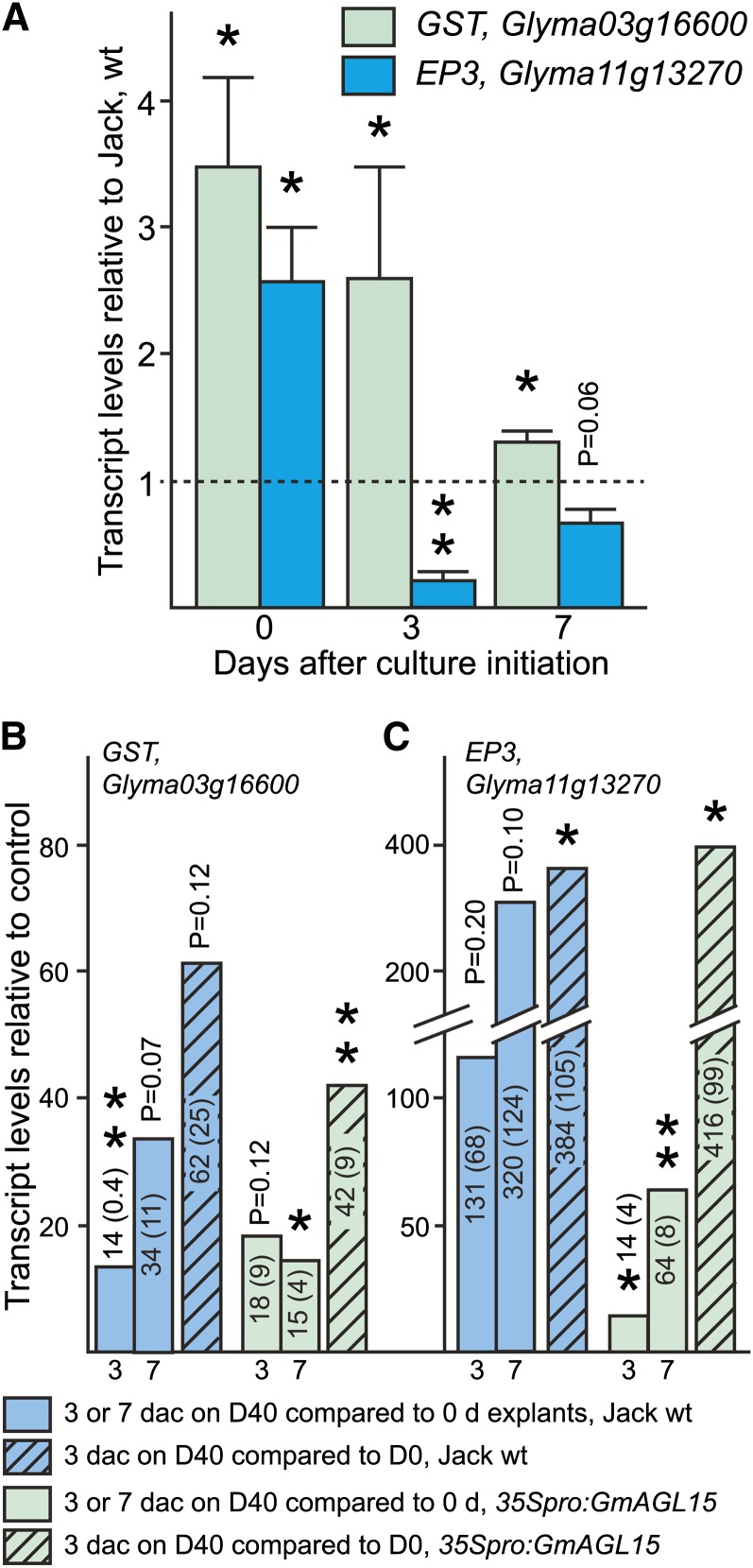
Considerable overlap exists between transcripts more abundant in 35Spro:GmAGL15 tissue than the nontransgenic control at 0 dac, and transcripts increased in response to culturing cv Jack wild type on D40 medium for 3 d (Fig. 7). This group of genes included at least seven GLUTATHIONE S-TRANSFERASE (GST) genes, some of which were increased in 35Spro:GmAGL15 tissue compared with the wild type as much as 17- to 26-fold (Supplemental Data Set S1). Transcript abundance also increased for 35Spro:GmAGL15 tissue when placed on D40 medium, but by 3 dac, there was no longer a significant difference between cv Jack and 35Spro:GmAGL15 tissue for this group of genes. Only one GST gene had decreased transcript abundance at 0 dac in the 35Spro:GmAGL15 tissue compared with the wild type.
Signal from the microarray plotted to show the difference between 35Spro:GmAGL15 and cv Jack and the time course for each is shown in Supplemental Figure S6A for one GST, Glyma03g16600 (signal shown is the average of biological duplicates at each time point per genotype). qRT-PCR confirmed the greatest difference between 35Spro:GmAGL15 and cv Jack at 0 dac for transcript accumulation from Glyma03g16600 (Fig. 8A). Figure 8B shows the changes between time points, comparing 3 or 7 dac on D40 medium with explants (0 dac; cv Jack or 35Spro:GmAGL15). While transcript in cv Jack wild type increased at 3 dac and further increased at 7 dac compared with explants, the GmAGL15 overexpressor tissue did not show a further increase at 7 dac, in agreement with the microarray results. All of the best matches to Arabidopsis genes for soybean genes with increased transcript in the 35S tissue compared with the wild type at 0 dac correspond to Arabidopsis genes in the τ family of GSTs that include auxin- and stress-responsive genes (for review, see Dixon and Edwards, 2010). To differentiate between changes in transcript due to time in culture and/or response to 2,4-D, we assessed transcript accumulation from Glyma03g16600 from tissue cultured 0 or 3 dac on D40 medium (which contains 2,4-D) or D0 medium. As shown in Figure 8B, a large increase in transcript was observed not only comparing 3 and 7 dac on D40 medium with explants but also comparing tissue cultured for 3 dac on D40 medium with tissue cultured for 3 dac on D0 medium (Fig. 8B, hatched bars). Tissue cultured for 3 dac on D0 compared with 0-dac explants showed a decrease in transcript (0.4; not significant). Thus, the presence of 2,4-D was necessary for increased transcript accumulation. In some cases, the fold change in transcript was not significant when data from the three to four biological replicates were assessed together, but this was due to large variations in the increases in transcript observed in the individual experiments. For example, for cv Jack wild type, 3 dac on D40 compared with 3 dac on D0 showed P = 0.12 for the data from all replicates, but the range in the individual experiments was P = 18.8 to 120.1, and each individual experiment showed significant differences from the control when the technical replicates within the experiment were assessed.
Figure 8.
Assessment of the response of the GST and EP3 genes to GmAGL15 and 2,4-D. A, Transcript accumulation from select genes associated with somatic embryogenesis in 35Spro:GmAGL15 compared with cv Jack wild type during culture. B and C, Transcript accumulation in response to time in culture on D40 medium comparing 3 and 7 dac with the 0-dac explants for cv Jack and 35Spro:GmAGL15 for the GST (B) and EP3 (C) genes. The hatched bars compare transcript from tissue cultured 3 dac on D40 medium with tissue cultured 3 dac on D0 medium (same as D40 but without 2,4-D). Means and (se) from three to four biological replicates are shown. Asterisks indicate significant differences at *P < 0.05 and **P < 0.01. [See online article for color version of this figure.]
Another gene that is relevant to SE for which intriguing patterns were observed in our microarray study corresponds to a chitinase gene that encodes a potential ortholog of carrot (Daucus carota) EXTRACELLULAR PROTEIN3 (EP3). In carrot, the addition of EP3 can rescue the production of somatic embryos in ts11, a temperature-sensitive line (van Hengel et al., 1998). The soybean gene Glyma11g13270 encodes a predicted protein that is 62.5% identical and 73.5% similar to the carrot protein and is represented by two overlapping probe sets on the array. The signal for these probe sets is shown in Supplemental Figure S6B. The fold change in 35Spro:GmAGL15 tissue compared with the wild type at 0 dac was approximately 16 (significant at P < 0.05; Supplemental Data Set S1). This gene also responded to culture on the D40 medium, and by 3 dac, the difference between genotypes was not significant in the microarray data. qRT-PCR on samples independent from those used for the array confirmed these results (Fig. 8A). Glyma11g13270 transcript was initially increased in 35Spro:GmAGL15 compared with cv Jack wild type (0 dac), but the fold increase in response to culture on D40 medium was greater in cv Jack than in 35Spro:GmAGL15, such that cv Jack “overtook” the overexpressors by 3 dac, even though in both cv Jack and 35Spro:GmAGL15 tissue transcript increased over the time course (Fig. 8, A and C; Supplemental Fig. S6B). As with the GST, Glyma11g13270 responded to 2,4-D in the medium (Fig. 8C); the fold change for 3 dac on D0 compared with explants was 0.03.
DISCUSSION
Soybean Orthologs of AGL15 and AGL18 Promote SE
Prior work documented an increase in transformants when soybean was biolistically transformed with 35Spro:GmAGL15 compared with controls (Thakare et al., 2008), but this enhancement could be through a variety of steps in the transformation process. One possible mechanism would be enhanced regeneration of the transformed cells by SE, because transformation potential has been reported to directly correlate with somatic embryogenic potential among 15 varieties of soybean (Ko et al., 2004; Kita et al., 2007; Klink et al., 2008). We report here that GmAGL15 significantly increases SE when expressed via the 35S promoter (Fig. 1; Supplemental Figs. S1 and S2). Therefore, the likely mechanism of the recovery of increased transformants reported by Thakare et al., (2008) is indeed due to the promotion of regeneration via SE.
In Arabidopsis, AGL15 could also, when ectopically expressed, promote SE in at least two systems (Harding et al., 2003; Thakare et al., 2008). In one system, immature embryos were cultured on medium lacking any exogenous hormones. Both wild-type and 35Spro:AGL15 embryos initiated secondary embryo production, but for the Wassilewskija (Ws) ecotype, a higher percentage of the 35Spro:AGL15 embryos produced secondary embryo tissue. This enhancement was not noted in the Columbia (Col) ecotype, but Col wild type was already more efficient at producing secondary embryos than was Ws. Two different alleles of loss of function of AGL15 showed reduction of somatic embryo production in Col, as did a loss-of-function allele in Ws. Because some SE systems in Arabidopsis induce SE by the placement of immature embryos on 2,4-D (Ikeda-Iwai et al., 2002) and because AGL15 is up-regulated by auxin (both synthetic 2,4-D and natural; Zhu and Perry, 2005), it is intriguing to consider that enhanced SE by 35Spro:AGL15 on hormone-free medium may be due to the transgene expression being independent of auxin. Soybean explants from the wild type or 35Spro:GmAGL15 show no SE development on D0 medium, but GmAGL15 does not appear to be expressed in response to 2,4-D (Fig. 4C). GmAGL18 does show increased transcript on medium containing 2,4-D (Fig. 4C).
Also in both Arabidopsis ecotypes, the presence of the 35Spro:AGL15 transgene promoted continued development as somatic embryo tissue longer than nontransgenic controls. In soybean, the frequency of initiation of somatic embryos does not necessarily correlate with continued proliferation (Meurer et al., 2001, and refs. therein). Thus, another important experiment is to test for the effect of the overexpression transgene on the continued proliferation of SE. Indeed, 35Spro:GmAGL15 increased the robustness (measured by green tissue that, in our hands, is what continues to proliferate in culture) of development when embryos induced on D40 were transferred to D20 medium (Fig. 1, D–F). Although tissue died without subculture, if proliferating tissue was routinely subcultured, 35Spro:GmAGL15 tissue continued producing abundant embryo tissue up to 1.5 years, when the cultures were terminated (Supplemental Fig. S2).
A gene encoding a protein related to AGL15 was isolated from soybean and found to be most similar to AGL18 (Fig. 2). In Arabidopsis, AGL15 and AGL18 have redundant roles in the promotion of the SAM SE system (Thakare et al., 2008) and in the control of flowering time (Adamczyk et al., 2007). In the SAM SE system, mature dry seeds were allowed to complete germination in medium containing 2,4-D (Mordhorst et al., 1998). A percentage of the callused seedlings had SE development from the shoot apical region, and the frequency with which this occurs was directly correlated with the accumulation of AGL15 in both Ws and Col ecotypes (Thakare et al., 2008). When ectopically expressed via the 35S promoter, AGL18 has many of the same phenotypes as 35S:AGL15, including enhancement of SE (Adamczyk et al., 2007). Similar to 35Spro:GmAGL15, 35Spro:GmAGL18 transgenic plants produced increased numbers of somatic embryos (Fig. 3A). While Thakare et al. (2008) did not find an increase in the number of transgenics for soybean expressing the Arabidopsis AGL15 and did not see an increase in SAM SE, or other overexpression phenotypes, for Arabidopsis carrying the 35Spro:GmAGL15 transgene, the 35Spro:GmAGL18 transgene produced weak overexpression phenotypes in Arabidopsis, including an increase in SAM SE (Fig. 3B). Interestingly, an LxLxL motif in the C-terminal domain of AGL15 was identified as important for interaction with components of histone deacetylase complexes to control gene expression (Hill et al., 2008). Other MADS proteins, including AGL18, have this motif. While GmAGL18 shows conservation in this region (Fig. 2A), the first Leu in GmAGL15 is changed to a Phe (Thakare et al., 2008), and possibly changes in protein interactions in this or other regions lead to changes in effect in heterologous systems.
Functional Classification of GmAGL15-Responsive Genes
The observation that 35Spro:GmAGL15 significantly increases SE compared with the control provided a unique opportunity to compare explant tissue before and during the early stages of induction of SE to investigate changes in the transcriptome that may increase competence for SE. Thus, to better understand how GmAGL15 promotes SE, a microarray experiment was conducted to identify genes responsive to GmAGL15 accumulation at 0, 3, and 7 dac compared with cv Jack wild type. Surprisingly, the number of genes with differential transcript accumulation in 35Spro:GmAGL15 tissue compared with the wild type was largest at 0 dac and progressively smaller at 3 and 7 d on D40 medium (Supplemental Data Set S1). AgriGO was used to assess categories that were overrepresented at different time points, and notably, genes associated with stress, in particular osmotic stress (0 dac) and oxidative stress (3 dac), were overrepresented in the GmAGL15 up-regulated lists. Stress is an important inducer of SE and can take many forms, including osmotic, salt, oxidative, and temperate stresses. Wounding, necessary to prepare the explants, causes stress, and it has been proposed that SE may be an extreme stress response of cells to culture conditions (Karami and Saidi, 2010). Wounding accelerates the appearance of SE in soybean (Santarem et al., 1997). Thibaud-Nissen et al. (2003) performed a time course separating the adaxial tissue, from which embryos develop, from the abaxial tissue of soybean immature cotyledon explants induced for SE and found an up-regulation of genes involved in oxidative stress preceding the appearance of embryos on the adaxial side. The synthetic auxin 2,4-D present in the culture medium is often considered to be a stressing agent, due to the much higher than endogenous levels used, and the generation of reactive oxygen species has been associated with exposure to 2,4-D (Karami and Saidi, 2010). Ethylene is involved in the stress response, and we recently reported that in soybean, GmAGL15 regulates genes involved in ethylene biosynthesis and response, with effects on SE (Zheng et al., 2013).
The pathway analysis using PathExpress also indicated changes in the stress response (Supplemental Table S4). For example, one pathway for which a significant number of genes was assigned (with P value correction) in the 3-dac list up-regulated in response to 35Spro:GmAGL15 was “phenylpropanoid biosynthesis.” Genes encoding enzymes in the phenylpropanoid pathway are up-regulated during barley (Hordeum vulgare) microspore embryogenesis upon stress treatment of the anthers (Jacquard et al., 2009), and alterations in this pathway have been correlated with the developmental potential of sessile oak (Quercus spp.) somatic embryos (Cvikrova et al., 1998, 2003). “Flavonoid biosynthesis” was one of the pathways overrepresented in superembryogenic Medicago truncatula (line 2HA) explants compared with its comparatively nonembryogenic progenitor Jemalong. “Ascorbate and aldarate metabolism” was one of the two pathways significantly overrepresented in Jemalong (Imin et al., 2008).
Interestingly, a large percentage of genes with increased (or decreased) transcript accumulation in response to 35Spro:GmAGL15 (compared with cv Jack wild type) at 0 d also show up-regulation (or down-regulation) in cv Jack wild type in response to culturing on D40 medium for 3 d. The genes that overlap in the expressed data set are overrepresented for those encoding products involved in the stress response, and this data set includes many GST genes. Over one-half-dozen GST genes, different from those showing altered transcript accumulation in response to overexpression of GmAGL15 at 0 dac, show increased transcript in the 35Spro:GmAGL15 tissue compared with the wild type at 3 dac. The closest Arabidopsis ortholog of some of these were, like all of the 0-dac up-regulated genes, in the τ GST family, but others were in other groups (Dixon and Edwards, 2010). In some cases, the genes are not expressed or are expressed at very low levels in the 0-dac tissue, and placement on D40 medium leads to increased transcript to a greater extent in 35Spro:GmAGL15 than in cv Jack (Glyma08g18660, Glyma03g33340, and Glyma20g24320). In other cases, there was expression in the 0-dac tissue of both cv Jack and 35Spro:GmAGL15, but the decrease in transcript accumulation was more rapid in cv Jack than in 35Spro:GmAGL15, such that at 3 dac, more transcript remained in the overexpressor tissue (Glyma01g04710 and Glyma10g02460). Studies have shown that transcripts from GST genes, which are involved in stress responses (for review, see Oztetik 2008), accumulate in response to auxin and in somatic embryos (Karami and Saidi, 2010). Products of GST genes may be involved in cells achieving embryogenic potential, possibly in part by buffering the effects of the high levels of auxin (Karami et al., 2009; Karami and Saidi, 2010). The expression of glutathione biosynthetic genes has been linked to somatic embryogenesis in wheat (Triticum aestivum; Bossio et al., 2013).
Other potential “markers” for cells with or acquiring embryogenic competence include heat-shock proteins, increased calcium, and chitinases (Fehér et al., 2003; Karami and Saidi, 2010). There are genes encoding HSPs that were up- or down-regulated at 0 and 3 dac, and genes encoding proteins involved in calcium binding were overrepresented at these time points (Fig. 6). Intriguingly a gene encoding a potential ortholog to carrot EP3 class IV endochitinase was up-regulated in the 35Spro:GmAGL15 tissue compared with the wild type at 0 dac (Supplemental Data Set S1). In carrot, the addition of EP3 can rescue the production of somatic embryos in the temperature-sensitive line ts11 (De Jong et al., 1992; Kragh et al., 1996; van Hengel et al., 1998). Both the carrot and an Arabidopsis ortholog (At3g54420) are not expressed in the somatic or zygotic embryos but rather in cells surrounding the embryos; therefore, they have been proposed to have a “nursing” function to promote embryogenesis (van Hengel et al., 1998; Passarinho et al., 2001). A potential ortholog of EP3 in orchard grass (Dactylis glomerata) is secreted in embryogenic but not nonembryogenic suspension cultures and has been suggested as a marker for embryogenic potential (Tchorbadjieva and Pantchev, 2006). The increased transcript for Glyma11g13270 in 35Spro:GmAGL15 compared with cv Jack wild type is lost at 3 and 7 dac due to the larger increase in transcript in cv Jack upon culturing on D40 medium compared with that observed in 35Spro:GmAGL15 (Fig. 8; Supplemental Fig. S6B; Supplemental Data Set S1). Because EP3 is present in supporting cells, but not in the actual embryos in carrot and Arabidopsis, the reduced level of increase of transcript in 35Spro:GmAGL15 compared with the wild type, such that by 3 dac the overexpressor tissue has less transcript than the wild type (although both increase over the time course; Supplemental Fig. S6B), is not inconsistent with 35Spro:GmAGL15 having increased SE. Too much as well as too little may inhibit the development of SE. EP3 is thought to promote SE by cleavage of arabinogalactan proteins (AGPs), thereby releasing GlcNAc signaling molecules (van Hengel et al., 2001; Passarinho and de Vries, 2002). Interestingly, some AGP genes showed increased transcript in 35Spro:GmAGL15 compared with the wild type at 0 dac, but others had decreased transcript. It has been reported that different AGPs have stimulatory or inhibitory effects on SE (Toonen et al., 1997).
Not only do the GSTs and the EP3-like protein show (1) increased transcript in 0-dac explants of 35Spro:GmAGL15 compared with the wild type and (2) increased transcript at 3 dac compared with 0 dac in the wild type, so do a large number of other genes (Fig. 7). Possibly, the AGL15-overexpressing tissue is more competent to respond to the 2,4-D induction due to “priming” of the tissue by changes in gene expression, such as stress-responsive genes, and systems to ameliorate excess auxin, resulting in more rapid, prolific, and robust SE.
Comparison of Control of Gene Regulation by (Gm)AGL15 in Soybean and Arabidopsis
Prior work examined the transcriptome in response to increased AGL15 in Arabidopsis using 10-d SAM SE tissue of Col wild type, 35Spro:AGL15 that by 21 d shows increased frequency of SAM SE, and agl15 agl18 that has a reduction in SAM SE (Zheng et al., 2009). This information was combined with ChIP-chip data to map in vivo binding sites for AGL15 to identify directly regulated genes from those indirectly regulated (Zheng et al., 2009). Comparing data obtained in Arabidopsis and soybean is a method to find shared components of the induction of SE as well as differences. As in Arabidopsis (Zheng et al., 2009), potential orthologs encoding the key embryo transcription factors ABI3 and FUS3 are directly regulated by GmAGL15 (Figs. 4B and 5). However, in soybean, no convincing ortholog of LEC2 was identified, and it will be interesting to see how interactions between genes are different in soybean, as LEC2 is central to embryo programs in Arabidopsis (Stone et al., 2001; To et al., 2006). Ethylene biosynthetic genes are expressed in response to 35Spro:GmAGL15 in soybean but not in Arabidopsis, and while components of signaling and response are regulated in a similar manner in the two species, others show the opposite pattern (Zheng et al., 2013). Comparing the expression data as a whole reveals roughly equal numbers of genes that behave the same in response to AGL15 in both species as genes that behave oppositely (e.g. up-regulated in one species but repressed in the other). If genes that are up-regulated at least at one time point in soybean for which an Arabidopsis ortholog has been identified that is also up-regulated by AGL15 (Supplemental Table S5) are analyzed using the Arabidopsis Genome Initiative identifier in DAVID (Huang et al., 2009), genes involved in transcription and in response to organic substances, endogenous stimuli, and abiotic stresses (false discovery rate < 0.05) are overrepresented. For the data set of genes repressed in both species (Supplemental Table S6), genes associated with cell membranes and cell walls including glycoproteins and lipoproteins are overrepresented. For genes expressed in soybean in response to GmAGL15 but repressed in Arabidopsis, genes associated with photosynthesis are enriched. However, for genes repressed in soybean but expressed in Arabidopsis, genes encoding products involved in the cytoskeleton are significantly overrepresented. This last observation could reflect an earlier time point in the overall scheme of SE sampled for soybean than for Arabidopsis and the origin of the SE in each species. In soybean, samples were assessed at 0, 3, and 7 dac, and it takes at least 21 to 30 dac for embryos. In Arabidopsis, samples were assessed at 10 d, and SAM SE are well developed by 21 d.
CONCLUSION
GmAGL15 and the closely related GmAGL18 are soybean MADS domain transcriptional regulators that significantly increase SE. Understanding the mechanism for this enhancement is relevant because genetic engineering for the improvement of agriculture requires regeneration of the transformed cell to a transgenic plant. SE is one mechanism of regeneration. Many plants or desirable cultivars of plants are recalcitrant to this process, making the generation of transgenics difficult or impossible. By understanding how GmAGL15 enhances SE, we will be able to promote this process in recalcitrant plants. Genes encoding key embryo transcription factors are directly expressed in response to GmAGL15. Transcriptome results indicate that explants from 35Spro:GmAGL15 share many genes that show changes in gene expression by the induction of SE of cv Jack for 3 d.
MATERIALS AND METHODS
Plant Material
Soybean (Glycine max) wild-type, 35Spro:GmAGL15, and 35Spro:GmAGL18 plants (all cv Jack) were grown in a greenhouse under a 15-h/9-h photoperiod in a sterilized mixture of 2:2:1 soil:Promix:sand and fed weekly. Transformation of soybean was carried out as described by Thakare et al. (2008), except that proliferation of the SE tissue on D20 was for 1 to 2 months prior to transformation. Recovered plants were tested by PCR to confirm their transgenic nature. For the lines used for subsequent culture and transcriptome studies, segregation of the transgene (as tested by PCR) indicated a probable single insertion per line.
Arabidopsis (Arabidopsis thaliana) wild-type and 35Spro:GmAGL18 plants (all Col ecotype) were grown as described previously (Thakare et al., 2008). Seeds for SAM SE were allowed to develop to dry seed, and culture for SAM SE was performed as described by Mordhorst et al. (1998).
Plasmid Constructs
For the generation of cDNA and gDNA constructs of GmAGL18 from chromosome 2, two fragments of GmAGL18 (cDNA and gDNA) were amplified by PCR with the same primers, 5′-CACCATGCTGGTGGTGGGTTCAGT-3′ (forward) and 5′-ATGTGAAGCCACTTGACTCCCTGA-3′ (reverse), from cDNA and gDNA, respectively. The underlined bases (CACC) were added to the forward primer for Gateway entry clone reactions. The PCR products of GmAGL18 were cloned into pENTR/D-TOPO vector (Invitrogen). After confirming the sequence, the LR recombination reaction with the Gateway destination vector GWB20 (provided by Dr. Tsuyoshi Nakagawa; Nakagawa et al., 2007) was performed according to the manufacturer’s protocol (Invitrogen). The vectors 35S:cGmAGL18-10xcmyc:NOS and 35S:gGmAGL18-10xcmyc:NOS were used for biolistic transformation for soybean. For transformation of Arabidopsis, Agrobacterium tumefaciens strain GV3101 and the floral dip method were used (Clough and Bent 1998). The details of the cDNA GmAGL15 construct from chromosome 11 were reported by Thakare et al. (2008).
Immature Embryo Culture and Scoring of Proliferating Embryos
Immature pods with 4- to 5-mm embryos were harvested from cv Jack wild-type and transgenic plants (35Spro:GmAGL15, line 8981 [cDNA], 35Spro:GmAGL18, line 9976 [gDNA] and line 1006 [cDNA]), rinsed briefly with water with a few drops of Liquinox, sterilized by immersing in 70% (v/v) isopropanol (30 s) and 25% (v/v) bleach (11 min), and washed twice for 5 min each with sterile distilled water. Immature zygotic embryos were aseptically excised, and the embryonic axis was removed. Approximately 21 to 25 individual cotyledons were cultured abaxial side down on D40 induction medium under diffused light at room temperature (Murashige and Skoog salts [Murashige and Skoog, 1962] containing B5 vitamins, 3% [w/v] Suc, 40 mg L−1 2,4-D, and 0.2% [w/v] phytagel, pH 7.0). Each cotyledon was visually scored using the index described by Meurer et al. (2001). The score for each plate was the average for all explants on the plate. Means and se were calculated from the entire set of plates. Controls for a particular experiment were cultured at the same time as the experiment.
To score proliferation after SE induction, proliferating tissue (identified by green color and globular morphology) from D40 medium were transferred to D20 medium (Murashige and Skoog salts containing B5 vitamins, 3% [w/v] Suc, 20 mg L−1 2,4-D, and 0.2% [w/v] phytagel, pH 5.8). Proliferation was scored at 30, 45, and 60 dac after subculture. The scoring system was as follows: 0, no embryo proliferation; 1, transferred material proliferated but less than 25% remained as green embryo tissue; 2, transferred material had 25% to 50% embryo tissue after proliferation; 3, more than 50% of transferred material was embryo tissue after proliferation; 4, the entire proliferating transferred tissue was covered with embryos. Representative images are shown in Supplemental Figure S7. Values were averaged per plate, and an overall average was determined.
Enrichment Test for DNA Bound by AGL15
Young embryos (4–8 mm, approximately 5 g) were harvested from 35Spro:GmAGL15 plants, sliced into small pieces, fixed with formaldehyde as described by Zheng et al. (2009), and flash frozen in liquid nitrogen. ChIP experiments were performed as described by Zheng and Perry (2011) with some modifications. Crude nuclei were extracted following Bowler et al. (2004), and the pellet was resuspended in a minimal amount of sonication buffer with reduced sarkosyl from that described by Zheng and Perry (2011; 10 mm potassium phosphate, pH 7, 0.1 mm NaCl, 0.25% [w/v] sarkosyl, 10 mm EDTA, and phenylmethylsulfonyl fluoride added fresh to 1 mm) and sonicated. After centrifugation, the solubilized chromatin was equally divided to two tubes, 3 volumes of modified 1× immunoprecipitation buffer lacking SDS was added (50 mm Tris, pH 7.5, 150 mm NaCl, 1.2 mm EDTA, and 1% [v/v] Triton X-100), and 2 µL of immune serum (BnAGL15) or preimmune serum was added to one each tube. The solubilized chromatin was gently mixed on a rotating wheel overnight at 4°C followed by 5 min at top speed in a microcentrifuge. The supernatant was moved to a new tube, and protein A-Sepharose 4B beads (Invitrogen) were added and incubated at 4°C for 3 h, with gentle mixing. Washing was as described by Zheng and Perry (2011) but using the immunoprecipitation buffer described above. Elution was repeated twice, the combined eluents were centrifuged for 2 min, and the top 375 μL was using for DNA analysis.
qPCR enrichment tests to assess the level of in vivo association of AGL15 with DNA fragments was performed and analyzed as described (Zheng et al., 2009). The specific oligonucleotides used for qPCR are listed in Supplemental Table S1.
Expression Arrays and qRT-PCR
Wild-type cv Jack and transgenic line 8981 were used for expression arrays. For cv Jack, cotyledon explants from 4- to 5-mm embryos were collected at 0, 3, and 7 dac on D40 medium, frozen in liquid nitrogen, and stored at −80°C until RNA extraction. For the transgenic line 8981, the two cotyledons were cultured and tracked to allow PCR on one cotyledon per embryo to confirm whether the embryo was transgenic or not, because the transgenic line 8981 was not, at the time of this experiment, a homozygote. Only explants confirmed as transgenic were collected for RNA extraction.
Total RNA was isolated from about 50 mg of embryo tissues according to the manufacturer’s protocol using the RNeasy plant mini kit (Qiagen), but the extraction buffer contained 1.06% (w/v) polyethylene glycol (Sigma). Two biological replicates of each genotype at each time point (0, 3, and 7 dac) were prepared and sent to the University of Kentucky Microarray Core Facility for probe generation and hybridization to the Affymetrix Soybean Genome Array. These time points allow us to test for changes in transcript accumulation in response to 35Spro:GmAGL15 in explants prior to culture (0 dac) and shortly after culture began but prior to any obvious embryo development (3 and 7 dac). Partek GS was used for analysis as described by Zheng et al. (2009).
For real-time PCR, 1 µg of total RNA was treated with DNase I (Invitrogen) and used for first-strand cDNA synthesis using the avian myeloblastosis virus reverse transcriptase system (Promega). Depending on the experiment, qRT-PCR was performed using the SsoAdvanced SYBR Green Supermix kit (Bio-Rad) or as described by Zheng et al. (2009). Amplification was performed in an iCycler (Bio-Rad) with the PCR system described by Zheng et al. (2009) or in a CFX Connect Real-time System as follows: 30 s at 95°C followed by 40 cycles of 5 s at 95°C and 30 s at 55°C. Immediately after amplification, a melting-curve protocol was performed by increasing each cycle by 0.5°C starting from 65°C and ending at 95°C. Oligonucleotides are shown in Supplemental Table S1. Data analysis was performed using REST software (Pfaffl et al., 2002).
Sequence data from this article can be found in The Arabidopsis Information Resource and Phytozome under the accession numbers listed in Table I as well as in the Gene Expression Omnibus (Barrett and Edgar, 2006; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE54523.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Overexpression of GmAGL15 enhances the production of somatic embryos on immature cotyledon explants of soybean.
Supplemental Figure S2. Overexpression of GmAGL15 allows prolonged somatic embryo development from immature cotyledon explants of soybean.
Supplemental Figure S3. Transcript accumulation profile of Glyma02g33040 from the Soybean eFP Browser at bar.utoronto.ca using the data set of Severin et al. (2010).
Supplemental Figure S4. Example images of plates of cultures on D40 medium for 60 d.
Supplemental Figure S5. An example of a seedling with somatic embryo development from the shoot apical region.
Supplemental Figure S6. Average signal from the microarrays for GST (Glyma03g16600) and a potential ortholog of EP3 (Glyma11g13270) plotted to show differences between cv Jack wild type and 35Spro:GmAGL15 over the time course.
Supplemental Figure S7. Representative images to describe the scoring system for embryo proliferation after somatic embryos from explants on D40 were moved to D20 medium.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Genes for which transcript is decreased in 35Spro:GmAGL15 compared with cv Jack wild type at 0, 3, and 7 d after culture of immature cotyledon explants on D40 medium.
Supplemental Table S3. Genes for which transcript is increased in 35Spro:GmAGL15 compared with cv Jack wild type at 0, 3, and 7 d after culture of immature cotyledon explants on D40 medium.
Supplemental Table S4. Pathway analysis for genes responsive to GmAGL15 as identified using PathExpress (http://bioinfoserver.rsbs.anu.edu.au/utils/PathExpress/).
Supplemental Table S5. Genes for which transcript is increased at least at one stage in soybean for 35Spro:GmAGL15 compared with cv Jack and for which a putative Arabidopsis ortholog is also increased in response to 35S:AGL15.
Supplemental Table S6. Genes for which transcript is decreased at least at one stage in soybean for 35Spro:GmAGL15 compared with cv Jack and for which a putative Arabidopsis ortholog is also decreased in response to 35S:AGL15.
Supplemental Data Set S1. Soybean expression array results.
Acknowledgments
We thank Jeanne Hartman, Olivia Jones, and Rachel Mueller (University of Kentucky) for assistance with cultures; Donna Wall and the University of Kentucky Microarray Core Facility for probe generation, array hybridization, and data collection; Dr. Tsuyoshi Nakagawa (Shimane University) for the gift of the GWB vectors; and Dr. Guojun Wang (University of Kentucky) and our anonymous reviewers for valuable comments on the manuscript.
Glossary
- SE
somatic embryogenesis
- cDNA
complementary DNA
- gDNA
genomic DNA
- 2,4-D
2,4-dichlorophenoxyacetic acid
- dac
days after placement in culture
- SAM SE
shoot apical meristem somatic embryo
- qRT
quantitative reverse transcription
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
- DSO
differential site occupancy
- GO
Gene Ontology
- Ws
Wassilewskija
- Col
Columbia
- AGP
arabinogalactan protein
Footnotes
This work was supported by the United Soybean Board (grant no. 0282/1282/2822), by the National Science Foundation (grant no. IOS–0922845), and by the University of Kentucky Microarray Pilot Grant Program. This is article 12–06–113 of the Kentucky Agricultural Experiment Station.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Barrett T, Edgar R. (2006) Gene Expression Omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol 411: 352–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossio E, Paleo AD, del Vas M, Baroli I, Acevedo A, Rios RD. (2013) Silencing of the glutathione biosynthetic pathway inhibits somatic embryogenesis in wheat. Plant Cell Tissue Organ Cult 112: 239–248 [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cvikrova M, Mala J, Eder J, Hrubcova M, Vagner M. (1998) Abscisic acid, polyamines and phenolic acids in sessile oak somatic embryos in relation to their conversion potential. Plant Physiol Biochem 36: 247–255 [Google Scholar]
- Cvikrova M, Mala J, Hrubcova M, Eder J, Zon J, Machackova I. (2003) Effect of inhibition of biosynthesis of phenylpropanoids on sessile oak somatic embryogenesis. Plant Physiol Biochem 41: 251–259 [Google Scholar]
- De Jong AJ, Cordewener J, Lo Schiavo F, Terzi M, Vandekerckhove J, Van Kammen A, De Vries SC. (1992) A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Edwards R (2010) Glutathione transferases. The Arabidopsis Book 8: e0131, /10.1199/tab.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A, Pasternak TP, Dudits D. (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult 74: 201–228 [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Goffard N, Frickey T, Weiller G. (2009) PathExpress update: the enzyme neighbourhood method of associating gene-expression data with metabolic pathways. Nucleic Acids Res 37: W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffard N, Weiller G. (2007) PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res 35: W176–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE. (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck GR, Perry SE, Nichols KW, Fernandez DE. (1995) AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE. (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Satoh S, Kamada H. (2002) Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J Exp Bot 53: 1575–1580 [DOI] [PubMed] [Google Scholar]
- Imin N, Goffard N, Nizamidin M, Rolfe BG. (2008) Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol 8: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquard C, Mazeyrat-Gourbeyre F, Devaux P, Boutilier K, Baillieul F, Clément C. (2009) Microspore embryogenesis in barley: anther pre-treatment stimulates plant defence gene expression. Planta 229: 393–402 [DOI] [PubMed] [Google Scholar]
- Karami O, Aghavaisi B, Mahmoudi Pour A. (2009) Molecular aspects of somatic-to-embryogenic transition in plants. J Chem Biol 2: 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami O, Saidi A. (2010) The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol Biol Rep 37: 2493–2507 [DOI] [PubMed] [Google Scholar]
- Kita Y, Nishizawa K, Takahashi M, Kitayama M, Ishimoto M. (2007) Genetic improvement of the somatic embryogenesis and regeneration in soybean and transformation of the improved breeding lines. Plant Cell Rep 26: 439–447 [DOI] [PubMed] [Google Scholar]
- Klink VP, MacDonald MH, Martins VE, Park SC, Kim KH, Baek SH, Matthews BF. (2008) MiniMax, a new diminutive Glycine max genotype with a rapid life cycle, embryogenic potential and transformation capabilities. Plant Cell Tissue Organ Cult 92: 183–195 [Google Scholar]
- Ko TS, Nelson RL, Korban SS. (2004) Screening multiple soybean cultivars (MG 00 to MG VIII) for somatic embryogenesis following Agrobacterium-mediated transformation of immature cotyledons. Crop Sci 44: 1825–1831 [Google Scholar]
- Kragh KM, Hendriks T, de Jong AJ, Lo Schiavo F, Bucherna N, Højrup P, Mikkelsen JD, de Vries SC. (1996) Characterization of chitinases able to rescue somatic embryos of the temperature-sensitive carrot variant ts 11. Plant Mol Biol 31: 631–645 [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Liu WN, Moore PJ, Collins GB. (1992) Somatic embryogenesis in soybean via somatic embryo cycling. In Vitro Cell Dev Biol Plant 28P: 153–160 [Google Scholar]
- Meurer CA, Dinkins RD, Redmond CT, McAllister KP, Tucker DT, Walker DR, Parrott WA, Trick HN, Essig JS, Frantz HM, et al. (2001) Embryogenic response of multiple soybean Glycine max (L.) Merr. cultivars across three locations. In Vitro Cell Dev Biol Plant 37: 62–67 [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC. (1998) Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Norman C, Runswick M, Pollock R, Treisman R. (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55: 989–1003 [DOI] [PubMed] [Google Scholar]
- Oztetik E. (2008) A tale of plant glutathione S-transferases: since 1970. Bot Rev 74: 419–437 [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarinho PA, de Vries SC. (2002) Arabidopsis chitinases: a genomic survey. The Arabidopsis Book 1: e0023, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarinho PA, Van Hengel AJ, Fransz PF, de Vries SC. (2001) Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 212: 556–567 [DOI] [PubMed] [Google Scholar]
- Perry SE, Lehti MD, Fernandez DE. (1999) The MADS-domain protein AGAMOUS-Like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol 120: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Nichols KW, Fernandez DE. (1996) The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell 8: 1977–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative Expression Software Tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarem ER, Pelissier B, Finer JJ (1997) Effect of explant orientation, pH, solidifying agent and wounding on initiation of soybean somatic embryos. In Vitro Cell Dev Biol Plant 33: 13–19 [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltrán JP, Huijser P, Pape H, Lönnig WE, Saedler H, Schwarz-Sommer Z. (1990) Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J 9: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen I, Mäkelä P, Heino P, Palva ET. (2001) Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana. Plant J 25: 1–8 [DOI] [PubMed] [Google Scholar]
- Tchorbadjieva MI, Pantchev IY. (2006) Secretion of a chitinase-like protein in embryogenic suspension cultures of Dactylis glomerata L. Biol Plant 50: 142–145 [Google Scholar]
- Thakare D, Tang W, Hill K, Perry SE. (2008) The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol 146: 1663–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen FO, Shealy RT, Khanna A, Vodkin LO. (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132: 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen MAJ, Schmidt EDL, van Kammen A, de Vries SC. (1997) Promotive and inhibitory effects of diverse arabinogalactan proteins on Daucus carota L. somatic embryogenesis. Planta 203: 188–195 [Google Scholar]
- van Hengel AJ, Guzzo F, van Kammen A, de Vries SC. (1998) Expression pattern of the carrot EP3 endochitinase genes in suspension cultures and in developing seeds. Plant Physiol 117: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC. (2001) N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol 125: 1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Zhang XL. (2010) Regulation of somatic embryogenesis in higher plants. Crit Rev Plant Sci 29: 36–57 [Google Scholar]
- Zheng Q, Zheng Y, Perry SE. (2013) AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol 161: 2113–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Perry SE (2011) Chromatin immunoprecipitation to verify or to identify in vivo protein-DNA interactions. Methods Mol Biol 754: 277–291 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Perry SE. (2005) Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J 41: 583–594 [DOI] [PubMed] [Google Scholar]