The C-linker domain of a K+ channel is required for the control of channel gating via its first α-helix located just below the channel pore and for the proper folding of the channel.
Abstract
Shaker K+ channels form the major K+ conductance of the plasma membrane in plants. They are composed of four subunits arranged around a central ion-conducting pore. The intracellular carboxy-terminal region of each subunit contains several regulatory elements, including a C-linker region and a cyclic nucleotide-binding domain (CNBD). The C-linker is the first domain present downstream of the sixth transmembrane segment and connects the CNBD to the transmembrane core. With the aim of identifying the role of the C-linker in the Shaker channel properties, we performed subdomain swapping between the C-linker of two Arabidopsis (Arabidopsis thaliana) Shaker subunits, K+ channel in Arabidopsis thaliana2 (KAT2) and Arabidopsis thaliana K+ rectifying channel1 (AtKC1). These two subunits contribute to K+ transport in planta by forming heteromeric channels with other Shaker subunits. However, they display contrasting behavior when expressed in tobacco mesophyll protoplasts: KAT2 forms homotetrameric channels active at the plasma membrane, whereas AtKC1 is retained in the endoplasmic reticulum when expressed alone. The resulting chimeric/mutated constructs were analyzed for subcellular localization and functionally characterized. We identified two contiguous amino acids, valine-381 and serine-382, located in the C-linker carboxy-terminal end, which prevent KAT2 surface expression when mutated into the equivalent residues from AtKC1. Moreover, we demonstrated that the nine-amino acid stretch 312TVRAASEFA320 that composes the first C-linker α-helix located just below the pore is a crucial determinant of KAT2 channel activity. A KAT2 C-linker/CNBD three-dimensional model, based on animal HCN (for Hyperpolarization-activated, cyclic nucleotide-gated K+) channels as structure templates, has been built and used to discuss the role of the C-linker in plant Shaker inward channel structure and function.
In plants, potassium channels from the Shaker family dominate the plasma membrane (PM) conductance to K+ in most cell types and play crucial roles in sustained K+ transport (Blatt et al., 2012; Hedrich, 2012; Sharma et al., 2013). Plant Shaker channels, like their homologs in animals (Craven and Zagotta, 2006; Wahl-Schott and Biel, 2009), belong to the six transmembrane-one pore (6TM-1P) cation channel superfamily. Functional channels are tetrameric proteins arranged around a central pore (Daram et al., 1997; Urbach et al., 2000; Dreyer et al., 2004). These channels can result from the assembly of Shaker subunits encoded by the same gene (homotetramers) or by different Shaker genes (heterotetramers). Heterotetramerization has been extensively reported within the inwardly rectifying Shaker channel group (five members in Arabidopsis [Arabidopsis thaliana]) and increased channel functional diversity (Jeanguenin et al., 2008; Lebaudy et al., 2008a).
Based on in silico sequence analyses, plant Shaker subunits display a short cytosolic N-terminal domain, followed by the 6TM-1P hydrophobic core, and a long C-terminal cytosolic region in which several domains can be identified. The first one, named C-linker (about 80 amino acids in length), is followed by a cyclic nucleotide-binding domain (CNBD), an ankyrin domain (involved in protein-protein interaction; Lee et al., 2007, Grefen and Blatt, 2012), and a domain named KHA (Ehrhardt et al., 1997) rich in hydrophobic and acidic residues. Sequence analysis of plant Shaker channels indicates that, among these cytosolic domains, the highest levels of similarity are displayed by the C-linker and the CNBD domains. Interestingly, both domains are also highly conserved in some members from the animal K+ channel superfamily, like Hyperpolarization-activated, cyclic nucleotide-gated K+ channel (HCN), K+ voltage-gated channel, subfamily H (KCNH), and Cyclic-nucleotide-gated ion channel (CNGC). In these animal 6TM-1P channels, the roles of C-linker and CNBD domains have been extensively investigated via crystal structure analyses (Zagotta et al., 2003; Brelidze et al., 2012), whereas plant Shaker channels are still poorly characterized at the structural level (Dreyer et al., 2004; Gajdanowicz et al., 2009; Naso et al., 2009; Garcia-Mata et al., 2010).
Aiming at investigating the structure-function relationship of plant Shaker channels, we have used the Arabidopsis Shaker subunit K+ channel in Arabidopsis thaliana2 (KAT2) as a model. We developed a subdomain-swapping strategy between KAT2 and another Shaker subunit displaying distinctive features, Arabidopsis thaliana K+ rectifying channel1 (AtKC1). The KAT2 subunit can form homomeric or heteromeric inwardly rectifying K+ channels at the PM and has been shown to be strongly expressed in guard cells, where it provides a major contribution to the membrane conductance to K+ (Pilot et al., 2001; Lebaudy et al., 2008b). In contrast, the behavior of AtKC1 is more complex. In planta, this subunit is coexpressed with other inwardly rectifying Shaker subunits, including KAT2, in different plant tissues (Jeanguenin et al., 2011), and in roots, direct evidence has been obtained that AtKC1 is involved in functional heterotetrameric channel formation with AKT1 (Reintanz et al., 2002; Honsbein et al., 2009). However, experiments performed in tobacco (Nicotiana tabacum) mesophyll protoplasts have revealed that when expressed alone, AtKC1 is entrapped in the endoplasmic reticulum (ER). However, in tobacco protoplasts and Xenopus laevis oocytes, coexpression of AtKC1 with KAT2 or other inwardly rectifying Shaker subunits (AKT1, KAT1, or AKT2) gives rise to functional heteromeric channels (Duby et al., 2008; Jeanguenin et al., 2011). In Arabidopsis, it is interesting that evidence of the AtKC1 retention in the ER compartment, in the absence of other Shaker subunits, is lacking, since in the native tissues, AtKC1 is always expressed with its inward partners, with which it is able to form heteromeric channels.
Here, we took advantage of the unique behavior of AtKC1 when expressed in heterologous systems to investigate the structure-function relationship of the C-linker of KAT2 by sequence exchange between these two channel subunits and by site-directed mutagenesis. The C-linker domain, which, to our knowledge, had never been studied as such in plant Shaker channels before, could be predicted to play crucial roles in channel properties due to its strategic location between the channel transmembrane core and the cytoplasmic CNBD domain. The resulting KAT2-AtKC1 chimeras were expressed in tobacco mesophyll protoplasts and in X. laevis oocytes for investigating their subcellular localization and measuring their activity at the cell membrane. Here, we show that two amino acids present in the C-linker are important for channel subcellular location and that a stretch of nine amino acids forming a short helix just below the membrane, downstream of the sixth transmembrane segment of the channel hydrophobic core, is involved in channel gating. The obtained experimental results are discussed in relation with a KAT2 C-linker/CNBD three-dimensional (3D) model based on animal HCN channels as structure templates.
RESULTS
The C-Linker Plays a Role in Channel Location and Activity at the PM
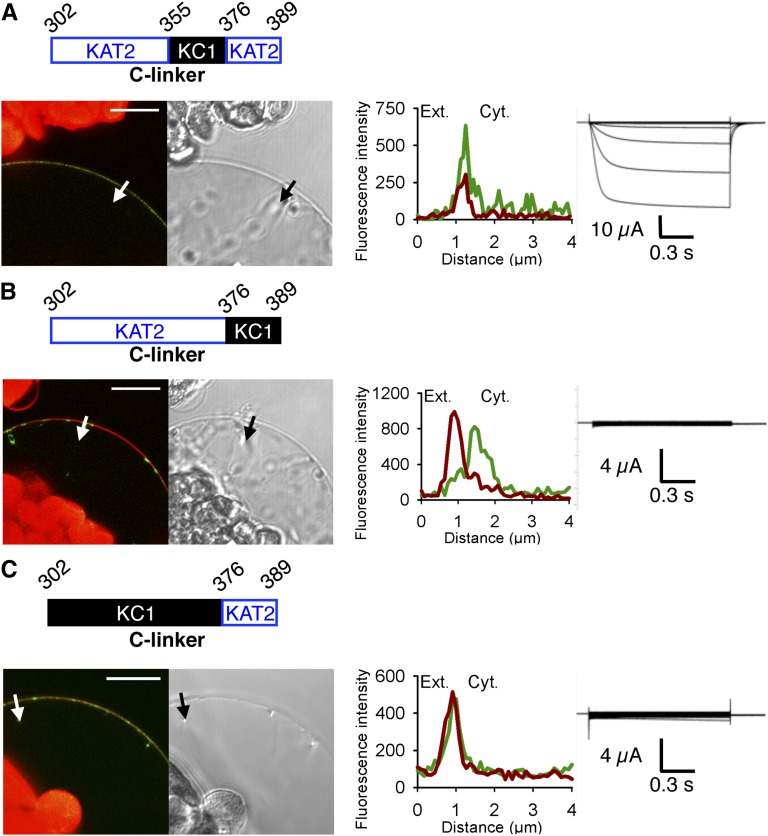
Aiming at investigating the role of the C-linker domain in plant Shaker channels, we performed an exchange of the C-linker sequence between KAT2 and AtKC1. The subcellular localization of each construct fused to GFP was assessed by confocal microscopy after transient expression in tobacco protoplasts. Protoplast PM was stained with FM4-64, which is a specific marker of this membrane during short incubations (Hosy et al., 2005). By performing GFP and FM4-64 fluorescence intensity profiles across the protoplast PM, it can be concluded that the chimera fused to GFP was targeted to the PM if GFP (in green) and FM4-64 (in red) signal peaks coincide. If the two peaks do not overlap, the Shaker channel tagged with GFP was not localized to the PM but detected in intracellular compartments. Further analysis of the latter by 3D reconstruction revealed that GFP signal was accumulated in a network-like structure characteristic of the ER. In parallel, the functionality of each chimeric subunit (without GFP tag) was checked by heterologous expression in X. laevis oocytes and current recordings.
The native KAT2 channel subunit that forms functional homotetramers when expressed in tobacco protoplasts (Xicluna et al., 2007) as well as in X. laevis oocytes (Pilot et al., 2001) was first used as a control. As expected, the analyses clearly indicated that the KAT2 channel was both targeted to the PM when expressed in tobacco protoplasts (96% of GFP-positive protoplasts [n = 219] with overlapping GFP and FM4-64 fluorescence profiles) and functional at the PM when expressed in oocytes (Fig. 1A1). Moreover, western analyses performed on oocyte membrane fractions and confocal imaging analyses provided confirmation that KAT2 is expressed at the PM in this latter system, as expected (Fig. 1A2).
Figure 1.
The C-linker domain is important for PM expression and channel functionality of the Shaker K+ channel KAT2 from Arabidopsis. A, Analyses of the native KAT2 subunit after transient expression in tobacco protoplasts and X. laevis oocytes. A1, The native KAT2 was studied by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes. Shown from left to right are a schematic representation of the C-linker sequence of the KAT2 subunit; a representative confocal microscopy image of GFP and FM4-64 fluorescence in a protoplast expressing the channel-GFP construct, together with the bright-field image of the corresponding protoplast (bar = 10 µm; chlorophyll autofluorescence in chloroplasts [red] is also seen in the confocal images); GFP (green) and FM4-64 (red) fluorescence intensity profiles across the protoplast membrane (and a pocket of cytoplasm; the white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm [Ext. and Cyt. indicate external and cytosolic sides, respectively]); and representative current traces recorded in oocytes expressing the channel construct (without GFP tag) in 100 mm K+ bath solution. Applied activation membrane voltages ranged from +40 to −170 mV (increments of 15 mV; holding potential, 0 mV; deactivation potential, −40 mV). A2, Further analyses were performed on oocytes expressing KAT2-GFP. Shown from left to right are linear Z stacks obtained at the upper pole of the oocyte (green signals correspond to GFP fluorescence and red signals correspond to FM4-64 fluorescence); GFP (green) and FM4-64 (red) fluorescence intensity profiles across the oocyte membrane (the white arrow on the confocal image marks the position of the analyzed section crossing the PM and the cytoplasm; bar = 10 µm); and protein gel blot analysis of PMs and internal membranes (IM) from Shaker-GFP-injected oocytes probed with a monoclonal antibody to GFP. Note the presence of monomers (108 kD) and dimers (216 kD) in the PM fraction. B, Alignment of the C-linker sequences of Shaker subunits KAT2 and AtKC1 from Arabidopsis. Identical residues are displayed with a gray background. C, Left, amino acid sequences of the C-linker upstream junction with the channel sixth transmembrane segment (S6) and downstream junction with the CNBD in the KAT2*AtKC1-C-linker chimera. Blue and black colors refer to KAT2 and AtKC1 sequences, respectively. Right, pictogram of the KAT2*AtKC1-C-linker chimera structure in which the sequences from KAT2 (blue) and AtKC1 (black) are indicated. D to F, Surface expression and activity at the PM of the chimeric channels (obtained by sequence swapping between KAT2 and AtKC1): KAT2*AtKC1-C-linker chimera (D), KAT2*AtKC1(302-354) (E), and KAT2*AtKC1(355-389) (F), investigated by the expression of Shaker-GFP fusions in tobacco mesophyll protoplasts and Shaker subunits (without GFP tag) in X. laevis oocytes as described in A1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.
Sequence alignment of KAT2 and AtKC1 C-linker domains, which are about 80 amino acids in length, revealed 45% amino acid identity (Fig. 1B). In a first experiment, the C-linker domain of KAT2 was replaced by the corresponding domain from AtKC1 (Fig. 1C). This chimera, named KAT2*AtKC1-C-linker, did not reach the PM in tobacco protoplasts (0% of GFP-positive protoplasts [n = 75]) and did not give rise to any significant exogenous currents when expressed in oocytes (Fig. 1D). This suggested that the AtKC1 C-linker contained determinants that were not compatible with KAT2 surface expression. This hypothesis was further investigated by subdividing the KAT2 C-linker into shorter regions and replacing these regions by the corresponding ones from AtKC1. The chimera containing the N-terminal half of the AtKC1 C-linker, which corresponds to the amino acid stretch 302 to 354 of KAT2 (Fig. 1E), was observed at the PM in 81% of the GFP-positive protoplasts (n = 57). However, K+-dependent currents were not observed in oocytes expressing this chimera in the tested voltage interval (+40 to −170 mV; Fig. 1E). On the other hand, a chimera harboring the second half of the AtKC1 C-linker, corresponding to amino acids 355 to 389 of KAT2, displayed intracellular localization in protoplasts (PM localization observed in 0% of GFP-positive cells [n = 55]; Fig. 1F). In agreement with such an absence of localization to the PM, the chimeric channel did not display activity at the PM when expressed in oocytes (Fig. 1F). Thus, the whole set of results suggested that the C-terminal part of the wild-type KAT2 C-linker domain contains residues playing an important role in channel surface expression, while the N-terminal part of this domain contains residues of crucial importance for channel functionality.
Identification of C-Linker Residues Playing a Role in Channel Localization to the PM
Residues playing a role in channel surface expression were looked for in the C-terminal part of the KAT2 C-linker (amino acids 355–389) by KAT2-AtKC1 sequence swapping and site-directed mutagenesis (Fig. 2). A KAT2 chimera in which the N-terminal part (amino acids 355–375) of this region was replaced by the corresponding sequence from AtKC1 was found to be localized to the PM (in 89% of the GFP-positive protoplasts [n = 85]) and to give rise to KAT2-like K+ currents in oocytes (Fig. 2A). In contrast, a KAT2 chimera in which the C-terminal part (amino acids 376–389) of this region was replaced by the corresponding sequence from AtKC1 neither reached the PM (n = 88) nor mediated K+ currents in oocytes (Fig. 2B). In agreement with this result, a KAT2 chimera in which the whole C-linker region (amino acids 302–375) upstream from this extreme C-terminal sequence (amino acids 376–389) was replaced by the corresponding region from AtKC1 (Fig. 2C) displayed localization to the PM (71% of the GFP-positive protoplasts [n = 77]). The absence of exogenous currents within the range of membrane potentials applied (+40 to −170 mV) in oocytes expressing this chimera (Fig. 2C) could be ascribed to the fact that the N-terminal part of the C-linker region contains elements that are crucial for channel functionality, a conclusion already supported by the data from Figure 1E.
Figure 2.
The extreme C-terminal part of the C-linker plays a role in KAT2 surface expression. Three different channel chimeras (A–C) were obtained by sequence swapping between KAT2 and AtKC1. The regions from KAT2 and AtKC1 are depicted in blue and black, respectively. Surface expression and activity at the PM of each chimeric channel were investigated by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes as described in the legend to Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.
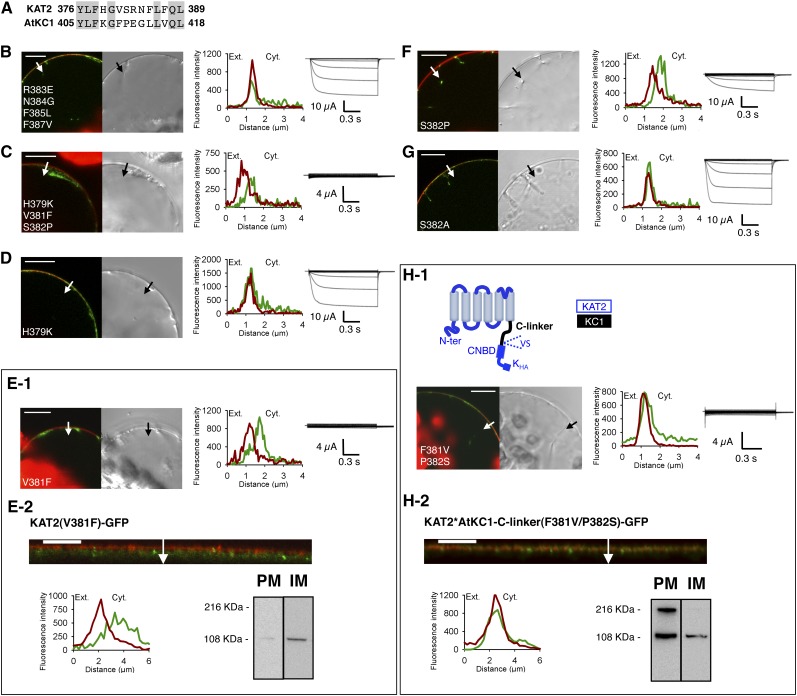
Altogether, these results indicated that the extreme C terminus of the C-linker from AtKC1, which differs from the corresponding region of KAT2 by only seven different amino acids (Fig. 3A), caused intracellular retention. To identify which mutations, among the seven that the swapping of the distal region of the C-linker introduced in KAT2, were responsible for the absence of localization to the PM, two KAT2 mutant proteins were generated by directed mutagenesis and named KAT2(R383E/N384G/F385L/F387V) and KAT2(H379K/V381F/S382P). KAT2(R383E/N384G/F385L/F387V) was found to be localized to the PM (80% of GFP-positive protoplasts [n = 92]) and to mediate large voltage-dependent K+ currents when expressed in oocytes (Fig. 3B). In contrast, KAT2(H379K/V381F/S382P) displayed intracellular localization (n = 97) and absence of K+ transport activity in oocytes (Fig. 3C). This indicated that at least one of the three mutated residues in this construct, H379K, V381F, and S382P, was crucial for proper channel trafficking.
Figure 3.
Point mutations V381F and S382P in the KAT2 C-linker result in impaired surface expression of KAT2. A, Alignment of the amino acid stretch 376 to 389 of KAT2 (identified as involved in KAT2 trafficking; Fig. 2) with the corresponding stretch (amino acids 405–418) of AtKC1. Gray background denotes identical residues. B to G, Search for amino acids of the KAT2 C-linker region affecting KAT2 channel surface expression when substituted by the amino acids at the corresponding positions in the AtKC1 C-linker. Amino acid substitution was carried out by site-directed mutagenesis based on the sequence alignments shown in A. Six KAT2 mutants were obtained: KAT2(R383E/N384G/F385L/F387V) (B), KAT2(H379K/V381F/S382P) (C), KAT2(H379K) (D), KAT2(V381F) (E), KAT2(S382P) (F), and KAT2(S382A) (G). H, Introduction of the two mutations F381V and P382S in the channel chimera KAT2*AtKC1-C-linker in order to replace the AtKC1 Phe and Pro residues by the KAT2 corresponding residues Val and Ser leads to the PM localization of the mutated chimera. A pictogram describing the mutated chimera, named KAT2*AtKC1-C-linker(F381V/P382S), is provided at the top of H1 (with sequences from KAT2 and AtKC1 in blue and sequence from AtKC1 in black). In B to D, E1, F, G, and H1, surface expression and activity at the PM of mutated channels were investigated by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes, as described in the legend to Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm. In E2 and H2, further analyses were performed in oocytes expressing KAT2(V381F) and KAT2*AtKC1-C-linker(F381V/P382S) fused to GFP to check their subcellular localization by confocal imaging and protein gel-blot analysis of cellular fractions, as described in the legend to the Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.
The three mutations H379K, V381F, and S382P were then individually introduced in KAT2 by site-directed mutagenesis, and the effect of each of them on channel localization to the PM was analyzed. The KAT2(H379K) mutant behaved similarly to KAT2 when expressed in protoplasts (PM expression observed in 96% of GFP-positive cells [n = 91]) as well as in oocytes (large K+ currents recorded in the oocytes; Fig. 3D). These results indicated that the H379K mutation was without any significant effect on channel localization to the PM.
In contrast, the KAT2(V381F) mutant exhibited intracellular retention in tobacco protoplasts (n = 96) and did not give rise to any exogenous K+ currents in oocytes (Fig. 3E1). To further analyze the subcellular localization of KAT2(V381F)-GFP fusion in tobacco protoplasts, a 3D reconstruction of the GFP fluorescence was carried out, showing that the mutant subunit was accumulated in a network-like structure characteristic of the ER (compare Supplemental Video S1, which shows a tobacco protoplast expressing KAT2-GFP located to the PM, and Supplemental Video S2). The subcellular localization of KAT2(V381F)-GFP was also checked in oocytes. This was achieved both by confocal microscopy of the fluorescence signal and by western-blot analysis of membrane fractions enriched either in PM or in internal membranes (Fig. 3E2). Both approaches support the conclusion that the KAT2(V381F) construct was retained in internal membranes, in agreement with the absence of K+ exogenous current in oocytes expressing this mutant subunit.
The third mutant, KAT2(S382P), was not detectable at the PM (n = 69) but gave rise to some currents when expressed in oocytes (Fig. 3F). The fact that the currents recorded in oocytes expressing the KAT2(S382P) mutant were of smaller amplitude than those recorded in oocytes expressing the wild-type KAT2 channel (compare Figs. 1A1 and 3F) was consistent with the data obtained in protoplasts showing that the S382P mutation hampered channel localization to the PM. In contrast, mutating the Ser in position 382 to an Ala in KAT2 was found to be without any significant effect on channel localization to the PM in protoplasts (97% of GFP-positive tobacco cells [n = 199]) and channel activity (current magnitude) in oocytes (Fig. 3G), suggesting that phosphorylation of KAT2 Ser-382 is not required for efficient targeting of the channel to the membrane. In conclusion, these experiments indicated that KAT2 localization to the PM was strongly impaired by the V381F mutation and probably also by the S382P mutation.
Once the two mutations V381F and S382P were identified as preventing KAT2 surface expression, we tested whether the inverse mutations F381V and P382S in the KAT2*AtKC1-C-linker chimera (previously shown to be unable to reach the PM when expressed in tobacco protoplasts; compare Fig. 1, C and D) could restore localization to the PM. The two mutations F381V and P382S were thus introduced into this chimera, and the resulting construct was named KAT2*AtKC1-C-linker(F381V/P382S). This chimera was localized to the PM in protoplasts (Fig. 3H1; 85% of GFP-positive tobacco cells [n = 144]; for the 3D reconstruction of the transformed protoplast, see Supplemental Video S3), providing further evidence that the two residues at positions 381 and 382 are essential for proper channel localization to the PM. It should be noted that expression of the KAT2*AtKC1-C-linker(F381V/P382S) chimera in X. laevis oocytes did not give rise to exogenous K+ currents (Fig. 3H1). Moreover, since confocal microscopy and western-blot analyses provided evidence that this chimera was localized to the PM in oocytes as well (Fig. 3H2), we concluded that it did not form functional channels in the tested voltage interval (+40 to −170 mV). Such an absence of activity was consistent with the previous data (Figs. 1E and 2C), supporting the hypothesis that the N-terminal part of the C-linker contains residues of crucial importance for channel functionality.
The Aromatic Ring Side Chain of Phe Introduced at Position 381 of KAT2 Prevents KAT2 Surface Expression
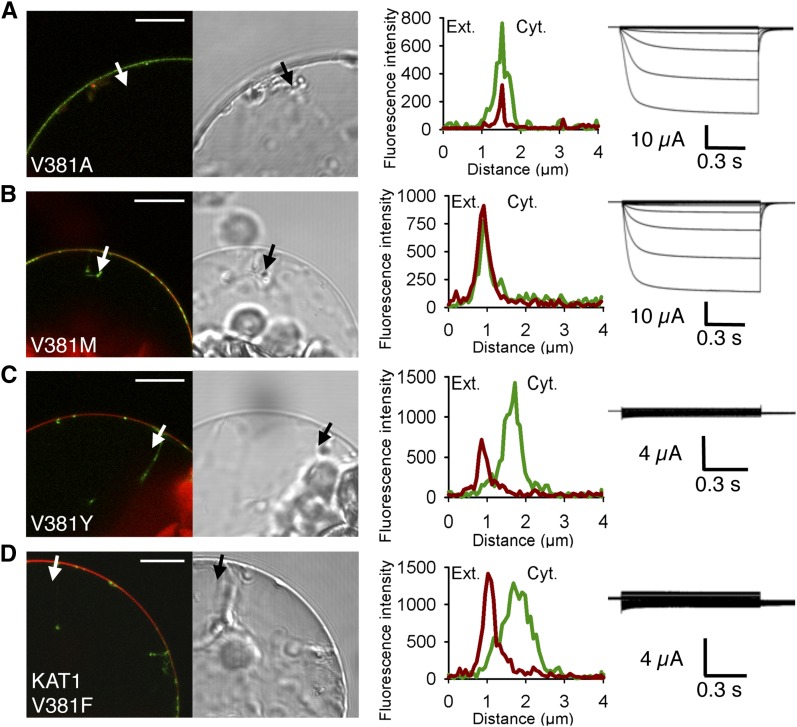
The dramatic effect of the V381F mutation on channel localization to the PM raised the question of whether any mutation introduced at this position results in similar deleterious effects. Different amino acids with different sizes and functional groups were thus introduced in place of Val-381. First, Val-381 was replaced by the much smaller aliphatic residue, Ala. The KAT2(V381A) mutant was well targeted to the PM (97% of GFP-positive protoplasts studied [n = 183]) and mediated large K+ currents in oocytes (Fig. 4A). Then, to substantially increase the volume of the side chain, we performed a Val-to-Met mutation (V381M). Again, the mutated channel was expressed to the PM (92% of GFP-positive protoplasts studied [n = 73]) and functional (electrically active) in oocytes (Fig. 4B). We then replaced Val-381 by Tyr, an amino acid that displays an aromatic ring like Phe, the amino acid initially identified as inducing strong intracellular retention of the channel when substituted for Val-381 in KAT2. Like the V381F mutation, V381Y totally inhibited KAT2 localization to the PM (n = 72; Fig. 4C), indicating that the presence of any aromatic ring at position 381 impaired KAT2 trafficking.
Figure 4.
Substitution of the C-linker Val residue by the aromatic amino acid Tyr in KAT2 and another Shaker channel from Arabidopsis, KAT1, results in impaired channel targeting to the PM. A to C, Substitution of Val-381 by Ala (A), Met (B), or Tyr (C) in KAT2. D, Substitution of Val-381 by Tyr in KAT1. Surface expression and activity at the PM of the mutated channels were investigated by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes, as described in the legend to Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.
Interestingly, such a V381F mutation was found to also impact the surface expression of another inwardly rectifying Shaker subunit, KAT1. Indeed, the mutated channel KAT1(V381F) showed intracellular localization (n = 64) in tobacco protoplasts and did not give rise to exogenous K+ currents in oocytes (Fig. 4D).
The Diacidic Motif Present Just Downstream of the C-Linker Domain Is Not Essential for KAT2 Channel Localization to the PM
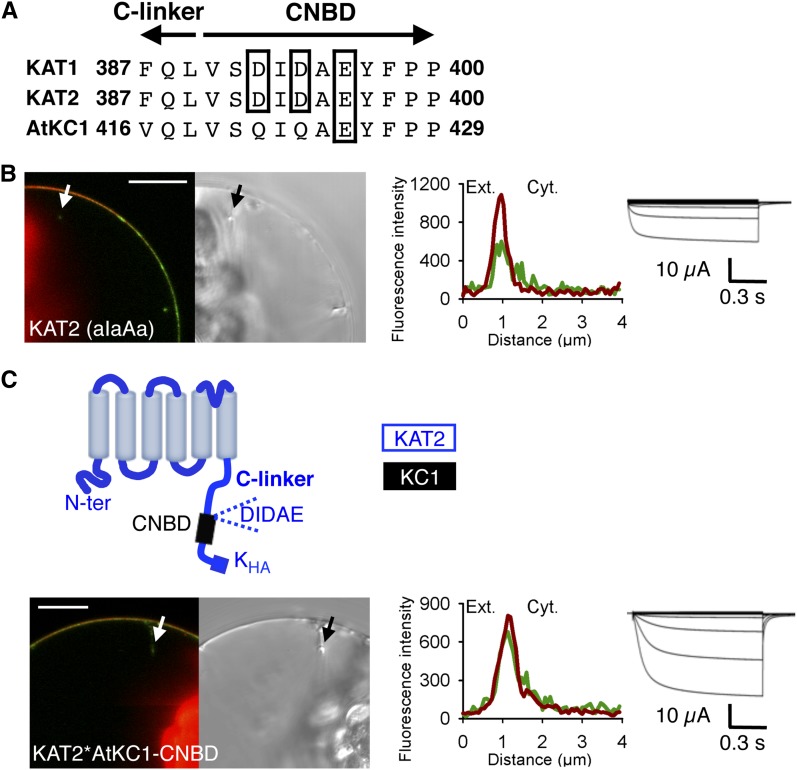
Diacidic motifs are ER export signals involved in the targeting of many PM proteins (Mikosch and Homann, 2009). Like KAT1, KAT2 displays a diacidic (triacidic) motif, DIDAE, just downstream of the C-linker, at the beginning of the channel CNBD (Fig. 5A). In KAT1, the DIDAE motif has been identified as essential for channel targeting to the PM (Mikosch et al., 2009). Results consistent with this conclusion (Supplemental Fig. S1, A and B) were obtained in our experiments, following replacement of each of the acidic residues of the KAT1 DIDAE motif by Ala (DIDAE mutated into aIaAa). The same mutations were then introduced in KAT2, giving a mutant named KAT2(aIaAa), in order to assess the contribution of the DIDAE motif to KAT2 surface expression. In contrast to the KAT1(aIaAa) mutant channel (Supplemental Fig. S1B), KAT2(aIaAa) was localized to the PM in tobacco protoplasts (81% of the GFP-positive protoplasts analyzed [n = 98]) and active at the PM in oocytes (Fig. 5B).
Figure 5.
KAT2 surface expression is not abolished by deletion of its triacidic motif DIDAE or substitution of its CNBD by that of AtKC1. A, Alignment of KAT1, KAT2, and AtKC1 sequences at the junction between the C-linker and the CNBD domains. Acidic amino acids of the triacidic motif DIDAE are boxed. Arrows indicate the C-linker and CNBD sequences. B, Mutation of the triacidic motif DIDAE into aIaAa in KAT2 does not prevent channel surface expression. C, Surface expression of the chimera obtained by replacing, in KAT2, the CNBD sequence downstream of the DIDAE motif by the corresponding sequence from AtKC1. A pictogram of the resulting chimera, displaying the sequences from KAT2 (blue) and AtKC1 (black), is provided at the top. Surface expression and activity at the PM of the KAT2(aIaAa) mutant (B) and the KAT2*AtKC1-CNBD chimera (C) were investigated by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes, as described in the legend to Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.
We then investigated whether the DIDAE motif could act as an ER export signal in the Shaker subunit AtKC1, which does not display this motif and is strongly retained in the ER when expressed in tobacco protoplasts (Duby et al., 2008; Jeanguenin et al., 2011). Sequence alignment indicates that AtKC1 displays QIQAE in place of the DIDAE motif (Fig. 5A). This QIQAE stretch was mutated into DIDAE, and the resulting mutant was named AtKC1(DIDAE). In agreement with previous analyses, our experiments indicated that the native AtKC1 subunit is indeed unable to reach the PM in tobacco protoplasts (n = 90; Supplemental Fig. S1C; for the 3D reconstruction of the transformed protoplast, see Supplemental Video S4). Like the native AtKC1, the AtKC1(DIDAE) mutant did not display surface expression (n = 58; Supplemental Fig. S1D), indicating that the DIDAE motif is not sufficient to target the channel to the PM in tobacco protoplasts.
In a subsequent experiment aimed at investigating the role of the channel CNBD independently of the DIDAE motif present at the N-terminal extremity of this domain, an 84-amino acid sequence from AtKC1 comprising the whole CNBD of this channel excepting its N-terminal QIQAE motif was introduced into KAT2 in place of the corresponding region (amino acids 398–481 in KAT2; Fig. 5C). The resulting chimera, named KAT2*AtKC1-CNBD, was found to be targeted to the PM in tobacco protoplasts (in 59% of GFP-positive tobacco cells [n = 63]) and to give rise to KAT2-like K+ transport activity in oocytes (Fig. 5C). Altogether, these results suggested that the CNBD does not play any decisive role in the property of AtKC1 to remain in the ER or in that of KAT2 to be targeted to the PM in tobacco protoplasts.
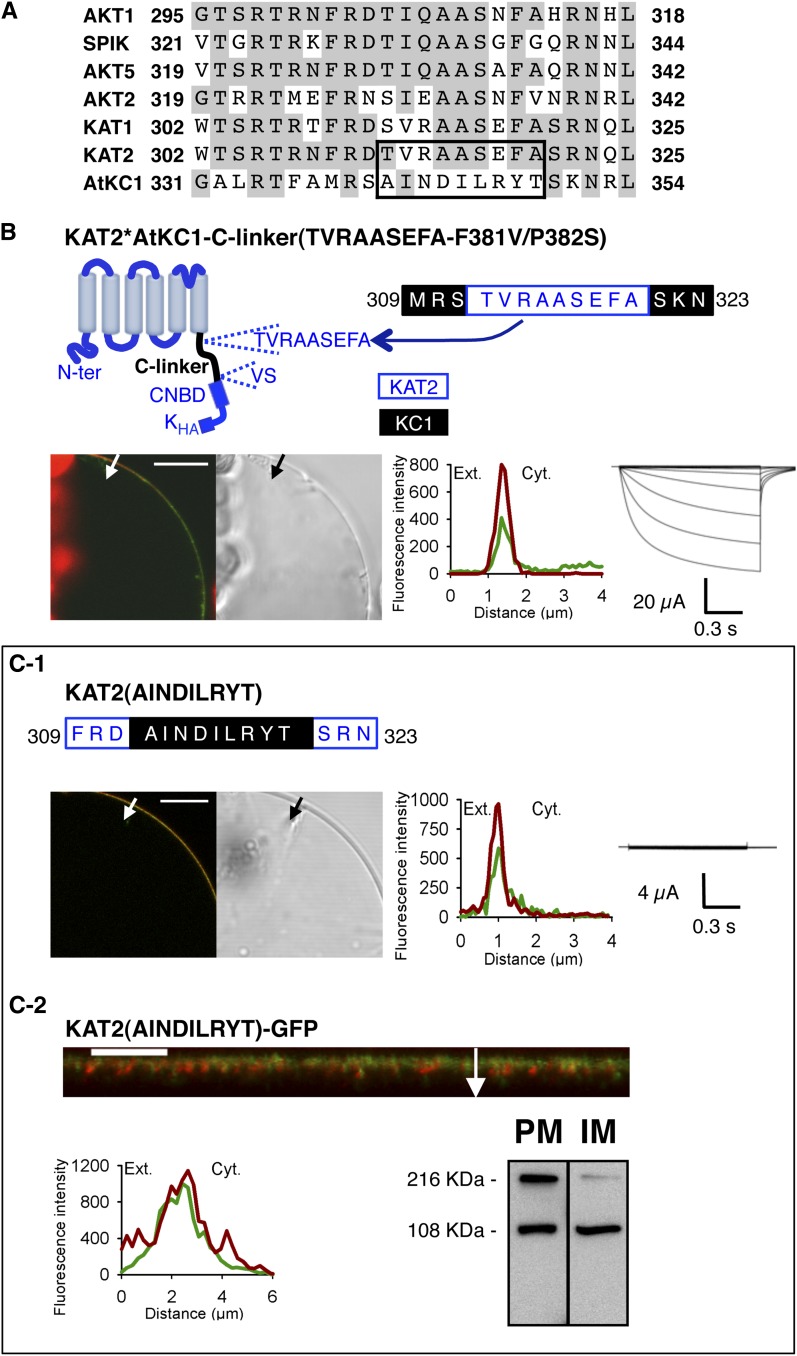
Identification of a Stretch of Nine Amino Acids Essential for Channel Activity
The previous hypothesis that the N-terminal part of the C-linker contains residues essential for channel functionality was checked by using the KAT2*AtKC1-C-linker(F381V/P382S) chimera, which is localized to the PM but does not form functional channels giving rise to K+ currents within the tested range of membrane potentials, +40 to −170 mV (Fig. 3H). An alignment of all Arabidopsis Shaker inward subunits comprising the N-terminal 24 amino acids of the C-linker was performed (Fig. 6A). This alignment pointed to a stretch of nine amino acids in AtKC1, 341AINDILRYT349, corresponding in KAT2 to 312TVRAASEFA320, which is poorly conserved in AtKC1 when compared with the other Shaker inwardly rectifying channel subunits. Two experiments were carried out to investigate the role of the nine-amino acid sequence in channel functionality. First, the KAT2*AtKC1-C-linker(F381V/P382S) chimera was further mutated in order to replace the AINDILRYT stretch of the C-linker present in this chimera (introduced from AtKC1) by the corresponding TVRAASEFA sequence displayed by the KAT2 C-linker. The resulting mutant, named KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S), was targeted to the PM in tobacco protoplasts (91% of the GFP-positive protoplasts [n = 89]; Fig. 6B), like the initial KAT2*AtKC1-C-linker(F381V/P382S) chimera (Fig. 3H). In contrast to the initial chimera, it was functional at the PM, mediating inward K+ currents (Fig. 6B). In a second set of experiments, the KAT2 sequence was mutated to change the 312TVRAASEFA320 motif into AINDILRYT. The resulting mutant, named KAT2(AINDILRYT), was targeted to the PM in tobacco protoplasts (89% of the GFP-positive protoplasts studied [n = 91]; Fig. 6C1; Supplemental Video S5) and as well as in X. laevis oocytes (Fig. 6C2) but did not display K+ transport activity in the tested voltage range in oocytes (Fig. 6C1). Altogether, these results indicate that the nine-amino acid stretch 312TVRAASEFA320 present in the KAT2 C-linker is a crucial determinant of KAT2 channel activity. It is also worth noting that replacing the AINDILRYT stretch by the TVRAASEFA one directly in the KAT2*AtKC1-C-linker chimera, with the resulting construct being named KAT2*AtKC1-C-linker(TVRAASEFA), did not restore channel surface expression (Supplemental Fig. S2). This result is consistent with the hypothesis that the KAT2 TVRAASEFA stretch plays an essential role in channel gating but not in channel targeting.
Figure 6.
The TVRAASEFA stretch present in the N-terminal region of the KAT2 C-linker plays an essential role in KAT2 channel functionality. A, Alignment displaying the first 24 amino acids of the C-linker from all Arabidopsis inward Shaker channel subunits. Identical residues in at least three members are displayed in gray background. The TVRAASEFA stretch of KAT2 (amino acids 312–320) and its counterpart in AtKC1 (amino acids 341–349) are boxed. B, The KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera is active at the PM. Top left, pictogram of this chimera showing the sequences from KAT2 (in blue) and AtKC1 (in black). This chimera was obtained from the KAT2*AtKC1-C-linker(F381V/P382S) one (Fig. 3H) by replacing the AINDILRYT stretch from the AtKC1 C-linker by the KAT2 TVRAASEFA sequence. The position of this substitution in the C-linker sequence of the resulting KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera is indicated at the top right (KAT2 amino acids in blue and AKC1 amino acids in black). C, Substituting the AtKC1 sequence AINDILRYT for the TVRAASEFA motif in KAT2 renders the channel inactive. The position of this substitution in the C-linker sequence of the resulting mutant, named KAT2(AINDILRYT), is indicated at the top in C1 (KAT2 amino acids in blue and AKC1 amino acids in black). In B and C1, surface expression and activity at the PM of the two constructs were investigated by subcellular localization of GFP fusions in tobacco protoplasts and current recordings in X. laevis oocytes, as described in the legend to Figure 1. In C2, further analyses were performed in oocytes expressing KAT2(AINDILRYT)-GFP to check its subcellular localization by confocal imaging and protein gel-blot analysis of cellular fractions, as described in the legend to Figure 1. A white arrow on the confocal image marks the position of the analyzed section crossing the PM and pockets of cytoplasm. Bars = 10 μm.
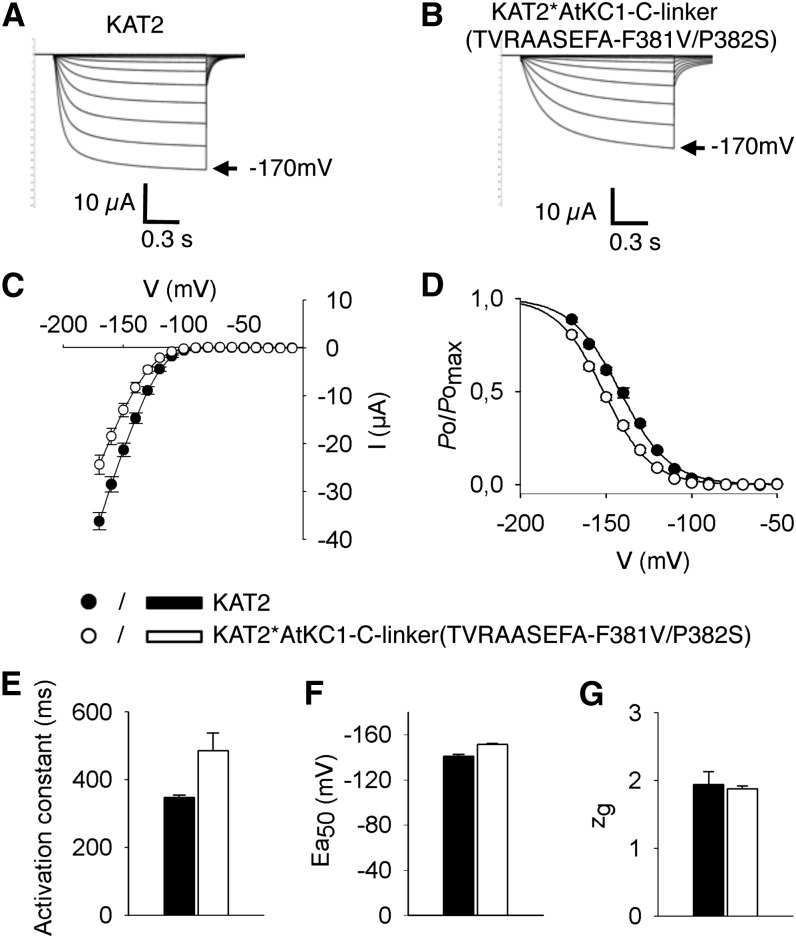
Electrophysiological Analysis of the KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) Chimera
Experiments were performed in X. laevis oocytes in order to compare the channel activity of the KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera with that of the wild-type KAT2 channel (Pilot et al., 2001) expressed in the same batches of oocytes. Like KAT2, the chimera mediated slowly activating voltage-gated inwardly rectifying channel activity (Fig. 7). The channel activation kinetics was fairly slower in the chimera (Fig. 7B) than in KAT2 (Fig. 7A) by about 40% (Fig. 7E), but an analysis of channel sensitivity to voltage using the Boltzmann model revealed very similar parameters: the same gating charge (close to 2; Fig. 7G) and close half-activation potentials (approximately −150 and −140 mV in the chimera and in KAT2, respectively; Fig. 7F).
Figure 7.
Electrophysiological characterization of the KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera in X. laevis oocytes reveals KAT2-like functional properties. Oocytes were injected with 15 ng of either KAT2 or KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) cRNA. Currents were recorded in 100 mm K+ external solution. A and B, Representative current traces in oocytes expressing KAT2 (A) or KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) (B). C to G, Steady-state current (I)/voltage (V) curves (C), relative open probabilities (D), activation time constants (E), half-activation potentials (Ea50; F), and equivalent gating charge (zg; G) of channel activity in oocytes expressing KAT2 (black bars, black circles) or the KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) chimera (white bars, white circles). Means ± se (n = 10–15) were obtained using the same oocyte batch. Applied activation membrane voltages ranged from +0 to −170 mV (increments of 10 mV; holding potential, 0 mV; deactivation potential, −60 mV).
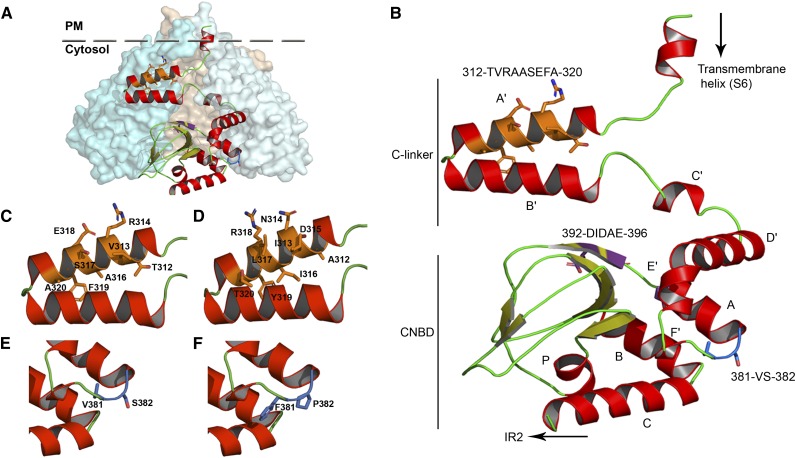
3D Model of the KAT2 C-Linker and CNBD Domains
Plant Shaker channels share extensive sequence similarities with animal members from the 6TM-1P cation channel superfamily in the transmembrane hydrophobic core and C-terminal domain (Derst and Karschin, 1998). Phylogenic analyses performed on the region extending from the beginning of the C-linker to the end of the CNBD sequence, without including the post-CNBD regions, revealed that the highest levels of similarity between KAT2 and animal channels was with the HCN, KCNH, and CNGC channel families (Supplemental Fig. S3). A 3D model of this region of KAT2 was built in order to discuss the experimental results obtained in this study. The best structure templates to obtain such a model were two HCN channels, mouse (Mus musculus) HCN2 (Protein Data Bank [PDB] identifier 3BPZ; Zagotta et al., 2003; Craven et al., 2008) and Strongylocentrotus purpuratus (sea urchin) hyperpolarization-activated channel (SpIH; PDB identifier 2PTM; Flynn et al., 2007; for sequence alignments, see Supplemental Fig. S4). The 3D model obtained from these templates for the KAT2 C-linker CNBD region is shown in Figure 8.
Figure 8.
Homology model of the C-linker and CNBD domains of KAT2. A, Assembly of the KAT2 C-linker and CNBD domains in a tetrameric configuration. One subunit is shown in ribbon display, while the other three are depicted in globular display, with each subunit colored differently. B, Ribbon representation of the KAT2 region (Met-295 to Gly-499) encompassing the end of the last transmembrane segment (S6), the C-linker domain (six α-helices [A′ to F′] separated by short loops), and the CNBD (one α-helix [A], a β-roll formed by β-strands, followed by two α-helices [B and C]). The α-helices are shown in red, and the β-strands are shown in green-yellow. The positions of the three sequences identified in this article (312TVRAASEFA320, 381VS382, and 392DIDAE396) are highlighted in orange, blue, and purple, respectively, and their corresponding side chains are displayed. C and D, Closeups of A′ and B′ α-helices of the KAT2 C-linker containing the native 312TVRAASEFA320 (C) and the mutated 312AINDILRYT320 (D) sequences. E and F, Closeups of the linker residues between F′ and A α-helices containing the native 381VS382 (E) and the mutated V381F/S382P (F) sequences.
DISCUSSION
The structure-function relationship of plant Shaker K+ channels is still poorly understood. This study aimed at analyzing the roles of the C-linker domain in the inwardly rectifying Shaker channel subunit KAT2 by sequence swapping between KAT2 and the Shaker subunit AtKC1 from Arabidopsis. For modeling the structure of the C-linker domain of KAT2 (and that of the downstream CNBD domain), we used two homologous HCN channels as crystal structure templates: the mouse HCN2 (Zagotta et al., 2003; Craven et al., 2008) and a sea urchin HCN named SpIH (Flynn et al., 2007; Fig. 8; Supplemental Figs. S3 and S4). All results are discussed in relation to the obtained structure model (Fig. 8).
V381F and S382P Mutations in the C-Terminal Part of the C-Linker Prevent KAT2 Localization to the PM
Here, we identified two contiguous amino acids present in the C-linker domain, Val-381 and Ser-382, as able to prevent KAT2 surface expression when replaced by the equivalent native residues from AtKC1, Phe-410 and Pro-411 (Fig. 3, E and F). The results can be summarized as follows. The former substitution, V381F, totally blocked the surface expression of KAT2 in tobacco protoplasts and in X. laevis oocytes (Fig. 3E). Interestingly, such a blockage was also observed when the Val-to-Phe mutation was introduced at the same place in another plant Shaker channel subunit, KAT1 (Fig. 4D). The introduction of Phe rather than the loss of Val has been identified as the cause of the defect in KAT2 channel targeting (Figs. 3E and 4, A–C). The inhibiting effect of the latter substitution, S382P, on KAT2 localization to the PM is less pronounced than that of the V381F substitution, since currents were still recorded in oocytes (Fig. 3F). It should also be noted that the involvement of Ser-382 in channel targeting to the membrane cannot be ascribed to the presence of a phosphorylable residue at position 382, since the KAT2(S382A) mutant channel is found to be localized to the PM (Fig. 3G).
Taken as a whole, these results indicated that Val-381 and Ser-382 were involved in proper targeting of the KAT2 channel. However, since each of the two amino acids could be replaced by another residue without any obvious effect on channel targeting to and activity in the PM, they were not likely to constitute an intrinsic trafficking motif. The hypothesis that we propose is that Val-381 and Ser-382 play a role in the proper folding of the protein, as discussed hereafter. In our 3D model of the KAT2 C-terminal part (Fig. 8B), Val-381 and Ser-382 are located at the junction between the last helix of the C-linker, named the F′ helix, and the first helix of the CNBD, named the A helix. This zone has been described in HCN channels to be flexible and able to absorb conformational changes (Taraska et al., 2009). Furthermore, evidence is available in HCN channels that the C-linker E′ and F′ helices play a role in proper folding of the CNBD (Santoro et al., 2011). It is also relevant that, in HCN channels expressed in mouse brain, the C-linker E′ and F′ helices and the CNBD form a site that binds auxiliary subunits, named TRIP8b or PEX5R, which play a role in the surface expression (and activity) of the channel (Santoro et al., 2011; Han et al., 2011; Bankston et al., 2012). We thus hypothesize that, in KAT2, the short sequence that connects the C-linker F′ helix to the CNBD and comprises the Val-381 and Ser-382 residues plays a role in the folding of the structure formed by the C-linker and CNBD (as in HCN channels) and that substituting Val-381 by a Phe or Ser-382 by a Pro compromises the proper folding of this structure. Indeed, the aromatic side chain of Phe is known to be involved in stacking interactions with other aromatic side chains, and Pro is described as an amino acid with strong conformational rigidity, unable to occupy many of the main-chain conformations (Betts and Russell, 2003). In other words, the Val-to-Phe and Ser-to-Pro substitutions could affect the folding of the C-linker and the CNBD region, and this would alter the presentation of a targeting motif at the protein surface or impair the interaction of this region with proteins involved in channel trafficking.
KAT2 Targeting to the PM and the DIDAE Triacidic Export Motif Present in KAT2 CNBD
Diacidic D/E-X-D/E motifs have been shown to function as discrete ER export signals in different membrane proteins from animal, yeast, and plant cells (Nishimura and Balch, 1997; Votsmeier and Gallwitz, 2001; Hanton et al., 2005). For instance, animal K+ channels from the Kir (for K+ inwardly-rectifying channel) family, which are active in the PM, harbor a six-amino acid stretch, FCYENE, containing a diacidic motif and constituting a discrete ER export signal (Ma et al., 2001). Search for such diacidic export signals in the plant Shaker channel KAT1 has led to the identification of the five-amino acid stretch DIDAE present at the frontier between the C-linker and the CNBD domains and required for efficient targeting of the channel to the PM (Mikosch et al., 2006). In this channel, progressive mutagenesis of the acidic amino acid of the DIDAE motif (by single, double, or triple mutations) resulted in the gradual reduction of ER export depending on the number of mutated acidic residues, the triple mutation (DIDAE mutated into aIaAa) resulting in the total absence of channel targeting to and activity in the PM (Mikosch et al., 2009). Thus, the complete ER export motif of KAT1 is most likely represented by the triacidic motif DIDAE. The role played by this motif in channel targeting would involve cooperative binding to the Coat protein II component Secretory pathway protein24 (Sieben et al., 2008).
As indicated above, a DIDAE stretch is present in KAT2 at the same relative position as in KAT1 (Fig. 5A). Deletion of the DIDAE motif [mutation into aIaAa; name of the mutated channel: KAT2(aIaAa)] resulted in a much less drastic effect on channel targeting to the PM in the case of KAT2 than in that of KAT1 (compare Fig. 5B and Supplemental Fig. S1B). The deletion reduced the percentage of transformed protoplasts displaying GFP fluorescence signals associated with the PM by about 15% in the case of KAT2 [see “Results”; percentages of GFP-positive protoplasts displaying surface expression: 96% and 80% for the wild-type KAT2 channel and the mutated KAT2(aIaAa) mutant, respectively] and by 100% in the case of KAT1 [we never observed fluorescence signals associated with the PM in protoplasts expressing the mutated KAT1(aIaAa) construct; Supplemental Fig. S1], in agreement with previous reports (Mikosch et al., 2009). Furthermore, electrophysiological recordings in oocytes indicated that the KAT2(aIaAa) mutant was targeted to and active in the PM (Fig. 5B). Hence, these results provided evidence that the triacidic DIDAE motif did not play any major role in KAT2 targeting to the PM, while they confirmed that this motif was required for KAT1 targeting to the PM.
Channel Gating and the C-Linker A′ Helix
The nine-amino acid stretch 312TVRAASEFA320 present in the N-terminal part of the KAT2 C-linker is a crucial determinant of channel activity. Replacement of this stretch by the corresponding sequence from AtKC1 (AINDILRYT) resulted in the total inhibition of KAT2 K+ transport activity within the range of membrane potentials tested (+40 to −170 mV; Fig. 6C). Furthermore, the inverse substitution in the KAT2*AtKC1-C-linker(F381V/P382S) chimera (replacement of the AtKC1 stretch AINDILRYT by the KAT2 TVRAASEFA stretch) rendered the chimera active (Fig. 6B). These results cannot be ascribed to an effect of the 312TVRAASEFA320 sequence on KAT2 trafficking, because protein surface expression was not affected upon substitution of this stretch by the corresponding sequence from AtKC1 (AINDILRYT; Fig. 6C; Supplemental Fig. S2).
In the 3D homology model of the KAT2 C terminus (Fig. 8), the TVRAASEFA sequence forms a large part of the C-linker A′ helix (Fig. 8B). In HCN channels, the A′ helix is parallel to the membrane, located just below the channel transmembrane core and in close proximity to the S4-S5 cytosolic loop, which links the fourth to the fifth transmembrane segment of the channel hydrophobic core just below the membrane. This position would allow the A′ helix to physically interact with the S4-S5 loop (Decher et al., 2004; Kwan et al., 2012). Conformational changes of the CNBD triggered by the binding of a cyclic nucleotide in HCN channels (Craven et al., 2008) or by the binding of a short endogenous β-sheet that acts as an intrinsic signal in KCNH channels (Brelidze et al., 2012) are transduced by the C-linker domain into conformational changes of channel regions present just below the membrane, displacing the A′ helix from its initial position. Displacement of the A′ helix away from the S4-S5 loop would favor opening of the channel permeation pathway, as a consequence of the ligand-loaded status of the CNBD (Decher et al., 2004; Kwan et al., 2012). In KAT2, the A′ helix is likely to play a similar role, since replacement of the native TVRAASEFA stretch by the AtKC1 corresponding sequence AINDILRYT resulted in a mutant channel present at the PM but unable to open and mediate K+ transport within the range of membrane potentials tested (+40 to −170 mV; Fig. 6C). Within the framework of this hypothesis, KAT2 gating would essentially depend on properties of the channel transmembrane core and the A′ helix, since the chimeric construct KAT2*AtKC1-C-linker(TVRAASEFA-F381V/P382S) behaved as a voltage-gated inwardly rectifying channel giving rise to K+ currents very similar to those observed with the wild-type KAT2 channel (Fig. 7). It is also tempting to speculate that the new gating properties conferred to heteromeric channels by the presence of AtKC1 subunits, resulting in channel activation at more negative potentials (Dreyer et al., 1997; Duby et al., 2008; Geiger et al., 2009; Honsbein et al., 2009; Grefen et al., 2010; Wang et al., 2010; Jeanguenin et al., 2011), could involve interactions of the AtKC1 AINDILRYT stretch with channel voltage sensor domains. Similarly, the chimeras containing the AINDILRYT stretch, localized to the PM but remaining electrically silent in our experiments, could be active at more negative voltages than those that can be reached in oocytes without the activation of endogenous conductances.
In conclusion, this study provides new information on the structure-function relationship of the C-linker domain in the plant family of Shaker K+ channels. In the Arabidopsis KAT2 channel, we have identified two contiguous residues as major structural determinants of channel localization to the PM and a stretch of nine amino acids as a crucial piece of the channel-gating machinery. This information, gained in the Arabidopsis KAT2 channel, is likely to be of value in analyzing the structure-function relationship of channels from both group 2 (KAT1 relatives) and group 1 (AKT1 relatives) of the plant Shaker family, due to sequence conservation in the regions pinpointed by this study (Supplemental Fig. S3), and also from the animal HCN and KCNH families, due to their sequence and structural similarities with plant Shaker channels.
MATERIALS AND METHODS
Construction of Chimeric Shaker Channel Subunits
Subdomain exchange between Arabidopsis (Arabidopsis thaliana) AtKC1 and KAT2 complementary DNAs (cDNAs) was performed by the bridge-overlap-extension PCR method (Mehta and Singh, 1999), with overlapping primers listed in Supplemental Table S1. Point mutations were generated by PCR mutagenesis using sense and antisense mutated primers (Supplemental Table S2) and Pfu ultra polymerase (Stratagene; http://www.genomics.agilent.com). Final chimeric Shaker channel subunits were cloned in pDonor 207 vector (Invitrogen; http://www.lifetechnologies.com). All constructs were verified by sequencing of both strands.
Subcellular Localization by GFP Imaging in Tobacco Protoplasts
All tested Shaker subunits were transferred into pLOC-GW-GFP in frame with GFP (Hosy et al., 2005; Jeanguenin et al., 2011). Transient expression in tobacco (Nicotiana tabacum) mesophyll protoplasts was performed as described by Hosy et al. (2005). Every construct was tested at least in three independent experiments 24 h after transfection. Confocal observations were made with an LSM780 microscope (Zeiss; www.zeiss.com) using a Plan-Apochromat X63/1,4 oil-immersion objective. GFP was excited at 488 nm, and a Band Pass 499- to 534-nm filter allowed the recovery of the emitted signal. To label the PM, 5 min of incubation with the fluorescent dye FM4-64 (20 μm) was carried out. This dye was excited at 561 nm, and a Band Pass 580- to 630-nm filter selected the emitted signal. Confocal acquisitions were set in a sequential mode (alternative switch between laser lines 488 and 561 nm) to record GFP and FM4-64 fluorescence. Fluorescence intensity profiles (obtained by profile analysis using LSM software) were determined along lines crossing the PM and pockets of cytoplasm.
Functional Characterization in Xenopus laevis Oocytes
To express chimeric and native Shaker channel subunits in X. laevis oocytes, in vitro transcription of the corresponding cDNA was performed with the mMESSAGE mMACHINE T7 Ultra kit (Ambion; http://www.ambion.com). Fifteen nanograms of complementary RNA (cRNA) of each construct in a volume of 50 nL was injected in X. laevis oocytes, and electrophysiological recordings were performed (72 h after injection) with the two-electrode voltage-clamp technique. pClamp10 software (Axon Instruments; http://www.moleculardevices.com) was used to record and analyze the data. For each construct, 10 to 15 oocytes were analyzed in three independent experiments, and representative traces are shown. Bath solutions contained (in mm): 100 potassium gluconate, 1 CaCl2, 2 MgCl2, and 10 HEPES-NaOH, pH 6.5. Inhibition by cesium was tested by the addition of 20 mm CsCl to the K100 solution. Current-intensity comparisons were based on steady-state average values (n = 10–15). Applied activation membrane voltages ranged from +40 to −170 mV (increments of 15 mV; holding potential, 0 mV; deactivation potential, −40 mV).
The relative open probability (Po/Pomax) was calculated as follows: Po/Pomax = I/imax, where imax is the current registered at maximal open probability obtained from the Boltzmann equation: I = imax × [1/(1 + exp(zg × F × (V − Ea50)/(R × T)))], in which “leak-subtracted” initial tail currents were fitted as described by Jeanguenin et al. (2011). zg is the equivalent gating charge, V is the applied membrane potential, Ea50 is the channel half-maximal activation potential, and F, R, and T stand for the Faraday constant, the ideal gas constant, and the temperature, respectively. Activation time constants (the time at which half of the current intensity is observed) were obtained at the membrane potential where Po/Pomax = 0.3. For tail current and activation time constant analyses, applied activation membrane voltages ranged from +0 to −170 mV (increments of 10 mV; holding potential, 0 mV; deactivation potential, −60 mV).
Subcellular Localization by GFP Imaging in X. laevis Oocytes
Shaker-GFP fusions previously cloned in pLOC-GW-GFP were amplified by PCR, and T7 promoter and linker sequences were added at the 5′ and 3′ ends, respectively. After in vitro transcription of the amplified fragments, 15 ng of cRNA was injected in oocytes for fluorescence imaging experiments. Oocyte GFP and FM4-64 fluorescence signals were monitored as described above for transiently transformed tobacco protoplasts, with the exception that images were obtained by performing linear z-stacks at the upper pole of the oocyte. The stacks comprised 4 to 5 µm and 20 to 30 slices. Fluorescence plot profiles followed the z axis.
Preparation of PM- and ER-Enriched Fractions from X. laevis Oocytes and Western-Blot Analysis
Oocytes (100–150 per assay) were injected with cRNAs encoding Shaker-GFP subunits and incubated as described above. Then, oocytes were suspended in 1 mL of homogenization buffer at 4°C (5 mm phosphate, pH 7.4, 5 mm MgCl2, 250 mm Suc, 0.5 mm Pefablock, 10 µg of leupeptin, 10 µg of aprotinin, 10 µg of pepstatin, and 60 µg of chymostatin) and were homogenized by 50 strokes of a Teflon-coated pestle in a 2-mL glass Dounce homogenizer. Homogenates were then centrifuged at 500g for 5 min at 4°C. The supernatants were then overlaid on top of a discontinuous Suc gradient (50% and 20% Suc in the homogenization buffer) and centrifuged at 4°C at 50,000g for 4 h in a swinging SW60Ti Beckman rotor. The 20% interface (enriched in PM) and the 50% interface (enriched for ER; Hill et al., 2005) were removed with a Pasteur pipette, mixed to 5 volumes of homogenization buffer without Suc, and centrifuged at 4°C at 150,000g for 2 h. The resulting membrane pellets were resuspended in homogenization buffer before protein determination by Bradford assay. For detection of GFP-tagged Shaker channels, about 50 µg of membrane proteins was solubilized with 2× Laemmli solubilization buffer, and proteins were separated by SDS-PAGE and further electrotransferred on a Millipore Immobilon-P polyvinylidene difluoride 0.45-µm membrane. GFP-tagged Shaker channels were then detected using an anti-GFP antibody (1:5,000) directly coupled to horseradish peroxidase (Miltenyi Biotec).
Model-Building Methods
The structure of the C-terminal region of KAT2 was modeled using the homology modeling approach under the Discovery Studio version 3.1 (Accelrys). Sequences of KAT2 (Met-295 to Gly-499) and AtKC1 (Met-324 to Leu-530) were analyzed with the Sequence Analysis tools to identify optimal templates from the PDB. Two x-ray structures were selected (PDB identifiers 2PTM and 3BPZ) that correspond to the C-linker and CNBD regions of two HCN channels, the SpIH (sea urchin [Strongylocentrotus purpuratus]) and HCN2 (mouse [Mus musculus]) channels, respectively. A multiple sequence alignment of the two templates with the corresponding sequences of KAT2 and AtKC1 was generated using the Constraint-based Multiple Alignment Tool at the National Center for Biotechnology Information (Papadopoulos and Agarwala, 2007; Supplemental Fig. S4). An additional sequence of 10 residues corresponding to the end of the sixth transmembrane helix (S6) of a potassium channel structure from Streptomyces lividans (PDB identifier 1BL8) was added to guide the orientation of the last transmembrane helix of KAT2 (Supplemental Fig. S4). Based on the 3D structure analysis of the templates compared with the KAT2 sequence, no disulfide bridge pattern was added to the homology model generation performed with modeler and the refine loop option. All models were generated without cyclic nucleotide. Refined structures were evaluated by Verify 3D (Discovery Studio) and Qmean at expasy (Benkert et al., 2011). For subsequent models of KAT2 bearing residues of AtKC1 (AINDILRYT and FP), chimeric sequences were generated according to the alignment between KAT2 and AtKC1 (Supplemental Fig. S4) and then used in modeler.
Sequence data from this article can be found in The Arabidopsis Information Resource database (http://www.arabidopsis.org) under accession numbers At4g18290 (KAT2), At5g46240 (KAT1), and At4g32650 (AtKC1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Disruption of the triacidic motif DIDAE in KAT1 prevents channel targeting to the PM, while introduction of this motif in AtKC1 is not sufficient to allow the resulting AtKC1(DIDAE) mutant to be targeted to the PM.
Supplemental Figure S2. Introduction of the KAT2 C-linker TVRAASEFA motif in the KAT2*AtKC1-C-linker chimera does not restore PM targeting.
Supplemental Figure S3. Phylogenetic relationships between sequences encompassing the C-linker and CNBD domains in plant Shaker channels and in animal channels characterized by the presence of a CNBD in their cytosolic C-terminal part.
Supplemental Figure S4. Protein sequences used to create homology models of plant channel C-linker and CNBD regions.
Supplemental Table S1. Bridge-PCR primers used for chimera cloning and for subdomain exchange between AtKC1 and KAT2 cDNAs.
Supplemental Table S2. Primers used for site-directed mutagenesis.
Supplemental Video S1. 3D reconstruction of a tobacco protoplast expressing KAT2 fused to GFP.
Supplemental Video S2. 3D reconstruction of a tobacco protoplast expressing KAT2(V381F) fused to GFP.
Supplemental Video S3. 3D reconstruction of a tobacco protoplast expressing KAT2*AtKC1-C-linker(F381V/P382S) fused to GFP.
Supplemental Video S4. 3D reconstruction of a tobacco protoplast expressing AtKC1 fused to GFP.
Supplemental Video S5. 3D reconstruction of a tobacco protoplast expressing KAT2(AINDILRYT) fused to GFP.
Supplemental Methods S1. 3D reconstruction of tobacco protoplasts, and phylogenic analysis.
Acknowledgments
We thank Jean François Briat for critical comments on the manuscript.
Glossary
- PM
plasma membrane
- 6TM-1P
six transmembrane-one pore
- CNBD
cyclic nucleotide-binding domain
- ER
endoplasmic reticulum
- 3D
three-dimensional
- PDB
Protein Data Bank
- cDNA
complementary DNA
- cRNA
copy RNA
Footnotes
This work was supported by the Alfonso Martin Escudero Foundation (to M.N.-C.) and the Marie Curie Programme (grant no. FP7–IEF to M.N.-C.).
The online version of this article contains Web-only data.
References
- Bankston JR, Camp SS, DiMaio F, Lewis AS, Chetkovich DM, Zagotta WN. (2012) Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc Natl Acad Sci USA 109: 7899–7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. In MR Barnes, IC Gray, eds, Bioinformatics for Geneticists. Wiley, Chichester, UK, pp 289–316 [Google Scholar]
- Blatt MR, Hills A, Chen ZH, Wang Y, Papanatsiou M, Lew VL. (2012) The conceptual approach to quantitative modeling of guard cells. Plant Signal Behav 8: 173–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze TI, Carlson AE, Sankaran B, Zagotta WN. (2012) Structure of the carboxy-terminal region of a KCNH channel. Nature 481: 530–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven KB, Olivier NB, Zagotta WN. (2008) C-terminal movement during gating in cyclic nucleotide-modulated channels. J Biol Chem 283: 14728–14738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven KB, Zagotta WN. (2006) CNG and HCN channels: two peas, one pod. Annu Rev Physiol 68: 375–401 [DOI] [PubMed] [Google Scholar]
- Daram P, Urbach S, Gaymard F, Sentenac H, Chérel I. (1997) Tetramerization of the AKT1 plant potassium channel involves its C-terminal cytoplasmic domain. EMBO J 16: 3455–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decher N, Chen J, Sanguinetti MC. (2004) Voltage-dependent gating of hyperpolarization-activated, cyclic nucleotide-gated pacemaker channels: molecular coupling between the S4-S5 and C-linkers. J Biol Chem 279: 13859–13865 [DOI] [PubMed] [Google Scholar]
- Derst C, Karschin A. (1998) Evolutionary link between prokaryotic and eukaryotic K+ channels. J Exp Biol 201: 2791–2799 [PubMed] [Google Scholar]
- Dreyer I, Antunes S, Hoshi T, Müller-Röber B, Palme K, Pongs O, Reintanz B, Hedrich R. (1997) Plant K+ channel alpha-subunits assemble indiscriminately. Biophys J 72: 2143–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer I, Porée F, Schneider A, Mittelstädt J, Bertl A, Sentenac H, Thibaud JB, Mueller-Roeber B. (2004) Assembly of plant Shaker-like K(out) channels requires two distinct sites of the channel alpha-subunit. Biophys J 87: 858–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Hosy E, Fizames C, Alcon C, Costa A, Sentenac H, Thibaud JB. (2008) AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J 53: 115–123 [DOI] [PubMed] [Google Scholar]
- Ehrhardt T, Zimmermann S, Müller-Röber B. (1997) Association of plant K+(in) channels is mediated by conserved C-termini and does not affect subunit assembly. FEBS Lett 409: 166–170 [DOI] [PubMed] [Google Scholar]
- Flynn GE, Black KD, Islas LD, Sankaran B, Zagotta WN. (2007) Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure 15: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdanowicz P, Garcia-Mata C, Gonzalez W, Morales-Navarro SE, Sharma T, González-Nilo FD, Gutowicz J, Mueller-Roeber B, Blatt MR, Dreyer I. (2009) Distinct roles of the last transmembrane domain in controlling Arabidopsis K+ channel activity. New Phytol 182: 380–391 [DOI] [PubMed] [Google Scholar]
- Garcia-Mata C, Wang J, Gajdanowicz P, Gonzalez W, Hills A, Donald N, Riedelsberger J, Amtmann A, Dreyer I, Blatt MR. (2010) A minimal cysteine motif required to activate the SKOR K+ channel of Arabidopsis by the reactive oxygen species H2O2. J Biol Chem 285: 29286–29294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Becker D, Vosloh D, Gambale F, Palme K, Rehers M, Anschuetz U, Dreyer I, Kudla J, Hedrich R. (2009) Heteromeric AtKC1·AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem 284: 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Blatt MR. (2012) Do calcineurin B-like proteins interact independently of the serine threonine kinase CIPK23 with the K+ channel AKT1? Lessons learned from a ménage à trois. Plant Physiol 159: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Chen Z, Honsbein A, Donald N, Hills A, Blatt MR. (2010) A novel motif essential for SNARE interaction with the K+ channel KC1 and channel gating in Arabidopsis. Plant Cell 22: 3076–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Noam Y, Lewis AS, Gallagher JJ, Wadman WJ, Baram TZ, Chetkovich DM. (2011) Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites. J Biol Chem 286: 20823–20834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton SL, Renna L, Bortolotti LE, Chatre L, Stefano G, Brandizzi F. (2005) Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R. (2012) Ion channels in plants. Physiol Rev 92: 1777–1811 [DOI] [PubMed] [Google Scholar]
- Hill WG, Southern NM, MacIver B, Potter E, Apodaca G, Smith CP, Zeidel ML. (2005) Isolation and characterization of the Xenopus oocyte plasma membrane: a new method for studying activity of water and solute transporters. Am J Physiol Renal Physiol 289: F217–F224 [DOI] [PubMed] [Google Scholar]
- Honsbein A, Sokolovski S, Grefen C, Campanoni P, Pratelli R, Paneque M, Chen Z, Johansson I, Blatt MR. (2009) A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E, Duby G, Véry AA, Costa A, Sentenac H, Thibaud JB. (2005) A procedure for localisation and electrophysiological characterisation of ion channels heterologously expressed in a plant context. Plant Methods 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanguenin L, Alcon C, Duby G, Boeglin M, Chérel I, Gaillard I, Zimmermann S, Sentenac H, Véry AA. (2011) AtKC1 is a general modulator of Arabidopsis inward Shaker channel activity. Plant J 67: 570–582 [DOI] [PubMed] [Google Scholar]
- Jeanguenin L, Lebaudy A, Xicluna J, Alcon C, Hosy E, Duby G, Michard E, Lacombe B, Dreyer I, Thibaud JB. (2008) Heteromerization of Arabidopsis Kv channel alpha-subunits: data and prospects. Plant Signal Behav 3: 622–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan DC, Prole DL, Yellen G. (2012) Structural changes during HCN channel gating defined by high affinity metal bridges. J Gen Physiol 140: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaudy A, Hosy E, Simonneau T, Sentenac H, Thibaud JB, Dreyer I. (2008a) Heteromeric K+ channels in plants. Plant J 54: 1076–1082 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Vavasseur A, Hosy E, Dreyer I, Leonhardt N, Thibaud JB, Véry AA, Simonneau T, Sentenac H. (2008b) Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc Natl Acad Sci USA 105: 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S. (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Zerangue N, Lin YF, Collins A, Yu M, Jan YN, Jan LY. (2001) Role of ER export signals in controlling surface potassium channel numbers. Science 291: 316–319 [DOI] [PubMed] [Google Scholar]
- Mehta RK, Singh J. (1999) Bridge-overlap-extension PCR method for constructing chimeric genes. Biotechniques 26: 1082–1086 [DOI] [PubMed] [Google Scholar]
- Mikosch M, Homann U. (2009) How do ER export motifs work on ion channel trafficking? Curr Opin Plant Biol 12: 685–689 [DOI] [PubMed] [Google Scholar]
- Mikosch M, Hurst AC, Hertel B, Homann U. (2006) Diacidic motif is required for efficient transport of the K+ channel KAT1 to the plasma membrane. Plant Physiol 142: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikosch M, Käberich K, Homann U. (2009) ER export of KAT1 is correlated to the number of acidic residues within a triacidic motif. Traffic 10: 1481–1487 [DOI] [PubMed] [Google Scholar]
- Naso A, Dreyer I, Pedemonte L, Testa I, Gomez-Porras JL, Usai C, Mueller-Rueber B, Diaspro A, Gambale F, Picco C. (2009) The role of the C-terminus for functional heteromerization of the plant channel KDC1. Biophys J 96: 4063–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Balch WE. (1997) A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277: 556–558 [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Cherel I, Boucherez J, Thibaud JB, Sentenac H. (2001) Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem 276: 3215–3221 [DOI] [PubMed] [Google Scholar]
- Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R. (2002) AtKC1, a silent Arabidopsis potassium channel alpha-subunit modulates root hair K+ influx. Proc Natl Acad Sci USA 99: 4079–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro B, Hu L, Liu H, Saponaro A, Pian P, Piskorowski RA, Moroni A, Siegelbaum SA. (2011) TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J Neurosci 31: 4074–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T, Dreyer I, Riedelsberger J. (2013) The role of K(+) channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front Plant Sci 4: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieben C, Mikosch M, Brandizzi F, Homann U. (2008) Interaction of the K(+)-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. Plant J 56: 997–1006 [DOI] [PubMed] [Google Scholar]
- Taraska JW, Puljung MC, Olivier NB, Flynn GE, Zagotta WN. (2009) Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods 6: 532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach S, Chérel I, Sentenac H, Gaymard F. (2000) Biochemical characterization of the Arabidopsis K+ channels KAT1 and AKT1 expressed or co-expressed in insect cells. Plant J 23: 527–538 [DOI] [PubMed] [Google Scholar]
- Votsmeier C, Gallwitz D. (2001) An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J 20: 6742–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl-Schott C, Biel M. (2009) HCN channels: structure, cellular regulation and physiological function. Cell Mol Life Sci 66: 470–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He L, Li HD, Xu J, Wu WH. (2010) Potassium channel alpha-subunit AtKC1 negatively regulates AKT1-mediated K(+) uptake in Arabidopsis roots under low-K(+) stress. Cell Res 20: 826–837 [DOI] [PubMed] [Google Scholar]
- Xicluna J, Lacombe B, Dreyer I, Alcon C, Jeanguenin L, Sentenac H, Thibaud JB, Chérel I. (2007) Increased functional diversity of plant K+ channels by preferential heteromerization of the shaker-like subunits AKT2 and KAT2. J Biol Chem 282: 486–494 [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E. (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425: 200–205 [DOI] [PubMed] [Google Scholar]