The mitochondrial deacetylase targets protein complexes involved in energy metabolism and regulates the activity of the ATP/ADP carrier.
Abstract
The posttranslational regulation of proteins by lysine (Lys) acetylation has recently emerged to occur not only on histones, but also on organellar proteins in plants and animals. In particular, the catalytic activities of metabolic enzymes have been shown to be regulated by Lys acetylation. The Arabidopsis (Arabidopsis thaliana) genome encodes two predicted sirtuin-type Lys deacetylases, of which only Silent Information Regulator2 homolog (SRT2) contains a predicted presequence for mitochondrial targeting. Here, we have investigated the function of SRT2 in Arabidopsis. We demonstrate that SRT2 functions as a Lys deacetylase in vitro and in vivo. We show that SRT2 resides predominantly at the inner mitochondrial membrane and interacts with a small number of protein complexes mainly involved in energy metabolism and metabolite transport. Several of these protein complexes, such as the ATP synthase and the ATP/ADP carriers, show an increase in Lys acetylation in srt2 loss-of-function mutants. The srt2 plants display no growth phenotype but rather a metabolic phenotype with altered levels in sugars, amino acids, and ADP contents. Furthermore, coupling of respiration to ATP synthesis is decreased in these lines, while the ADP uptake into mitochondria is significantly increased. Our results indicate that SRT2 is important in fine-tuning mitochondrial energy metabolism.
Mitochondria are central hubs of energy metabolism in plants and animals. In addition to a fine-tuned mitochondria-to-nuclear signaling that regulates transcription of nuclear gene expression (Rhoads and Subbaiah, 2007; Schwarzländer et al., 2012), posttranslational modifications of proteins are thought to be essential for the regulation of central metabolic pathways and thus determine the plasticity of plant metabolism (Hartl and Finkemeier, 2012). In mammalian mitochondria, the regulation of metabolic functions by posttranslational Lys acetylation of proteins was recently discovered to be of great importance (Newman et al., 2012; Rardin et al., 2013). The εN-acetylation of Lys side chains (subsequently referred to as Lys acetylation) is a reversible and highly regulated posttranslational modification of both prokaryotic and eukaryotic proteins (Sadoul et al., 2011; Xing and Poirier, 2012). Lys acetylation can have a strong impact on the biochemical function of proteins as the transfer of the acetyl group to Lys masks the positive charge, which is known to be important in many catalytic centers of enzymes, as well as for protein-protein and protein-DNA interactions. In plants, Lys acetylation was, until recently, mainly thought to occur on histone proteins as regulatory mechanism for transcription and chromatin functions (Hollender and Liu, 2008). However, several central metabolic enzymes of diverse subcellular compartments were recently discovered to be Lys acetylated in Arabidopsis (Arabidopsis thaliana), and in vitro deacetylation tests confirmed that Lys acetylation affects enzyme activities and protein functions (Finkemeier et al., 2011; Wu et al., 2011). Although even organellar-encoded proteins, such as the β subunit of the ATP synthase and the large subunit of Rubisco, have been identified as Lys acetylated in Arabidopsis, it is as yet unclear which enzymes regulate this modification in mitochondria and chloroplasts of plants. Generally, protein acetyltransferases and deacetylases are known to catalyze the reversible modification of the εN-group of Lys. In addition to the classical family of histone deacetylases, a second family of protein deacetylases exists, namely the sirtuins, which are conserved across bacteria, yeast (Saccharomyces cerevisiae), plants, and animals (Hollender and Liu, 2008). Sirtuins catalyze an NAD+-dependent deacetylation of acetyl-Lys in proteins and thereby produce a deacetylated Lys, as well as the metabolites nicotinamide and 2′-O-acetyl-ADP-ribose. Sirtuins have recently emerged as key regulators of life span, cell survival, apoptosis, and metabolism in different heterotrophic organisms (Sauve, 2010; Houtkooper et al., 2012; Sebastián et al., 2012). They are also of great interest with regard to energy metabolism, as they are NAD+ dependent and function in a nutrient- and redox-dependent manner (Guarente, 2011). Based on phylogenetic analysis, sirtuin-type proteins can be grouped into four different classes (class I–IV), not all of which solely possess a Lys deacetylase activity. Mammalian Sirtuin5 (SIRT5) for example (class III) possesses additional Lys succinylase and demalonylase activity (Du et al., 2011). The most studied enzymes so far are mainly derived from class I (Houtkooper et al., 2012), while the function of class II-type enzymes is still somewhat obscure. To date, only the mammalian SIRT4 was characterized from this class and possessed an ADP-ribosyltransferase activity on mitochondrial Glu dehydrogenase (Haigis et al., 2006). Only very recently, additional Lys deacetylase activity was also demonstrated for SIRT4 (Laurent et al., 2013; Rauh et al., 2013). The Arabidopsis genome encodes two sirtuins from two classes, with Silent Information Regulator1 homolog (SRT1; At5g55760) from class IV and SRT2 (At5g09230) from class II (Pandey et al., 2002). Because the actual biological function of type II sirtuins in mammalian organisms is still under some debate (Newman et al., 2012), and neither sirtuin-specific activity nor function of any plant sirtuin has been demonstrated so far, it was our goal to establish the function of the predicted mitochondrial class II sirtuin SRT2 of Arabidopsis.
RESULTS
Arabidopsis SRT2 Encodes Seven Splice Forms, of Which Two Are Degraded by Nonsense-Mediated Decay
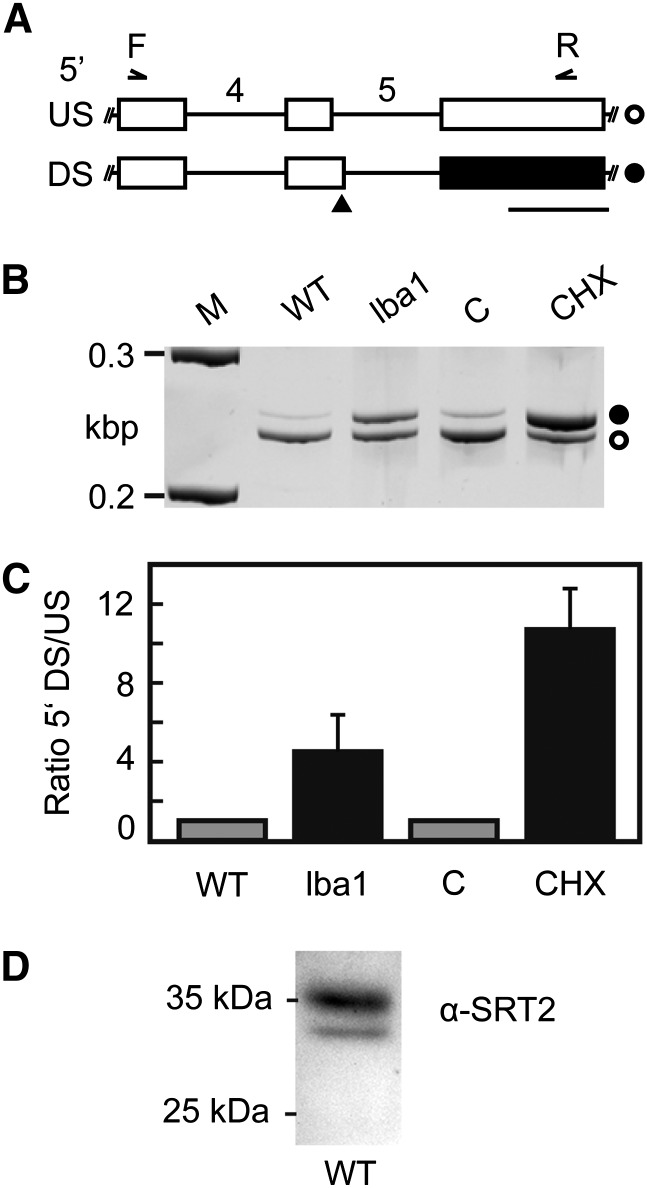
In contrast to seven sirtuin genes in the mammalian genome, Arabidopsis possesses only two genes encoding putative sirtuin-type proteins. However, seven transcript isoforms are predicted for the Arabidopsis SRT2 gene (At5g09230.1–At5g09230.7/SRT2.1–SRT2.7, TAIR10 [The Arabidopsis Information Resource 10]), which are generated from alternative splicing (AS) of the SRT2 precursor mRNA. Hence, we were curious to find out whether all of the annotated splice forms are expressed and whether different protein isoforms will be generated from these transcripts. We were able to detect all of the annotated as well as some additional splicing variants for SRT2 by an analysis of high-throughput RNA sequencing data from Arabidopsis seedlings (Supplemental Fig. S1). As the protein sequences of the annotated SRT2 isoforms are very similar and mainly differ in their N- or C-terminal regions (Supplemental Fig. S1; Supplemental Table S1), we had a closer look at an AS event in intron 5 occurring in both SRT2.4 and SRT2.6 transcripts (Fig. 1A; Supplemental Fig. S2). This splice form is predicted to result in N-terminal short forms of the protein according to the Arabidopsis genome annotation (TAIR10), due to an alternative translation start from a downstream codon. However, at the same time, the introduction of this splice site could also result in the introduction of a premature termination codon, which should result in degradation of this splicing variant by the RNA surveillance mechanism nonsense-mediated decay (NMD). To test this hypothesis, the ratio of splicing variants resulting from usage of the alternative 5′ splice sites in control and NMD-impaired samples was determined by reverse transcription (RT)-PCR (Fig. 1, B and C). Coamplification with oligonucleotides spanning the indicated region resulted in two amplification products of the expected sizes (Fig. 1B). Using the previously described missense mutant in a core NMD factor, low-beta-amylase1 (Yoine et al., 2006), and treatment with the translation inhibitor cycloheximide, which is known to suppress NMD, a relative increase of the splicing variant derived from the downstream 5′ splice site was found, clearly supporting its NMD target identity (Fig. 1C). Hence, the open reading frame annotation in TAIR10 needs to be revised for SRT2.4 and SRT2.6. To confirm our results and to analyze the presence and localizations of the predicted protein isoforms, we performed a western-blot analysis using an SRT2-specific antibody (αSRT2) raised against the recombinant protein of the SRT2.1 splice form. The specificity of the antiserum was confirmed by the absence of the immunosignal in SRT2 knockout plants (see below; Fig. 2B). Because all predicted SRT2 proteins (TAIR10) are highly similar in the core sequence and mainly differ in their N- or C-terminal regions, all isoforms should at least theoretically be detected in the western blot in the range between 20 and 40 kD (Supplemental Fig. S1; Supplemental Table S1). However, only two protein bands were detected, with the main signal at around 36 kD (Fig. 1D). This band matches the predicted size of SRT2.1, SRT2.2, SRT2.5, and SRT2.7 with cleaved N-terminal targeting presequences (Supplemental Table S1). Hence, this mature protein isoform is referred to as SRT2A in the following. A further weaker band was detected at around 31 kD, which matches the size of the mature C-terminal short-form SRT2.3, and is subsequently referred to as SRT2B. No protein band was detected for the predicted mature protein isoforms of SRT2.4 and SRT2.6 at 25 kD, and thus the western-blot analysis confirms the absence of these protein isoforms as predicted from our NMD target analysis.
Figure 1.
AS of SRT2 precursor mRNA generates an NMD target and is regulated by polypyrimidine tract-binding protein splicing factors. A, Partial models of SRT2 splicing variants resulting from usage of an upstream (US, top) or downstream (DS, bottom) 5′ splice site in intron 5 (introns numbered). Boxes and lines depict exons and introns, respectively. White boxes correspond to coding sequence, and black box indicates an untranslated region resulting from the introduction of a premature termination codon (black triangle) upon usage of the downstream 5′ splice site. White circle indicates SRT2 splice form 1,2,3, and 7, and black circle indicates splice form 4 and 6. Binding positions of primers used for coamplification of splicing variants in B and C are shown. Bar = length of 100 bp. B, RT-PCR analysis of AS event shown in A. For NMD impairment, low-beta-amylase1 (lba1) mutant or cycloheximide (CHX) versus mock (C) treatment were analyzed. M indicates ladder with 0.2- and 0.3-kb DNA fragments. C, Quantitative analysis of splicing ratios for samples depicted in B using a bioanalyzer. Displayed are mean values (n = 3–6, ± sd). Data are normalized to the wild type (WT) or mock treatment. D, Western-blot analysis of SRT2 protein in Arabidopsis leaves.
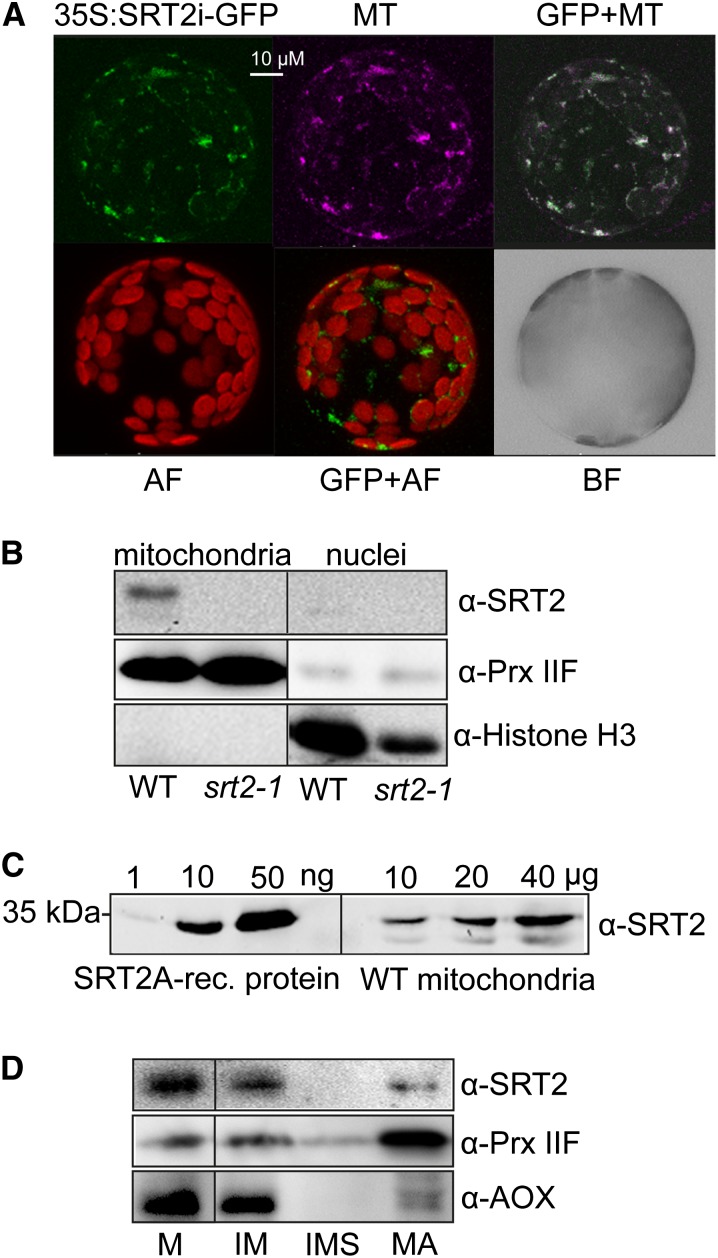
Figure 2.
SRT2-encoded proteins localize to mitochondria in Arabidopsis. A, Arabidopsis protoplasts showing GFP localizations (green) for a full-length SRT2 (plus introns)-GFP fusion construct (35S:SRT2i-GFP). Protoplasts were isolated from stable Arabidopsis transformants. Purple indicates the mitochondrial localization of MitoTracker (MT). GFP+MT indicates overlay image of 35S:SRT2i-GFP and MitoTracker; AF indicates autofluorescence of chloroplasts; and BF indicates bright-field image of protoplast. B, Western-blot analysis of SRT2 protein in mitochondrial and nuclear fractions. Antisera against PRX IIF was used as mitochondrial marker and histone H3 as nuclear marker, respectively. C, Western-blot analysis of SRT2 proteins in isolated mitochondria. Protein extracts of isolated mitochondria from wild-type (10, 20, and 40 µg) seedlings as well as recombinant 6× His-SRT2A protein (1, 10, and 50 ng) were analyzed by western blotting using SRT2 antiserum. D, Western-blot analysis of SRT2 protein in subfractionated Arabidopsis wild-type mitochondria. Ten micrograms of protein were loaded for each fraction. Antisera against Alternative Oxidase1 and PRX IIF were used as controls for the inner membrane and matrix proteins, respectively. M, Mitochondria; IM, inner mitochondrial membrane; IMS, intermembrane space; MA, matrix.
Arabidopsis SRT2-Encoded Protein Variants Are Located in Mitochondria
All annotated SRT2 splice forms encode a predicted N-terminal presequence of different length for targeting to mitochondria (Supplemental Table S1). To determine the subcellular localization of all expressed SRT2 isoforms, the full-length genomic sequence of SRT2 including introns and without stop codon (SRT2i) was cloned into the binary vector pK7WGF2 for C-terminal GFP fusion (Karimi et al., 2002). The 35S:SRT2i-GFP construct was stably transformed into Arabidopsis, and independent transformants were analyzed for their GFP fluorescence signal by confocal laser scanning microscopy. All transformants showed only mitochondria-localized GFP signals, which overlapped with the mitochondria- specific dye MitoTracker Red (Invitrogen; Fig. 2A), confirming the predictions of the TargetP algorithm (Supplemental Table S1). No GFP signal was detected in the nucleus for any of the transformants (neither after transient nor stable transformation) in contrast to the report by Wang et al. (2010). We confirmed the absence of SRT2 in the nucleus and its presence in mitochondria by a western-blot analysis of enriched nuclear and mitochondrial fractions using αSRT2 and compartment specific antisera (PRX IIF for mitochondrial type II peroxiredoxin F and histone H3 for nuclei) as control, respectively (Fig. 2B). Furthermore, SRT2 was identified as fairly low-abundance protein in Arabidopsis mitochondria, with only about 0.2 ng SRT2 µg–1 (0.02% [w/w]) of mitochondrial protein (Fig. 2C). Further subfractionation of mitochondria into matrix, inner membrane, and intermembrane space revealed that SRT2 is primarily localized at the inner mitochondrial membrane, while a weak signal was also detected in the matrix fraction (Fig. 2D).
Arabidopsis SRT2 Is a NAD+-Dependent Lys Deacetylase
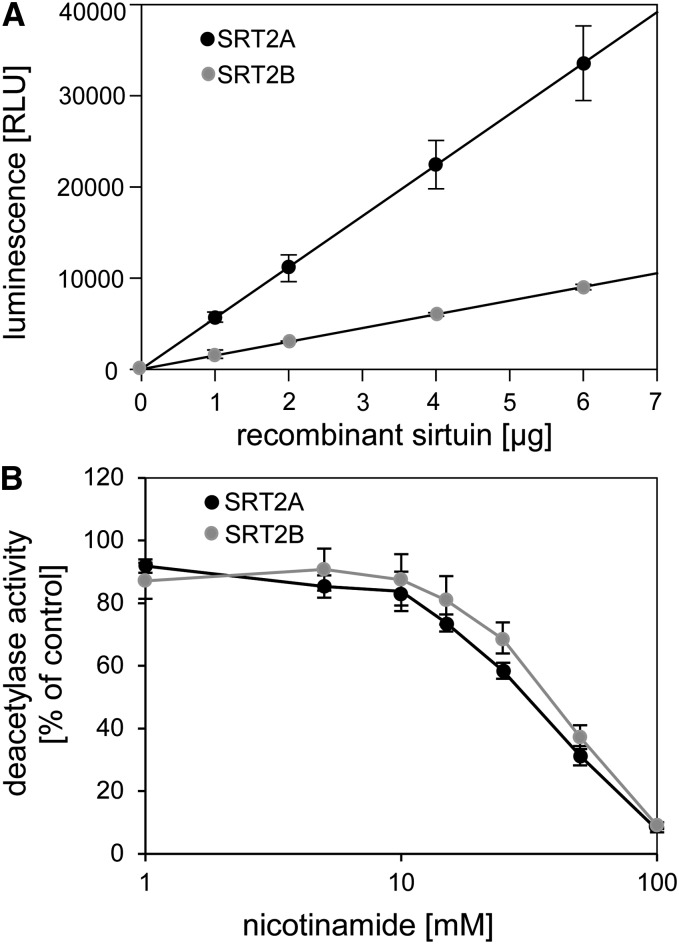
To investigate the catalytic activity of the two major SRT2 isoforms, we next overexpressed and purified both SRT2A and SRT2B proteins as N-terminally His-tagged proteins from Escherichia coli. A Lys-acetylated peptide of p53, which is a common substrate of different types of mammalian sirtuins, was used in a luminescence assay based on trypsin cleavage of the deacetylated peptide. Both SRT2A and SRT2B proteins were able to deacetylate the artificial p53 peptide in a concentration-dependent manner, indicating that both proteins can act as sirtuin-type deacetylases (Fig. 3A). The activity of both proteins was linear with respect to enzyme concentration in the range from 1 to 6 µg for both SRT2A and SRT2B. However, SRT2B (the C-terminally truncated form) was about 4 times less active than SRT2A (Fig. 3A). To further investigate sirtuin-specific enzyme characteristics, we tested the inhibition of both SRT2 proteins by its reaction product nicotinamide. A concentration-dependent effect of nicotinamide on SRT2 deacetylase activities was observed, which resulted in a half-maximum inhibition at about 30 mm (50% inhibition of initial activity [IC50] = 29.7 mm, fitted with the solver tool of Microsoft Excel 2010) with 100 µm acetylated p53-peptide as substrate and 1 µm SRT2 enzyme (Fig. 3B). A similar nicotinamide concentration was reported for half-maximum inhibition of human poly(ADP-ribose)polymerase enzyme activity (Ungerstedt et al., 2003), while classical sirtuins usually have much higher affinities for nicotinamide as inhibitor (IC50 around 100 µm) under comparable reaction conditions (Schmidt et al., 2004). However, it should be noted that for competitive inhibitors, IC50 values are not equivalent to inhibitor constant values, and thus IC50 values from different enzymes are not necessarily comparable (Brandt et al., 1987).
Figure 3.
Arabidopsis SRT2A and SRT2B proteins possess NAD+-dependent deacetylase activity. A, Linear range of SRT2A and SRT2B NAD+-dependent deacetylase activities on acetylated p53 peptide using the SIRT-Glo assay (Promega). The deacetylase activities were recorded as luminescent signal from deacetylated and trypsin-digested peptides using 1 to 6 µg SRT2 recombinant proteins. B, Potency of nicotinamide concentration (1–100 mm) on SRT2A and SRT2B (1 µm each) deacetylase activities. Data are expressed as means ± se from independent purifications of recombinant proteins (n = 3).
Increased Lys Acetylation of Mitochondrial Inner Membrane Complexes in srt2-1
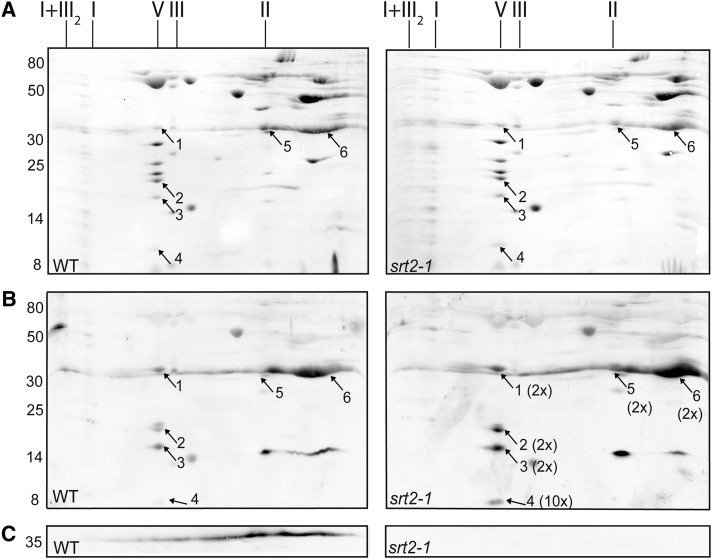
To identify in vivo SRT2 protein substrates, we used the srt2-1 knockout line (Wang et al., 2010), which is deficient in SRT2 protein (srt2-1; Fig. 2B), to detect changes in Lys acetylation levels of mitochondrial proteins in the mutant compared with the wild type. We also isolated a second mutant allele (srt2-2; Supplemental Fig. S3), which showed the same phenotype as the srt2-1 mutant, and thus srt2-1 was used for the further experiments. Given that SRT2 is primarily located at the inner mitochondrial membrane (Fig. 2C) and changes in Lys acetylation could potentially impact protein-protein interactions and complex formation, we isolated intact protein complexes from wild-type and srt2-1 mitochondria solubilized with digitonin and analyzed them by two-dimensional blue-native PAGE (Fig. 4). No changes in protein complex formation and composition or in protein abundances of the respiratory oxidative phosphorylation (OXPHOS) complexes were detected between the wild type and srt2-1 (Fig. 4A). However, in the western-blot analysis using an antibody directed against acetyl-Lys, we detected increased acetylation of the ATP synthase (complex V) subunits γ (1.9-fold ± 0.2, n = 6, spot 1), δ/ε (2.1-fold ± 0.6, n = 6, spot 2), subunit 8 (2.3-fold ± 0.7, n = 6, spot 3), and ATP17, which is a plant-specific subunit of the ATP synthase (10.2-fold ± 4.2, n = 6, spot 4) in srt2-1 compared with the wild type (Fig. 4B; Supplemental Table S2). Furthermore, a 1.6-fold-increased Lys acetylation (1.6-fold ± 0.2, n = 6) of two protein complexes primarily containing the ATP/ADP carrier proteins (AAC1 to AAC3; spots 5 and 6) as well as the voltage-dependent anion channel proteins (VDAC1 to VDAC4; spots 5 and 6) were detected in the srt2-1 mutant. However, as the VDAC proteins are localized to the outer mitochondrial membrane, it is more likely that the signal reflects the differential Lys acetylation of the AAC carrier proteins in the inner mitochondrial membrane where SRT2 is also located. To a lesser extent, the gel spot also contained two mitochondrial substrate carrier proteins of the inner mitochondrial membrane, A BOUT DE SOUFFLE (BOU; spot 6) and a di- and tricarboxylate transporter protein (spot 6; Fig. 4B; Supplemental Table S2), which possibly also partly contribute to the immune signal. To reveal whether SRT2 is present in one of the differentially Lys-acetylated protein complexes, we detected the SRT2 protein in the two-dimensional blue-native western blot using the purified αSRT2 antibody (Fig. 4C). In agreement with the multiprotein deacetylase function of sirtuins, the SRT2 protein was not present in a specific protein complex but rather widely distributed over a whole range of protein complexes with different sizes, starting from complex I at around 1 megadalton down to smaller protein complexes of around 100 kD (Fig. 4C). As expected, the detected immune signal was absent in the srt2-1 knockout line (Fig. 4C). Hence, the extent of Lys acetylation of the ATP synthase as well as of the protein complex containing the ATP/ADP carrier is dependent on the presence of SRT2 in plant mitochondria.
Figure 4.
Differences in Lys acetylation levels in the protein complexes of wild-type (WT) and srt2-1 mitochondria. A, PonceauS stain of wild type and srt2-1 two-dimensional blue-native SDS-PAGE of mitochondrial protein complexes. The identity of OXPHOS complexes is indicated above the gels. The molecular mass scale (in kilodaltons) is indicated on the left. Numbers (1–6) refer to proteins identified by LC-MS/MS. ATP synthase subunit γ (1), δ/ε (2), subunit 8 (3), ATP17 (At4g30010; 4), ATP/ADP carrier proteins AAC1 to AAC3 and VDAC1 to VDAC4 (5), and ATP/ADP carrier proteins AAC1 to AAC3, VDAC1 to VDAC4, A BOUT DE SOUFFLE, and di- and tricarboxylate transporter protein (6). For a full list with protein abundances, see Supplemental Table S2. I+III2, Supercomplex composed of complex I and dimeric complex III; I, complex I; V, complex V (ATP synthase); III, dimeric complex III. B, Differences in Lys acetylation levels of proteins from the wild type and srt2-1 were detected by western-blot analysis using the anti-acetyl-Lys antibody. Numbers (1–6) refer to proteins identified by LC-MS/MS. The average fold change increase in Lys acetylation in srt2-1 is indicated in brackets behind the numbers (n = 6). C, The presence and absence of SRT2 protein was detected by western-blot analysis using anti-SRT2 antiserum.
SRT2 Interacts with Several of Its Substrate Proteins
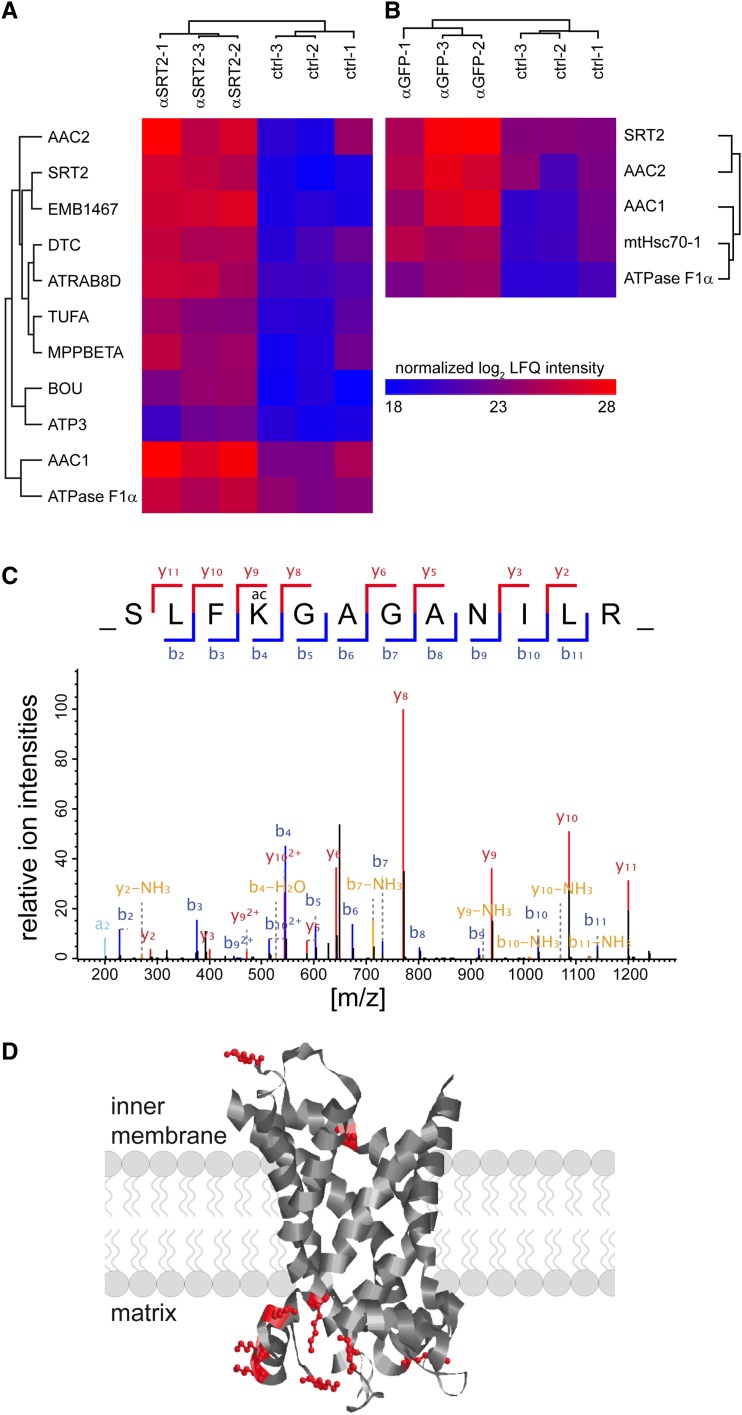
To confirm the interaction of SRT2 with the differentially Lys-acetylated protein complexes, we performed two different pull-down approaches. In the first approach, SRT2-interacting proteins were isolated by coimmunoprecipitation using the purified SRT2 antibody on isolated protein extracts from wild-type mitochondria (Fig. 5A). In the second approach, mitochondria from 35S:SRT2-GFP lines were isolated, and interacting proteins were copurified using GFPTrap-A beads (Chromotek; Fig. 5B). Only three different proteins were significantly enriched, alongside SRT2, in both coimmunoprecipitation approaches and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based label-free quantification compared with their respective controls for background correction (Cox and Mann, 2008; Hubner et al., 2010; Fig. 5; Supplemental Table S3). Strikingly, the AAC proteins, which were significantly more Lys acetylated in srt2-1 mitochondria (Fig. 4B), were among these proteins. Furthermore, the α subunit of the ATP synthase was more than 2-fold enriched using both approaches. Seven more proteins were identified in the αSRT2-coimmunoprecepitation analyses that were not enriched in the SRT2.1-GFP pull downs (Fig. 5; Supplemental Table S3). Among these were several additional interesting candidates, such as the 76-kD NADH dehydrogenase subunit of complex I, beta subunit of mitochondrial processing peptidase, an integral protein of complex III, which is responsible for the removal of presequences of imported precursor proteins, a GTP-binding elongation factor Tu family protein, and the member of the Ras superfamily GTPase homolog E1B, as well as three proteins, which were part of the complexes that showed increased Lys acetylation on the western blot in the srt2-1 lines: the γsubunit of the ATP synthase, the mitochondrial carrier protein BOU, and the di- and tricarboxylate transporter (Fig. 5; Supplemental Table S3). Hence, these proteins also have to be considered as potential in vivo SRT2-interacting proteins.
Figure 5.
SRT2 interacts with several proteins of the inner mitochondrial membrane. A and B, Two-way hierarchical clustering of SRT2-interacting proteins from two different coimmunoprecipitation experiments. A, Antibody specific for Arabidopsis SRT2 (αSRT2) was incubated with mitochondrial extracts from wild-type plants (n = 3). B, Antibody specific for GFP (αGFP) was used on mitochondrial extracts from transgenic plants expressing a SRT2.1:GFP fusion protein (αGFP; n = 3). Only proteins that were significantly enriched were selected for the clustering (Student’s t test with permutation-based false discovery rate < 0.05). The color code represents relative protein abundance, measured as the log-summed peptide intensities for each protein, after normalization by the label-free quantification algorithm in the MaxQuant software package. C, Tandem mass spectrometry fragmentation spectrum of the peptide containing the evolutionary conserved acetylated K345 (AAC1) site of the ATP/ADP carrier proteins. D, Scheme of the AAC1 to AAC3 protein topology in the inner mitochondrial membrane (after Klingenberg, 2008) with 11 Lys acetylation sites at the matrix- and intermembrane-exposed loops.
To identify the actual position of the Lys acetylation sites on the SRT2-interacting proteins, we performed a further pull-down experiment using the acetyl-Lys antibody. Several of the SRT2-interacting proteins contained one or more Lys-acetylated sites (Supplemental Table S4). First and foremost, 11 Lys acetylation sites were detected on the AAC1 to AAC3 proteins (Fig. 5C; Supplemental Table S4). Nine of these sites were positioned at the loops protruding into the matrix and two into the intermembrane space. Furthermore, three Lys acetylation sites were identified on the ATP synthase complex (α and γ subunits), three sites on the di- and tricarboxylate transporter protein, and two sites on the mitochondrial heat shock protein 70-1 (Supplemental Table S4).
Mitochondrial Respiratory Control and 14C-ADP Uptake Is Significantly Altered in srt2-1 Mitochondria
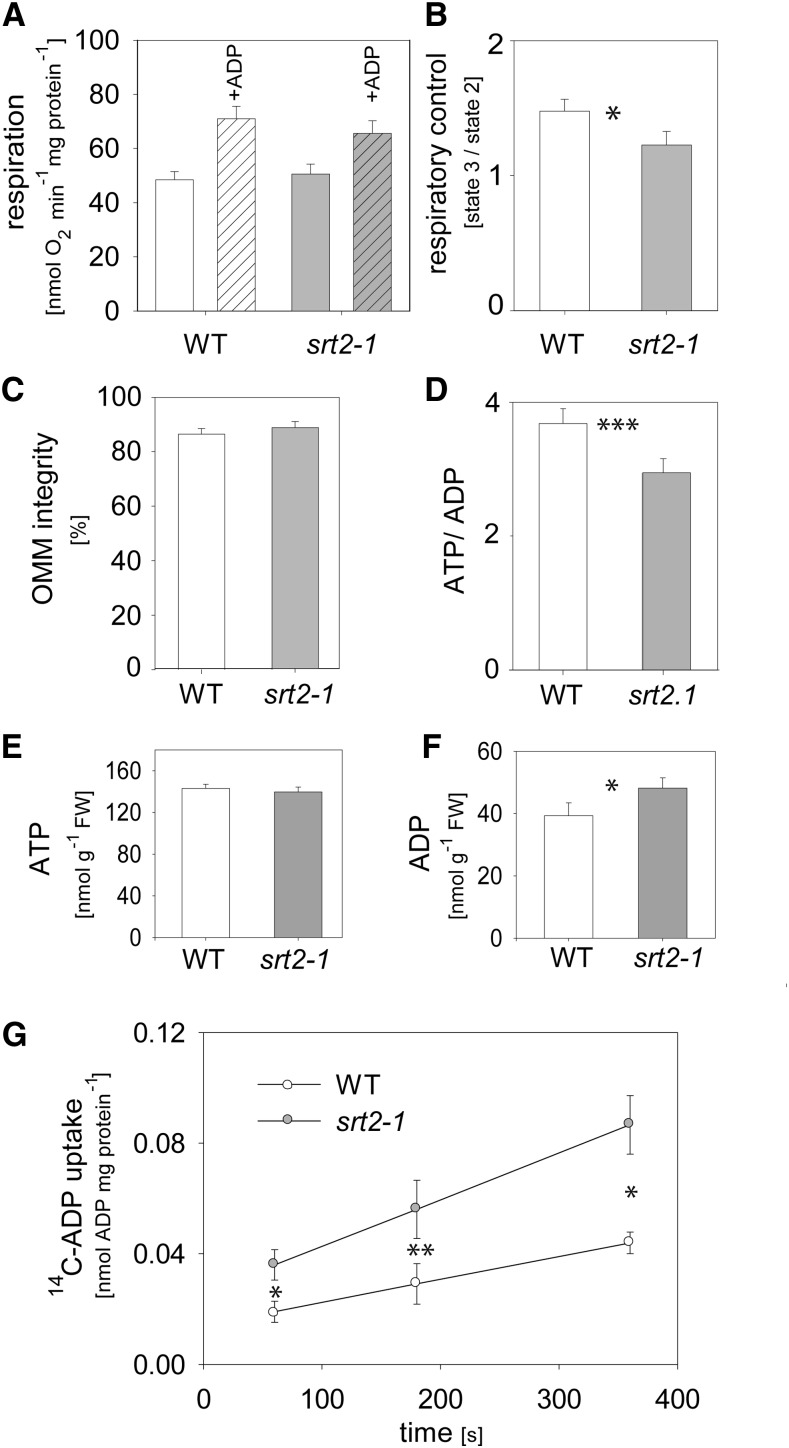
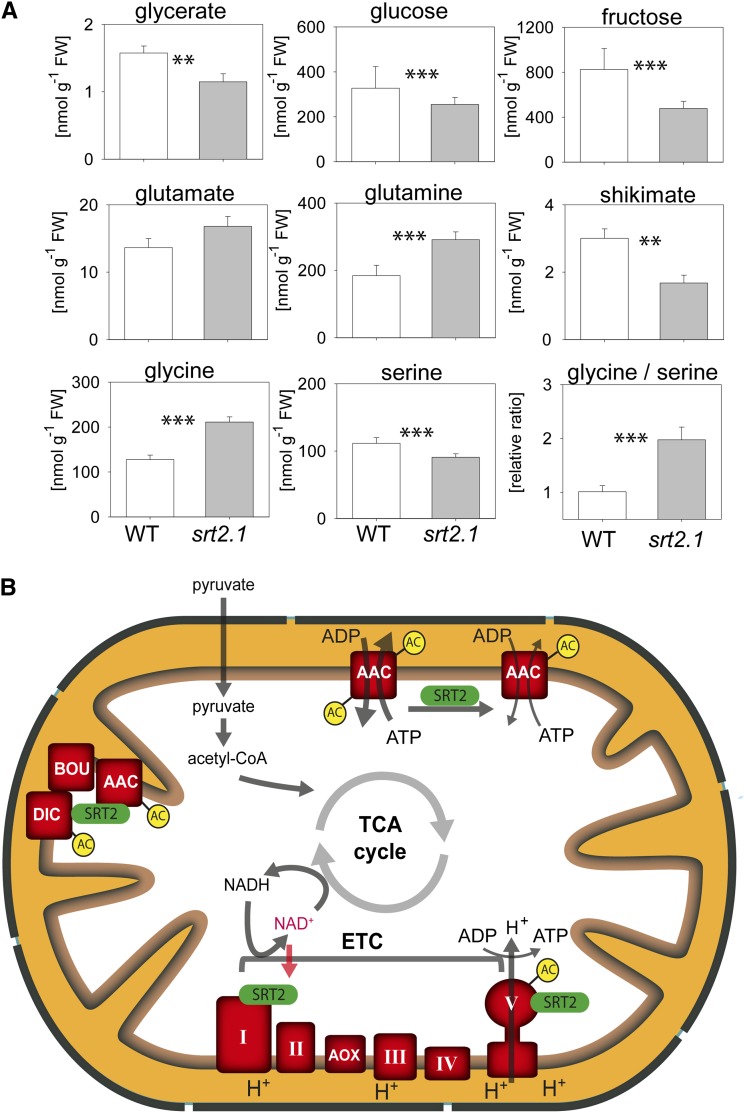
Because the ATP/ADP carrier proteins (AAC1–AAC3) as well as the ATP synthase complex were identified as most robust interaction partners of SRT2 (Fig. 5) and both protein complexes also showed increased acetylation in the absence of SRT2 (Fig. 4), we tested whether the loss of SRT2 affects OXPHOS and ADP uptake of mitochondria. However, no significant changes were observed in the respiratory activities of the OXPHOS complexes I and II driven by TCA cycle-dependent malate and pyruvate oxidation (Fig. 6A). Nevertheless, the respiratory control ratio, which indicates the coupling of respiration to ATP synthesis, was significantly decreased in srt2-1 mitochondria compared with those of the wild type (Fig. 6B). This was not due to a damage of the outer mitochondrial membrane, as the latency of cytochrome c oxidation before and after Triton-X100 addition was in both mutant and the wild type around 90% (Fig. 6C). Other explanations for a decreased coupling of respiration in the srt2-1 mutant could be due to a decreased ATP synthase activity or an enhanced uncoupling of srt2-1 mitochondria. When we measured the total ATP as well ADP levels in the srt2-1 seedlings compared with the wild type, we observed a decreased ATP to ADP ratio, which was due to increased ADP but not ATP levels (Fig. 6, D–F). To test whether the mitochondrial ADP uptake rates are affected in srt2-1, we measured 14C-ADP uptake on isolated mitochondria. Interestingly, the rate of 14C-ADP uptake into srt2-1 mitochondria was much more efficient and significantly increased compared with the wild type at each measured time point (60–360 s; Fig. 6G). In energized mitochondria, the AAC carriers usually transport matrix ATP against cytosolic ADP in a 1:1 exchange ratio, while they are unable to transport AMP (Haferkamp et al., 2002; Klingenberg, 2008). However, the AAC carrier activity is not necessarily coupled to mitochondrial ATP production, as the transport proceeds with high activity when mitochondria are completely uncoupled (Klingenberg, 2008). Hence, we conclude that SRT2 positively affects the ATP synthase activity by Lys deacetylation when NAD+ is available as substrate, while deacetylation of the ATP/ADP carrier proteins negatively impacts their activity. In addition to increased ADP levels in srt2-1 seedlings, also Gln and Gly levels were increased, while several metabolites of central metabolism, such as sugars (Fru, Glc, erythritol, and myoinositol), as well as some amino acids (Ser, Pro, Arg, Thr, Tyr, and Ala) and organic acids (shikimate, ascorbate, pyruvate, fumarate, and γ-aminobutyrate), were significantly decreased in abundance (Fig. 7A; Supplemental Table S5). Hence, SRT2 is most likely involved in the fine regulation of mitochondrial energy metabolism and thus indirectly also affects up- and downstream metabolic pathways in Arabidopsis (Fig. 7B).
Figure 6.
Loss of SRT2 affects coupling of ATP synthesis to mitochondrial respiration and ADP uptake into mitochondria. A, TCA cycle-dependent respiration (pyruvate, malate) in isolated mitochondria of wild-type (WT) and srt2-1 seedlings. Respiratory activities were measured in state II and state III (100 µm ADP) mitochondria (n = 6, + se). B, Respiratory control ratios calculated from respiratory activities before and after ADP addition (n = 6, ± se). C, Integrity of the outer membrane calculated by the latency of cytochrome c oxidation before and after addition of Triton-X100 to mitochondria (n = 6, ± se). D, ATP to ADP ratios in srt2-1 seedlings compared with the wild type (n = 5, ± se). E, ATP contents in srt2-1 seedlings compared with the wild type (n = 5, ± se). F, ADP contents in srt2-1 seedlings compared with the wild type (n = 5, ± se). G, Time course of 14C-ADP uptake into isolated mitochondria of wild-type (white bars) and srt2-1 seedlings (black bars; n = 5, + se). Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test).
Figure 7.
Metabolic phenotype of SRT2 knockout line and simplified working model for SRT2 functions in plant mitochondria. A, Metabolite contents in srt2-1 compared with the wild type (WT; n = 5, ± se). The full list of metabolites including the absolute values can be found in Supplemental Table S5. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test). B, SRT2 is dependent on NAD+ as substrate and interacts with the 76-kD subunit of complex I. SRT2 interacts and most likely deacetylates the ATP synthase (complex V) as well as a complex containing the AAC proteins AAC1 to AAC3, the putative di-and tricarboxylate transporter (DIC), and the metabolite transporter BOU. Lys acetylation sites of SRT2 interaction partners are indicated (AC). [See online article for color version of this figure.]
DISCUSSION
Herein, we established that Arabidopsis SRT2 functions as mitochondrial Lys deacetylase, and we identified its first substrates and interaction partners. While mammalian mitochondria contain three different types of sirtuins (SIRT3–SIRT5) with very diverse physiological functions (Verdin et al., 2010), the Arabidopsis genome only encodes a single sirtuin gene (SRT2) for mitochondrial targeting. According to the latest Arabidopsis genome annotation (TAIR10), SRT2 potentially expresses seven protein isoforms. Here, we have demonstrated that the two nearly identical splice forms SRT2.4 and SRT2.6, which are generated from an AS site in intron five, are both targets of NMD. Furthermore, we did not observe proteins corresponding to these two splicing variants in the western-blot analysis (Fig. 1) nor have we detected any of their sequence-specific peptides in the LC-MS/MS analysis (Supplemental Fig. S1).
The Native Arabidopsis SRT2 Isoforms Are 31 and 36 kD in Size and Are Localized in Mitochondria
All of the annotated SRT2 protein isoforms contain N-terminal presequences predicted for mitochondrial targeting (Supplemental Table S1). The subcellular localization analysis using a C-terminal GFP fusion to the SRT2 gene containing introns confirmed the mitochondrial localization of all expressed SRT2 isoforms (Fig. 2A). Importantly, we detected no nuclear GFP signal for SRT2, although Wang et al. (2010) exclusively observed a nuclear localization for SRT2 (Pandey et al., 2002). Furthermore, we confirmed the mitochondrial localization of the native SRT2 protein by western-blot analysis and in pull-down experiments using isolated mitochondria (Fig. 2; Supplemental Table S3). Two immune-reactive protein bands of 31 and 36 kD in size, respectively, were detected in our western-blot analysis (Figs. 1D and 2B). These two protein bands were named SRT2A and SRT2B, respectively, as they fitted to the size of the predicted processed isoforms of SRT2.1/SRT2.2/SRT2.5/SRT2.7 and SRT2.3 after removal of their presequences (Supplemental Table S1). The native SRT2 peptides identified in the LC-MS/MS analysis of the coimmunoprecipitation experiment further support this observation (Supplemental Fig. S1). Hence, we conclude that there are two mature SRT2 proteins present in plant mitochondria, which mainly differ in their C-terminal part.
The Arabidopsis Class II Sirtuin Is a Protein Deacetylase
Having confirmed that SRT2 is present in two isoforms in plant mitochondria, the question emerged what enzymatic activity these two isoforms have, because Arabidopsis sirtuins were to date merely annotated on the basis of sequence homology. Comparison of the amino acid sequence of the Arabidopsis SRT2 proteins to the mammalian sirtuins revealed that SRT2 shares the highest homology with human SIRT4 (42% amino acid sequence identity), followed by 31% identity with hSIRT3 and 25% identity with hSIRT5 (Supplemental Fig. S4). Because the true enzymatic function of the mammalian SIRT4 is still under debate (Feldman et al., 2012; Newman et al., 2012), we purified the two mature SRT2A and SRT2B proteins as N-terminally His-tagged proteins upon heterologous expression in E. coli. Strikingly, SRT2A and, to a slightly lesser extent, SRT2B were active as protein Lys deacetylases (Fig. 3).
SRT2 Interacts with Proteins of the Inner Mitochondrial Membrane Involved in Energy Metabolism and Metabolite Transport
In subfractionated Arabidopsis mitochondria, most of the SRT2 immunosignal was detected to be present in the inner membrane fraction and to a lesser extent in the matrix (Fig. 2). Given that the SRT2 protein contains no membrane-spanning domain, we assume that SRT2 is associated with the inner membrane via protein-protein interactions. Several of our identified interaction partners of SRT2 are integral proteins of the inner mitochondrial membrane, such as the ATP/ADP carriers, beta subunit of mitochondrial processing peptidase, and the metabolite carrier BOU, a putative carboxylate transporter, as well as two matrix-exposed subunits of the ATP synthase (complex V) and the 76-kD matrix-exposed complex I subunit (Fig. 5). Although we did not identify a Lys acetylation site on the 76-kD subunit of the Arabidopsis complex I in srt2-1 mitochondria (Supplemental Table S4) and complex I-dependent respiration was not affected in the mutant (Fig. 6A), it is possible that the constitutive knockout plants have been partially adapted to loss of SRT2. Binding of SRT2 to complex I is anyhow interesting, as the activity of sirtuins is generally regulated by its substrate NAD+. Thus, SRT2 could possibly act in concert with an active complex I oxidizing NADH. The interaction of SRT2 with complex V subunits is interesting because Arabidopsis SRT2 knockout mutant showed a decreased coupling of mitochondrial respiration to ATP synthesis, which indicates a decreased activity of complex V and which could explain the decreased in vivo Glc and Fru levels due to decreased respiratory energy conversion. Although total ATP levels were not changed, as a decrease in mitochondrial ATP production can possibly be compensated for by photosynthesis, the total ADP contents were increased, suggesting an adaptation of the cellular metabolism to the loss of SRT2. Similar observations were recently also reported for human cells deficient in SIRT4. Knockdown of SIRT4 in Human Embryonic Kidney 293 cells resulted in increased ADP/ATP ratios, while overexpression of SIRT4 showed the opposite effect (Ho et al., 2013). Furthermore, Ho et al. (2013) demonstrated that the activity of the mitochondrial ATP synthase was not affected by the loss of SIRT4 but that uncoupling of mitochondria was increased. Interestingly, uncoupling of respiration was relieved when the ATP/ADP carrier ADP/ATP Translocase2 was knocked down simultaneously with SIRT4 (Ho et al., 2013). However, Lys acetylation levels of mitochondrial proteins were not investigated in that study. It will be interesting to find out whether knockdown of the AAC carriers in the Arabidopsis srt2-1 background would have a similar effect.
The AAC Carriers as Well as the ATP Synthase Complexes Are the Major Targets of SRT2-Dependent Deacetylation in Arabidopsis Mitochondria
Two recent studies identified the first organellar Lys-acetylated proteins in Arabidopsis (Finkemeier et al., 2011; Wu et al., 2011). Among these proteins were several plastidial proteins but only a few mitochondrial proteins, such as cytochrome c, the α subunit of the ATP synthase complex, and AAC1. Here, we detected several additional mitochondrial Lys-acetylated proteins in western-blot analysis of the two-dimensional blue-native PAGE as well as in the protein pull downs of the SRT2-interacting proteins (Fig. 7; Supplementary Table S3). The most prominent Lys-acetylated proteins we detected were several subunits of the ATP synthase complex as well as two protein complexes containing the AAC carrier proteins AAC1 to AAC3. These protein complexes were also significantly increased in Lys acetylation in srt2-1 compared with the wild type (Fig. 4). Hence, we conclude that Arabidopsis SRT2 has very specific target proteins for deacetylation and that it does not act as universal mitochondrial protein deacetylase such as SIRT3 in mammalian mitochondria (Lombard et al., 2007; Rardin et al., 2013). Loss of mammalian SIRT3 results in the hyperacetylation of mitochondrial proteins, which is not observed upon deletion of the mouse mitochondrial SIRT4 and SIRT5 (Lombard et al., 2007). Similarly, no overall hyperacetylation of mitochondrial proteins was observed upon deletion of Arabidopsis SRT2 in our study.
The Activity of the ATP/ADP Carrier Proteins Is Dependent on SRT2
AAC carrier proteins have a central role in energy metabolism in eukaryotic cells, as they are the gateways for ATP supply to the cytosol (Haferkamp et al., 2011). Here, we identified 11 acetylation sites on the ATP/ADP carrier proteins AAC1 to AAC3 (Supplemental Table S4). Interestingly, the ADP/ATP carrier 2 was previously also identified as interacting protein of human SRT4-FLAG protein (Ahuja et al., 2007). However, Ahuja et al. (2007) did not investigate whether this interaction affected the acetylation status of ADP/ATP carrier. The AAC carrier proteins are highly conserved in sequence between species, and according to the topology model of Klingenberg (2008), most of the acetylated Lys residues can be found in the matrix-exposed loops (Fig. 5D). Only two of the identified acetylated Lys residues reside in the loops exposed to the intermembrane space. Seven of the 11 acetylated Lys residues are highly conserved between species and were also shown to be Lys acetylated in yeast, rat, mouse, and human tissue, respectively (Supplemental Fig. S5; Choudhary et al., 2009; Weinert et al., 2011; Henriksen et al., 2012; Lundby et al., 2012; Sol et al., 2012). Furthermore we detected a 1.6- to 2-fold increase in Lys acetylation of the AAC carriers in the srt2 mutants (Fig. 4). In addition, Lys acetylation levels of three of the matrix-exposed sites were recently identified as highly increased (up to 9-fold) in SIRT3 –/– mice (Rardin et al., 2013), confirming that the AAC proteins are universal targets of sirtuins in different species. While it has reported nearly 20 years ago that fatty acids can induce an uncoupling of the ATP/ADP carriers (Brustovetsky and Klingenberg, 1994), to our knowledge, no study has yet investigated whether Lys acetylation affects the activity of these carriers. Here, we have demonstrated that an increased Lys acetylation correlated with an increased 14C-ADP uptake rate in srt2-1 mitochondria (Fig. 6G), indicating that SRT2 regulates the Lys acetylation level and activity of the carrier proteins.
In summary, we have shown that Arabidopsis SRT2 is a mitochondrial Lys deacetylase acting on specific target proteins of the inner membrane such as the ATP synthase and the ATP/ADP carriers (Fig. 7B). Although loss of SRT2 resulted neither in major structural changes of the OXPHOS complex nor in differences in the activities of the mitochondrial electron transport complexes I and II, it did cause a decreased coupling of the ATP synthase complex as well as an increase of the mitochondrial ADP uptake via the ATP/ADP carrier proteins, thereby contributing to fine-tuning of mitochondrial energy metabolism.
MATERIALS AND METHODS
Plant Material and Growth Conditions
For mitochondria isolation, Arabidopsis (Arabidopsis thaliana; ecotype Columbia [Col-0]) seedlings were grown in liquid culture as described in Morgan et al. (2008). For growth on plates, Arabidopsis seeds were treated and grown the same way except that the medium was supplemented with 0.8% (w/v) phytoagar. For translation inhibition, 11-d-old seedlings were transferred to liquid one-half Murashige and Skoog medium containing 2% (w/v) Suc and 10 µg mL–1 cycloheximide and incubated for 4 h at room temperature (equivalent treatment for mock, but in absence of drug; Drechsel et al., 2013).
The srt2-1 (SALK_131994.45.80) and srt2-2 (SALK_149295.52.35) lines were obtained from the SALK collection (Ungerstedt et al., 2003). The screening for homozygous transfer DNA insertion lines was performed as described before (Finkemeier et al., 2005). Primer sequences are listed in Supplemental Table S6.
Plant Transformation
All vector constructs were verified by sequencing and transformed into Agrobacterium tumefaciens strain C58 followed by floral-dip transformation of Arabidopsis (Col-0) plants (Clough and Bent, 1998). Transformants were selected by germination of seeds on Murashige and Skoog-agar plates containing kanamycin (85 µm). Resistant plants were transferred to soil and propagated.
RNA Isolation and RT-PCR
Total RNA isolation and on-column DNaseI treatment was performed using the Universal RNA Purification Kit (EURx), according to the manufacturer’s instructions. Avian Myeloblastosis Virus reverse transcriptase native (EURx) was used for RT with dT20, followed by PCR amplification with listed oligonucleotides (Supplemental Table S6). RT-PCR products were run on 2% (w/v) agarose Tris-acetate (40 mm Tris, 20 mm acetate, 1 mm EDTA, pH 8.0) or 12% (w/v) native polyacrylamide gels (Tris-Gly native running buffer) and visualized by UV illumination upon ethidium bromide staining.
Analyses of AS Patterns
SRT2 splicing variants, derived from alternative 5′ splice site usage in intron 5, were PCR amplified from complementary DNA (primer sequences listed in Supplemental Table S6). PCR products had the expected sizes of 231 and 242 bp upon usage of the up- and downstream 5′ splice site, respectively. For PCR product quantitation, samples were analyzed with DNA 1000 chips using the Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. For sequence confirmation, RT-PCR products were cloned into pGEM-T vector (Promega) and sequenced (Eurofins).
GFP Construct
35S:SIR2i-GFP fusion construct was generated using the Gateway cloning system (Invitrogen) according to the manufacturer’s protocols. Entry clones were generated with the pENTR/SD/TOPO (Invitrogen) vector. For C-terminal GFP fusion, the TAIR10 annotated genomic sequence of SIR2.7 was amplified from Arabidopsis (Col-0) DNA using primers lacking stop codons as listed in Supplemental Table S6. LR reactions for in-frame recombination into pK7FWG2 vector (GFP fusion) were performed as described before (Karimi et al., 2002).
Protoplast Isolation and Confocal Laser Scanning Microscopy
Protoplast isolation was performed from 4-week-old Arabidopsis leaves after the tape sandwich method described by Wu et al. (2009). Staining with 200 nm MitoTracker Red (Invitrogen) was performed according to the manufacturer’s protocol. Imaging was performed with a spectral TCS SP5 MP confocal laser scanning microscope (Leica Microsystems) using an argon and DPSS laser, respectively, at an excitation wavelength of 488 (eGFP) and 561 nm (MitoTracker Red). The water immersion objective lens HCX PL APO 20.0 × 0.70 IMM UV was used for imaging in multitrack mode with line switching. eGFP and MitoTracker Red fluorescence was measured at 500 to 530 nm and 620 to 660 nm, respectively.
Heterologous Expression and Purification of Recombinant SRT2 Proteins
SRT2A and SRT2B complementary DNAs were amplified by PCR excluding the coding region for the 47-amino acid signal peptide. Primer sequences are listed in Supplemental Table S6. The PCR product was cloned after restriction digestion with BamHI and SalI in frame into the pQE30 vector (Qiagen), which allows expression and purification of the recombinant, N-terminally 6× His-tagged protein. Expression and purification of the recombinant protein were performed in BL21* cells (Invitrogen) as described in Finkemeier et al. (2005). Vector constructs were verified by sequencing.
Sirtuin Activity Assay
The SRT2 activity assay was performed as described in the manufacturer’s protocol of the SIRT-Glo assay (Promega) using 1 to 6 µg of recombinant SRT2 per reaction.
Antiserum Production
For antiserum production, a rabbit was immunized with 850 µg heterologously expressed and gel-purified SRT2A protein (Pineda). The antiserum was affinity purified against SRT2A protein after the protocol described in Tran et al. (2012).
Western-Blot Analyses
Proteins were blotted onto a nitrocellulose membrane and incubated overnight with primary antibodies as indicated in the text. Secondary anti-horseradish peroxidase antibody (ThermoFisher Scientific) was used in a 1:2,000 dilution followed by detection with SuperSignal West Dura enhanced chemiluminescent substrate (ThermoFisher Scientific).
Isolation of Organelles
Mitochondria were isolated from 10-d-old Arabidopsis seedlings grown in liquid cultures as described before (Morgan et al., 2008). The integrity of the outer membrane was determined by the latency of cytochrome c oxidation before and after addition of Triton-X100 (Sweetlove et al., 2007). Subfractionation of mitochondria was performed as described in detail in Finkemeier et al. (2005). Nuclei were isolated from 4-week-old Arabidopsis leaves as described previously (van Blokland et al., 1997).
Respiratory Measurements
Measurements of mitochondrial respiration were performed at 20°C in a Clark-type oxygen electrode (Oxygraph; Hansatech Instruments) in a basic medium (0.3 m mannitol, 5 mm KH2PO4, 3 mm MgSO4, 10 mm KCl, 0.1% [w/v] bovine serum albumin, pH 7.5) containing 10 mm pyruvate, 10 mm malate, 100 µm thiamine pyrophosphate, and 300 µm NAD+ as described before (Sweetlove et al., 2002). Oxygen consumption was recorded before and after addition of 1 mm ADP.
14C-ADP Uptake into Isolated Mitochondria
The uptake of 14C-ADP into isolated mitochondria was performed with slight modifications as described in Winkler et al. (1968) and Haferkamp et al. (2002). Mitochondrial protein (0.6 mg) was incubated for 5 min in the basic medium (0.3 m mannitol, 5 mm KH2PO4, 3 mm MgSO4, 10 mm KCl, 0.1% [w/v] bovine serum albumin, pH 7.5) containing 5 mm Gly, 10 mm malate, and 10 mm pyruvate to energize mitochondria. 14C-labeled ADP (0.1 µCi mL–1) was added together with unlabeled ADP to the sample in a final concentration of 0.2 mm. Samples were taken from the reaction precisely at the indicated time and rapidly separated from media by vacuum filtration on a filter (pore size, 0.22 µm; Millipore). The mitochondria were subsequently washed twice with basic medium. Incorporated radiolabel was determined by liquid scintillation counting.
ATP and ADP Contents
ATP and ADP contents of seedlings were determined luminometrically using the KinaseGlo reagent (Promega) for detection of ATP. Extraction of metabolites was performed as described before (Finkemeier et al., 2005). ADP was converted to ATP for 1 h at 37°C by addition of 1 µg pyruvate kinase (Sigma) and 32 mm phosphoenolpyruvate to the pH-adjusted (pH 7.5) metabolite extract.
Primary Metabolite Profiling by Gas Chromatography-Mass Spectrometry
Metabolite extraction, derivatization, and relative metabolite levels were determined using an established gas chromatography-mass spectrometry protocol as described previously (Roessner et al., 2001; Lisec et al., 2006). Metabolites were identified by comparison to database entries of authentic standards (Kopka et al., 2005; Schauer et al., 2005).
Two-Dimensional Blue-Native SDS-PAGE
Freshly isolated mitochondria (750 µg protein) were sedimented by centrifugation for 10 min at 23,700g. The pellet was resuspended in 75 µL of digitonin solubilization buffer. Blue-native gel electrophoresis and second gel dimension was carried out in a standard dual cooled gel electrophoresis chamber (Hoefer) with a gel dimension of 18 × 16 cm as described in Klodmann et al. (2011). Gels were either transferred to nitrocellulose membranes for western-blot analysis or stained with Coomassie colloidal blue. Protein spots were excised from gels and trypsin digested as described before (Morgan et al., 2008) and analyzed using LC-MS/MS as described below.
Protein Coimmunoprecipitation
For GFP coimmunoprecipitation, mitochondria isolated from full-length SRT2.1-GFP and SRT2.7-GFP (SRT2.7 presequence 1–67 amino acids; control) seedlings were sedimented and resuspended in immunoprecipitation buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10% [v/v] glycerol, 2 mm EDTA, 5 mm dithiothreitol, 1% [v/v] Triton-X100, 1 mm phenylmethylsulfonylfluoride, and protease inhibitor cocktail tablets [Roche]). The extract was precleared on Sepharose beads and subjected to immunoprecipitation with GFP-Trap Beads (Chromotek) overnight at 4°C. After incubation, the supernatant was discarded, and beads were washed thoroughly using IP buffer. Proteins were eluted using 0.1% (v/v) trifluoroacetic acid.
For antibody coimmunoprecipitation, affinity-purified SRT2 and citrate synthase (control) antibodies were covalently bound to protein A Sepharose (Invitrogen). Coimmunoprecipitation of SRT2 and interacting proteins was carried out as described above except that mitochondria isolated from wild-type seedlings were used. The eluted proteins were trypsin digested and analyzed by LC-MS/MS as described below.
Enrichment of Lys-Acetylated Peptides
Lys-acetylated peptides were enriched from trypsin-digested mitochondrial proteins as described before (Finkemeier et al., 2011).
Trypsin Digestion and LC-MS/MS Data Acquisition and Analysis
Proteins were denatured in 6 m urea/2 m thiourea, reduced with 1 mm dithiothreitol, and alkylated with 5 mm iodoacetamide. Proteins were digested overnight using 1 µg proteomics-grade trypsin (Roche) at 37°C. Digestion was stopped with 1% (w/v) formic acid. Peptides were desalted using Sep-Pak C18 cartridges (Waters), and peptide-containing eluates were evaporated using a speed vac. Dried peptides were redissolved in 2% (v/v) acetonitrile and 0.1% (v/v) trifluoroacetic acid before analysis. Samples were analyzed using either a Rheos Allegro UPLC (Flux Instruments) or an EASY-nLC 1000 (Thermo Fisher) coupled to an LTQ-Orbitrap XL or an Orbitrap Elite Mass Spectrometer (Thermo Fisher), respectively. Peptides were separated on 15-cm fritless, self-pulled fused silica emitters (15 cm × 0.75 µm) and packed in house with reversed-phase ReproSil-Pur C18-AQ 3-µm beads resin (Dr. Maisch GmbH). Peptides (1 µg for coimmunoprecipitations, 0.5 µg for blue-native PAGE gel spots) were loaded on the column and eluted for 120 min using a segmented linear gradient of 3% to 95% solvent B (80% [v/v] acetonitrile, 0.5% [v/v] acetic acid) at a flow rate of 250 nL min–1. Survey full-scan mass spectra were acquired in the Orbitrap analyzer. The scanned mass range was 300 to 1,650 m/z, at a resolution of 60.000 (Orbitrap XL) or 120.000 (Elite) at 400 m/z, respectively, and the seven (Orbitrap XL) or 20 (Elite) most intense ions were sequentially isolated, fragmented (collision-induced dissociation at 35 eV), and measured in the linear ion trap. Peptides with a charge of +1 or with unassigned charge state were excluded from fragmentation for tandem mass spectrometry, and dynamic exclusion of maximum 500 m/z values prevented repeated selection of selected masses for 90 s. Ions were accumulated to a target value of 106 for full Fourier transform-mass spectrometry in the Orbitrap and of 104 for tandem mass spectrometry in the linear ion trap.
Raw data were processed using MaxQuant software (version 1.3.0.5; http://www.maxquant.org/; Cox and Mann, 2008) and searched against The Arabidopsis Information Resource protein database (build TAIR10_pep_20101214; ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/), with trypsin specificity and a maximum of two missed cleavages at a protein and peptide false discovery rate of 1%. Carbamidomethylation of Cys residues were set as fixed, and oxidation of Met, N-terminal acetylation, and Lys acetylation were set as variable modifications. Label-free quantitation of coimmunoprecipitated proteins was also performed in MaxQuant, and subsequent quantitative statistical analyses were conducted similar to the method described by Hubner et al. (2010) in Perseus (version 1.3.0.4, http://www.maxquant.org/), using the parameters specified in Supplemental Table S7.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number SRT2 (At5g09230).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of the seven annotated SRT2 isoforms of Arabidopsis (TAIR10).
Supplemental Figure S2. Precursor mRNA of SRT2 is subject to alternative spicing.
Supplemental Figure S3. Characterization of SRT2 knockout lines.
Supplemental Figure S4. Amino acid sequence alignment of the different protein isoforms of Arabidopsis SRT2 with the mitochondrial human sirtuins (HsSIRT3, HsSIRT4, and HsSIRT5).
Supplemental Figure S5. Lys acetylation sites on the ADP/ATP translocator proteins AAC1 to AAC3.
Supplemental Table S1. Characteristics of putative proteins encoded by the seven SRT2 splice forms as annotated in TAIR10.
Supplemental Table S2. Proteins identified from excised spots from two-dimensional blue-native SDS PAGE (Fig. 7) as determined by LC-MS/MS.
Supplemental Table S3. Significantly enriched proteins in label-free coimmunoprecipitation experiments.
Supplemental Table S4. Lys acetylation sites of proteins.
Supplemental Table S5. Full gas chromatography-mass spectrometry metabolite profile of shoot material from wild-type and srt2-1 seedlings.
Supplemental Table S6. Primer list.
Supplemental Table S7. MaxQuant settings for data analysis and label-free quantification.
Acknowledgments
We thank Axel Imhof (Ludwig-Maximilians-Universität Munich) for measuring time at the LTQ Orbitrap when the machine at the Ludwig-Maximilians-Universität biocenter was defective.
Glossary
- AS
alternative splicing
- NMD
nonsense-mediated decay
- RT
reverse transcription
- IC50
50% inhibition of initial activity
- OXPHOS
oxidative phosphorylation
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- Col-0
ecotype Columbia
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft, Germany (Emmy Noether Programme FI–1655/1–1 and Research Unit 804 to I.F.) and the Max Planck Gesellschaft (to A.R.F., P.A.P., P.J.B., M.M.).
These authors contributed equally to the article.
Present address: Max Planck Institute for Plant Breeding Research, Carl von Linné Weg 10, 50829 Cologne, Germany.
Present address: Leibniz University Hannover, Institute of Botany, 30419 Hannover, Germany.
Present address: Eidgenössiche Technische Hochschule Zürich, Institute of Biochemistry, Schafmattstrasse 18, 8093 Zurich, Switzerland.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. (2007) Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem 282: 33583–33592 [DOI] [PubMed] [Google Scholar]
- Brandt RB, Laux JE, Yates SW. (1987) Calculation of inhibitor Ki and inhibitor type from the concentration of inhibitor for 50% inhibition for Michaelis-Menten enzymes. Biochem Med Metab Biol 37: 344–349 [DOI] [PubMed] [Google Scholar]
- Brustovetsky N, Klingenberg M. (1994) The reconstituted ADP/ATP carrier can mediate H+ transport by free fatty acids, which is further stimulated by mersalyl. J Biol Chem 269: 27329–27336 [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, Rätsch G, Wachter A. (2013) Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, Denu JM. (2012) Sirtuin catalysis and regulation. J Biol Chem 287: 42419–42427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Goodman M, Lamkemeyer P, Kandlbinder A, Sweetlove LJ, Dietz KJ. (2005) The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J Biol Chem 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Finkemeier I, Laxa M, Miguet L, Howden AJ, Sweetlove LJ. (2011) Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol 155: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2011) The logic linking protein acetylation and metabolism. Cell Metab 14: 151–153 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Fernie AR, Neuhaus HE. (2011) Adenine nucleotide transport in plants: much more than a mitochondrial issue. Trends Plant Sci 16: 507–515 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Hackstein JHP, Voncken FGJ, Schmit G, Tjaden J. (2002) Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur J Biochem 269: 3172–3181 [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126: 941–954 [DOI] [PubMed] [Google Scholar]
- Hartl M, Finkemeier I. (2012) Plant mitochondrial retrograde signaling: post-translational modifications enter the stage. Front Plant Sci 3: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen P, Wagner SA, Weinert BT, Sharma S, Bacinskaja G, Rehman M, Juffer AH, Walther TC, Lisby M, Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol Cell Proteomics 11: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E, et al. (2013) SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging 5: 835–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C, Liu Z. (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50: 875–885 [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. (2012) Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Klingenberg M. (2008) The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta 1778: 1978–2021 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP. (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, et al. (2013) SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 50: 686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, et al. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep 2: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MJ, Lehmann M, Schwarzländer M, Baxter CJ, Sienkiewicz-Porzucek A, Williams TC, Schauer N, Fernie AR, Fricker MD, Ratcliffe RG, et al. (2008) Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol 147: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, He W, Verdin E. (2012) Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem 287: 42436–42443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, et al. (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci USA 110: 6601–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CF, Zerweck J, Schutkowski M, et al. (2013) An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun 4: 2327. [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC. (2007) Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl C, Stauffer E, Kahles A, Wagner G, Drechsel G, Rätsch G, Wachter A. (2012) Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. Plant Cell 24: 4360–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K, Wang J, Diagouraga B, Khochbin S. (2011) The tale of protein lysine acetylation in the cytoplasm. J Biomed Biotechnol 2011: 970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA. (2010) Sirtuins. Biochim Biophys Acta 1804: 1565–1566 [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM. (2004) Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem 279: 40122–40129 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, König AC, Sweetlove LJ, Finkemeier I. (2012) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Sebastián C, Satterstrom FK, Haigis MC, Mostoslavsky R. (2012) From sirtuin biology to human diseases: an update. J Biol Chem 287: 42444–42452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, Deng CX, Choudhary C. (2012) Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS ONE 7: e50545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH. (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Taylor NL, Leaver CJ. (2007) Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol Biol 372: 125–136 [DOI] [PubMed] [Google Scholar]
- Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A, Moorhead GB. (2012) Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J 71: 263–272 [DOI] [PubMed] [Google Scholar]
- Ungerstedt JS, Blömback M, Söderström T. (2003) Nicotinamide is a potent inhibitor of proinflammatory cytokines. Clin Exp Immunol 131: 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blokland R, ten Lohuis M, Meyer P. (1997) Condensation of chromatin in transcriptional regions of an inactivated plant transgene: evidence for an active role of transcription in gene silencing. Mol Gen Genet 257: 1–13 [DOI] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LWS, Haigis MC. (2010) Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 35: 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CZ, Gao F, Wu JG, Dai JL, Wei CH, Li Y. (2010) Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51: 1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WR, Olsen JV, Jensen LJ, Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal 4: ra48. [DOI] [PubMed] [Google Scholar]
- Winkler HH, Bygrave FL, Lehninger AL. (1968) Characterization of the atractyloside-sensitive adenine nucleotide transport system in rat liver mitochondria. J Biol Chem 243: 20–28 [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Oh MH, Schwarz EM, Larue CT, Sivaguru M, Imai BS, Yau PM, Ort DR, Huber SC. (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol 155: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Poirier Y. (2012) The protein acetylome and the regulation of metabolism. Trends Plant Sci 17: 423–430 [DOI] [PubMed] [Google Scholar]
- Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. (2006) The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J 47: 49–62 [DOI] [PubMed] [Google Scholar]