A basic helix-loop-helix transcription factor antagonistically regulates growth and immunity.
Abstract
Plants need to finely balance resources allocated to growth and immunity to achieve optimal fitness. A tradeoff between pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and brassinosteroid (BR)-mediated growth was recently reported, but more information about the underlying mechanisms is needed. Here, we identify the basic helix-loop-helix (bHLH) transcription factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH IBH1 (HBI1) as a negative regulator of PTI signaling in Arabidopsis (Arabidopsis thaliana). HBI1 expression is down-regulated in response to different PAMPs. HBI1 overexpression leads to reduced PAMP-triggered responses. This inhibition correlates with reduced steady-state expression of immune marker genes, leading to increased susceptibility to the bacterium Pseudomonas syringae. Overexpression of the HBI1-related bHLHs BRASSINOSTEROID ENHANCED EXPRESSION2 (BEE2) and CRYPTOCHROME-INTERACTING bHLH (CIB1) partially inhibits immunity, indicating that BEE2 and CIB1 may act redundantly with HBI1. In contrast to its expression pattern upon PAMP treatment, HBI1 expression is enhanced by BR treatment. Also, HBI1-overexpressing plants are hyperresponsive to BR and more resistant to the BR biosynthetic inhibitor brassinazole. HBI1 is nucleus localized, and a mutation in a conserved leucine residue within the first helix of the protein interaction domain impairs its function in BR signaling. Interestingly, HBI1 interacts with several inhibitory atypical bHLHs, which likely keep HBI1 under negative control. Hence, HBI1 is a positive regulator of BR-triggered responses, and the negative effect of PTI is likely due to the antagonism between BR and PTI signaling. This study identifies a novel component involved in the complex tradeoff between innate immunity and BR-regulated growth.
The first layer of plant innate immunity is constituted by the recognition of pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns that act as distinctive microbial features betraying the presence of potentially infectious nonself (Dodds and Rathjen, 2010). Recognition of PAMPs by corresponding surface-localized plant pattern recognition receptors (PRRs) leads to PAMP-triggered immunity (PTI), which is sufficient to provide broad-spectrum disease resistance to most microbes. In the plant model Arabidopsis (Arabidopsis thaliana), the best-studied PRRs are the leucine-rich repeat receptor kinases (LRR-RKs) FLAGELLIN-SENSING2 (FLS2) and ELONGATION FACTOR TU RECEPTOR (EFR) that recognize the bacterial PAMPs flagellin (or its peptide surrogate flg22) and elongation factor Tu (or its peptide surrogates elf18 or elf26, respectively; Monaghan and Zipfel, 2012). Immediately after binding to their respective ligands, FLS2 and EFR heteromerize with the regulatory LRR-RK BRI1-ASSOCIATED KINASE1/SOMATIC-EMBRYOGENESIS RECEPTOR KINASE3 (BAK1/SERK3) as well as additional members of the SERK subfamily of LRR-RKs (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011; Sun et al., 2013). The receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1) is a direct substrate of FLS2, EFR, and BAK1 that is required for a number of flg22- and elf18-induced responses (Lu et al., 2010; Zhang et al., 2010). Notably, not all Arabidopsis PRRs are BAK1 dependent (Monaghan and Zipfel, 2012). For example, fungal chitin is perceived directly by the Lys motif receptor kinase CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) in Arabidopsis (Miya et al., 2007; Wan et al., 2008), and chitin-triggered responses are unaffected by mutations in BAK1 or other SERKs, unlike flg22- or elf18-triggered responses (Shan et al., 2008; Ranf et al., 2011). BIK1 also associates with CERK1 and is required for chitin-induced responses (Zhang et al., 2010), indicating that BIK1 may be the first convergent component shared by the FLS2, EFR, and CERK1 pathways, which lead to largely overlapping transcriptional changes (Wan et al., 2008).
Upon PAMP binding, PRR activation leads to a plethora of early (minutes to hours) immune responses comprising rapid and transient bursts of calcium and reactive oxygen species (ROS), activation of mitogen-activated protein kinases and calcium-dependent protein kinases, and transcriptional reprogramming of hundreds of genes (Boller and Felix, 2009; Monaghan and Zipfel, 2012). Later (hours to days) responses include increased production of the defense hormones ethylene, salicylic acid, and jasmonic acid, deposition of the β-1,3-glucan polymer callose at the cell walls, and induced resistance to pathogens (Boller and Felix, 2009). Notably, in certain cases, prolonged PAMP treatments (e.g. flg22 or elf18) also trigger growth inhibition of young seedlings (Gómez-Gómez et al., 1999). Although the exact molecular basis of this latter phenomenon is not clear, it indicates that sustained activation of immune responses, which are highly energy consuming, has a clear cost on plant growth.
Therefore, plants need to finely balance resources allocated to growth and immunity to maintain optimal growth while at the same time ensuring immunity to would-be microbial pathogens that are constantly presented in the phyllosphere and rhizosphere (Bulgarelli et al., 2013). Over the last few years, a number of examples have emerged illustrating that growth-promoting hormones (such as auxin, GA3, and cytokinin) or secreted peptides (such as phytosulfokine) can modulate PTI or other aspects of plant defenses (Robert-Seilaniantz et al., 2011; Igarashi et al., 2012; Mosher et al., 2013). One of the best-studied hormonal pathways in plants is the brassinosteroid (BR) signaling pathway (Clouse, 2011). BRs regulate numerous aspects of plant physiology, including cell elongation and photomorphogenesis. The main receptor for BRs is the LRR-RK BRASSINOSTEROID INSENSITIVE1 (BRI1; Hothorn et al., 2011; She et al., 2011). BRI1 is structurally related to FLS2 and EFR and also forms a ligand-induced complex with BAK1 (and other SERKs), which are key positive regulator(s) of the pathway (Clouse, 2011). Similar to the role of BIK1 in the FLS2/EFR pathways (Lu et al., 2010; Zhang, et al., 2010), BR binding leads to the phosphorylation-mediated activation and release of the receptor-like cytoplasmic kinases BRASSINOSTEROID SIGNALING KINASES (BSKs) and CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) from the BRI1-BAK1 complex (Clouse, 2011; Kim et al., 2011). BSKs and CDG1 then activate the phosphatase BRI1 SUPPRESSOR1 to dephosphorylate the glycogen synthase kinase3 BRASSINOSTEROID INSENSITIVE2 (BIN2). BIN2 acts as a central negative regulator of BR signaling by phosphorylating the basic helix-loop-helix (bHLH) transcription factors BRASSINAZOLE-RESISTANT1 (BZR1) and BZR2/BRI1-ETHYL METHANESULFONATE-SUPPRESSOR1 (BES1) that are master switches of BR-mediated responses (Clouse, 2011; Kim et al., 2011). Active dephosphorylation of BZR1 and BES1 by protein phosphatase 2A relieves their 14-3-3-mediated retention in the cytoplasm, enabling nuclear translocation of BZR1 and BES1 and binding to their target promoters (Clouse, 2011; Tang et al., 2011). Interestingly, BIK1 also interacts with BRI1, but in contrast to its role in PTI signaling, BIK1 is a negative regulator of BR signaling (Lin et al., 2013). In contrast, BSK1 associates with both BRI1 and FLS2 and is a positive regulator for both pathways (Tang et al., 2008; Shi et al., 2013).
Given the obvious similarities in the components involved and overall mechanisms between the BRI1-mediated and FLS2/EFR-mediated pathways, cross talk between these two pathways has often been postulated, potentially at the level of the shared coreceptor BAK1. Recent work in Arabidopsis has demonstrated that activation of BRI1, either by exogenous BR treatment or genetically by increasing endogenous BR levels or enhancing BRI1 outputs, leads to the inhibition of PTI responses triggered by flg22 and elf18 (Albrecht et al., 2012; Belkhadir et al., 2012). The fact that BAK1 overexpression partially reverts the effect of BRI1 overexpression on PTI responses suggested that the antagonistic effect on PTI might be caused by competition for BAK1 between BRI1 and FLS2/EFR (Belkhadir et al., 2012). However, in wild-type plants, BAK1 does not appear to be rate limiting and BR treatment does not affect FLS2-BAK1 complex formation or FLS2 or BIK1 activation upon flg22 perception (Albrecht et al., 2012). In addition, responses to chitin, which are BAK1 independent, are also affected by BR treatment (Albrecht et al., 2012). Therefore, while it is currently clear that activation of the BR pathway has an inhibitory effect on PTI signaling, the actual mechanisms underlying or regulating this antagonism are still unclear (Vert and Chory, 2011; Choudhary et al., 2012; Wang et al., 2012).
RESULTS AND DISCUSSION
The Expression of the Predicted bHLH Transcription Factor HBI1 Is Down-Regulated upon PAMP Treatments
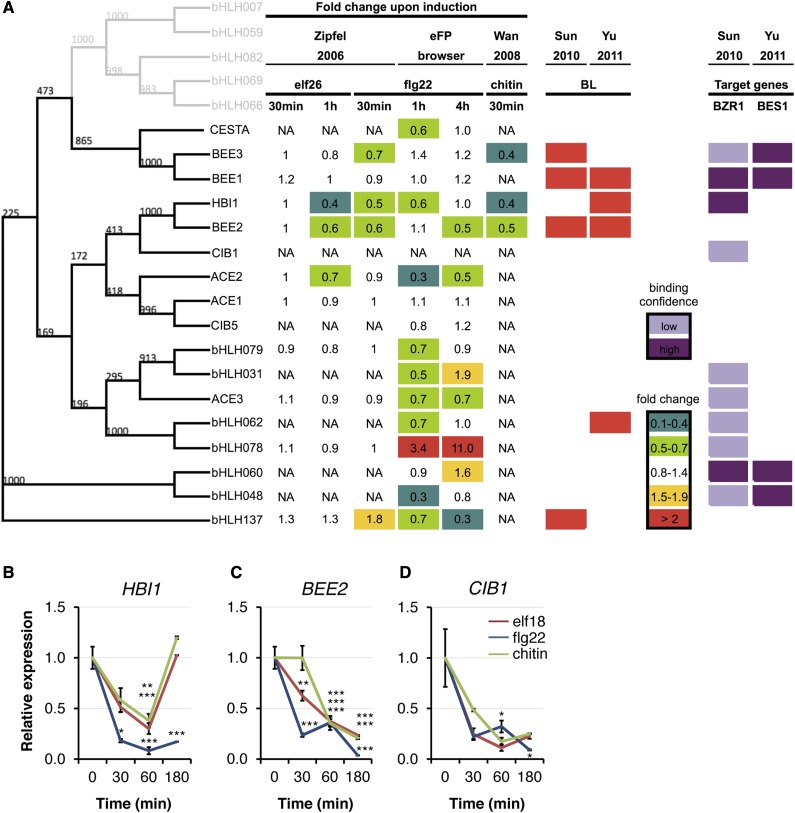
Several PRRs and positive regulators of PTI signaling have previously been identified based on the up-regulation of their transcript levels in response to PAMP treatment (Navarro et al., 2004; Zipfel et al., 2006; Chinchilla et al., 2007; Wan et al., 2008; Lu et al., 2010; Singh et al., 2012). Therefore, we hypothesized that genes down-regulated by several PAMPs could represent potential negative regulators of PTI signaling. Analysis of publicly available microarray data identified only 15 genes that were commonly down-regulated at either 30 or 60 min after treatment with the PAMPs flg22, elf26, or chitin (Zipfel et al., 2006; Wan et al., 2008; Supplemental Table S1). We initially focused on At2g18300, which encodes the putative bHLH transcription factor (also named HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH IBH1 [HBI1]; Bai et al., 2012). Previous microarray analyses (Wan et al., 2008; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Fig. 1A) indicated that HBI1 transcript levels were down-regulated more than 2-fold over 60-min time-course treatments of wild-type Arabidopsis seedlings with elf26, flg22, or chitin (Fig. 1A). We confirmed this down-regulation independently by quantitative real-time-PCR (RT-qPCR; Fig. 1B).
Figure 1.
The expression of the related bHLH transcription factors HBI1, BEE2, and CIB1 is commonly down-regulated by different PAMPs. A, Phylogenetic tree of bHLH transcription factors belonging to clade 18 (nomenclature from Toledo-Ortiz et al. [2003]). Expression profiles of clade 18 bHLHs are shown in response to PAMPs (Zipfel et al., 2006; Winter et al., 2007; Wan et al., 2008; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and BL (Sun et al., 2010; Yu et al., 2011). Direct binding of BZR1 and BES1 to promoters of clade 18 bHLHs is shown by chromatin immunoprecipitation chip assays (Sun et al., 2010; Yu et al., 2011). NA, Data not available. B to D, Expression kinetics of HBI1 (At2g18300; B), BEE2 (At4g36540; C), and CIB1 (At4g34530; D) in response to PAMP treatment monitored by RT-qPCR. Two-week-old Col-0 seedlings were treated with 100 nm elf18, 100 nm flg22, or 1 mg mL−1 chitin. Transcript levels were normalized to U-box (At5g15400) gene expression and relative to time zero. Results are averages ± 2× se (n = 3). One-way ANOVA/Dunnett: *P < 0.05, **P < 0.01, ***P < 0.001. The experiments were performed at least twice with similar results.
HBI1 Negatively Regulates Innate Immunity
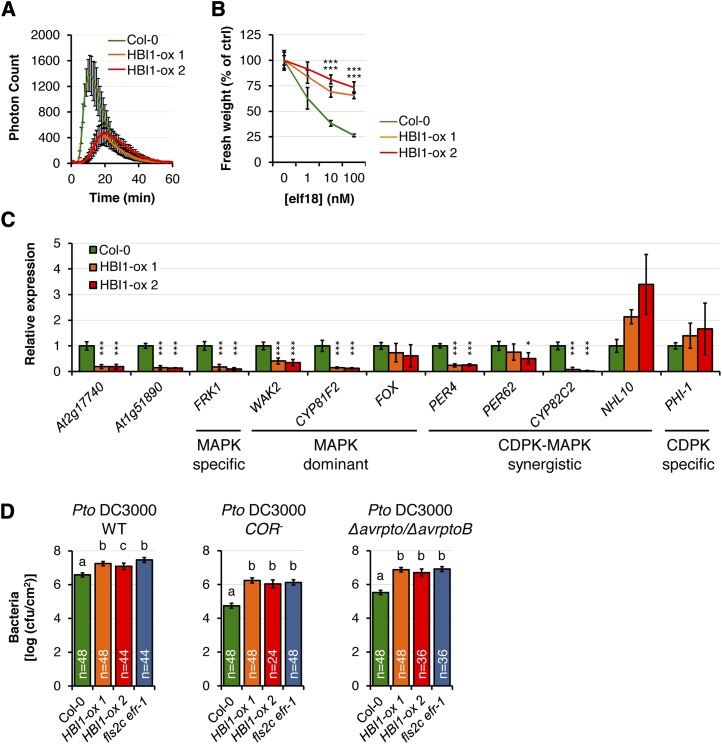
To test if HBI1 could be a negative regulator of PTI signaling, we generated transgenic Arabidopsis lines overexpressing HBI1 under the control of the constitutive cauliflower mosaic virus 35S promoter. Two independent homozygous lines strongly overexpressing HBI1 (HBI1-ox 1 and HBI1-ox 2; Supplemental Fig. S1) were selected for detailed analysis. Strikingly, both HBI1-ox 1 and HBI1-ox 2 leaves exhibited significantly reduced elf18-triggered ROS burst when compared with wild-type Columbia-0 (Col-0) leaves (Fig. 2A). This effect on the ROS burst was also apparent following treatment with flg22 or chitin (Supplemental Fig. S2). In addition to the ROS burst, which corresponds to an early PTI response, we also assayed the HBI1-ox lines for PAMP-induced seedling growth inhibition, a response that occurs within days of treatment with some PAMPs, such as flg22 and elf18 (Gómez-Gómez et al., 1999; Kunze et al., 2004). Similar to the effect observed in the ROS burst assay, both HBI1-ox lines were strongly impaired in elf18-triggered seedling growth inhibition (Fig. 2B).
Figure 2.
HBI1 overexpression inhibits immunity. A, Oxidative burst measured in leaf discs of 4-week-old plants in response to 100 nm elf18. Results are averages ± 2× se (n = 8). B, Seedling growth inhibition triggered by elf18. Fresh weights of 2-week-old seedlings were measured 1 week after the addition of elf18. Values are represented relative to untreated plants. Results are averages ± 2× se (n = 6). One-way ANOVA/Holm-Sidak: ***P < 0.001. C, Steady-state transcript levels of PTI marker genes measured by RT-qPCR in 2-week-old seedlings. Transcript levels were normalized to U-box gene expression and relative to Col-0. Results are averages from three independent biological repeats ± 2× se (n = 9). Two-way ANOVA/Holm-Sidak: *P < 0.05, ***P < 0.001. D, Spray infection (inoculum, 107 colony-forming units [cfu] mL−1) of 4-week-old plants with Pto DC3000 wild type (WT; left), COR− (middle), or Δavrpto/ΔavrptoB (right). Bacterial populations were quantified as colony-forming units cm−2 at 3 d post inoculation. Results are averages ± 2× se across four independent experiments. Two-way ANOVA/Holm-Sidak: a≠bP < 0.001, a≠cP < 0.015. All experiments were repeated at least three times with similar results.
HBI1 is a predicted bHLH transcription factor. Because we observed an inhibition of both early and late PAMP-induced responses (Fig. 2, A and B; Supplemental Fig. S2), we hypothesized that HBI1 overexpression may affect the steady-state expression of PTI marker genes. Indeed, we found that the steady-state transcript levels of seven out of 11 tested PTI marker genes were lower in HBI1-ox lines compared with wild-type Col-0 (Fig. 2C).
Next, we tested if HBI1 overexpression leads to enhanced disease susceptibility to phytopathogenic bacteria. Consistent with the reduced PAMP-triggered responses and steady-state expression of PTI marker genes (Fig. 2, A–C; Supplemental Fig. S2), HBI1-ox plants were more susceptible than wild-type Col-0 plants to spray infection with the virulent strain Pseudomonas syringae pv tomato (Pto) DC3000 as well as to the isogenic hypovirulent strains COR− and Δavrpto/ΔavrptoB (Fig. 2D). Notably, the increased susceptibility of the HBI1-ox plants to these bacterial strains was comparable to the enhanced disease susceptibility of double null mutant fls2c efr-1 plants (Fig. 2D).
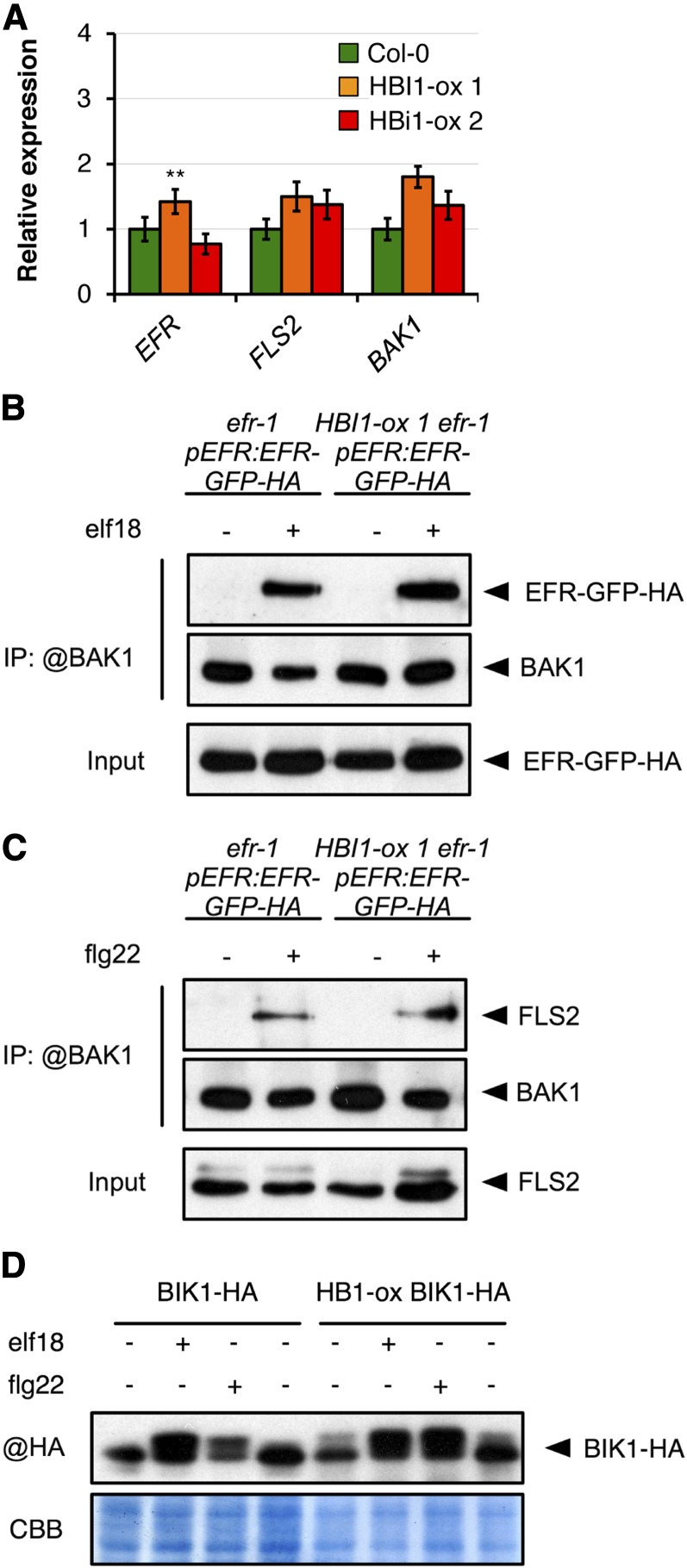
As both early and late PAMP-induced responses were impaired, we tested if HBI1 overexpression may affect the accumulation of PRRs or BAK1, the ligand-induced complex formation between PRRs and BAK1, or BIK1 activation. FLS2, EFR, and BAK1 accumulated to wild-type levels in HBI1-ox lines (Fig. 3, A–C). Furthermore, elf18- or flg22-induced complex formation between EFR-GFP and BAK1 or FLS2 and BAK1, respectively, was not affected in the HBI1-ox lines (Fig. 3, B and C). Moreover, HBI1 overexpression did not impair the elf18- or flg22-induced phosphorylation of BIK1-HEMAGGLUTININ (HA; Fig. 3D), although HBI1 overexpression seemed to induce a slight phosphorylation of BIK1-HA in the absence of PAMP treatment (Fig. 3D).
Figure 3.
HBI1 overexpression does not affect the accumulation, formation, or activity of the PRR complex. A, Steady-state transcript levels of the individual receptor genes measured by RT-qPCR in 2-week-old seedlings. Transcript levels were normalized to U-box gene expression and relative to Col-0. Results are averages from three independent biological repeats ± 2× se (n = 9). Two-way ANOVA/Holm-Sidak: **P < 0.01. B, elf18-induced EFR-BAK1 complex formation. Coimmunoprecipitation of BAK1 and EFR-GFP-HA was performed on total protein extracts from 2-week-old Arabidopsis seedlings (efr-1/35S:EFR-GFP-HA or HBI1-ox 1 efr-1/35S:EFR-GFP-HA) treated (+) or not (−) with 100 nm elf18 for 5 min. Total proteins (Input) were subjected to immunoprecipitation (IP) with anti-BAK1 antibodies cross linked to TrueBlot beads, followed by immunoblot analysis with anti-BAK1 or anti-GFP antibodies to detect native BAK1 or EFR-GFP-HA, respectively. C, flg22-induced FLS2-BAK1 complex formation. Coimmunoprecipitation of BAK1 and FLS2 was performed on total protein extracts from 2-week-old Arabidopsis seedlings (efr-1/35S::EFR-GFP-HA or HBI1-ox 1 efr-1/35S:EFR-GFP-HA) treated (+) or not (−) with 100 nm flg22 for 5 min. Total proteins (Input) were subjected to immunoprecipitation with anti-BAK1 antibodies cross linked to TrueBlot beads, followed by immunoblot analysis with anti-BAK1 or anti-FLS2 antibodies to detect native BAK1 or FLS2, respectively. D, Phosphorylation of BIK1-HA. Two-week-old Arabidopsis seedlings (35S:BIK1-HA or HBI1-ox 1 35S:BIK1-HA) were treated (+) or not (−) with 100 nm flg22 or elf18 for 5 min. Immunoblot analysis was performed on total protein extracts with anti-HA antibodies to observe BIK1-HA bandshift. CBB, Coomassie Brilliant Blue. All experiments were performed at least twice independently with similar results.
Together, these results show that HBI1 overexpression negatively impacts innate immunity without affecting the accumulation of the PRR complex, or its ligand-induced dynamic and activation, but rather by affecting the steady-state expression of a subset of immune marker genes.
The HBI1 Homologs BEE2 and CIB1 Partially Inhibit Immunity
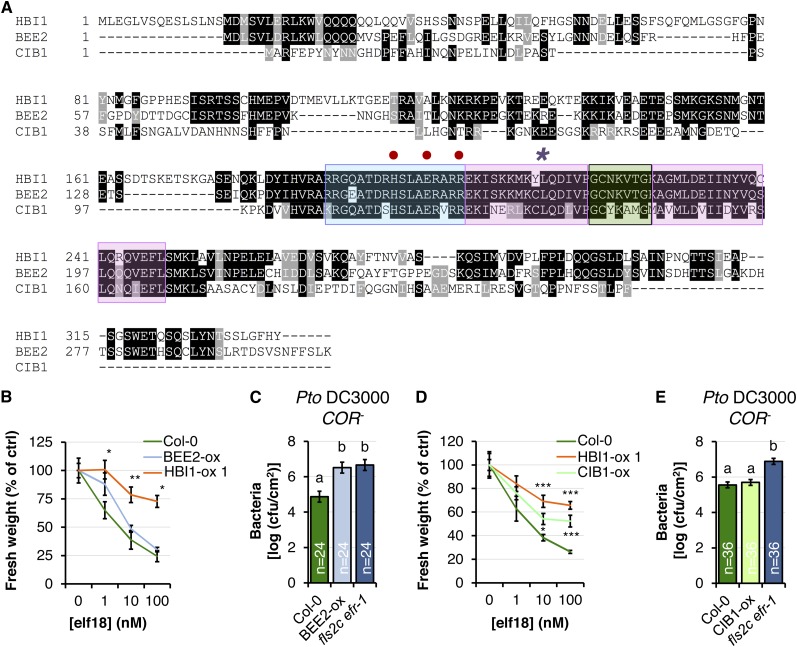
HBI1 is closely related to the bHLHs BRASSINOSTEROID ENHANCED EXPRESSION2 (BEE2 [At4g36540]; Friedrichsen et al., 2002) and CRYPTOCHROME-INTERACTING BHLH (CIB1 [At4g34530]; Figs. 1 and 4A), which are involved in responses to BR and blue light, respectively (Wang et al., 2002; Kim et al., 2005). Interestingly, HBI1 and CIB1 seem to have evolved through an interchromosomal duplication event (Toledo-Ortiz et al., 2003). Therefore, we tested if BEE2 and/or CIB1 are regulated similarly to HBI1. BEE2 was also found to be down-regulated by PAMP treatment in microarray experiments (Wan et al., 2008), while no data were available for CIB1 (Wan et al., 2008; Fig. 1A). Using RT-qPCR, we measured the transcript accumulation of BEE2 and CIB1 after treatment with elf18, flg22, or chitin and found that the expression of both genes was down-regulated by these PAMPs (Fig. 1, C, D, and F).
Figure 4.
Overexpression of the HBI1 homologs BEE2 and CIB1 partially inhibits immunity. A, Multiple alignment of HBI1 and the two closest homologs, BEE2 and CIB1. Blue, pink, and green colors indicate the basic domain, helices, and the loop region of the bHLH domain, respectively. B, Seedling growth inhibition triggered by elf18. Fresh weights of 2-week-old seedlings were measured 1 week after the addition of elf18. Values are represented relative to the untreated plants. Results are averages ± 2× se (n = 6). One-way ANOVA/Holm-Sidak: **P < 0.01, *P < 0.05. C, Spray infection (inoculum, 107 colony-forming units [cfu] mL−1) of 4-week old plants with Pto DC3000 COR−. Bacterial populations were quantified as colony-forming units cm−2 at 3 d post inoculation. Results are averages ± 2× se across two independent experiments (n = 24). Two-way ANOVA/Holm-Sidak: a≠bP < 0.002. D, Seedling growth inhibition triggered by elf18, as in B. Results are averages ± 2× se (n = 6). One-way ANOVA/Holm-Sidak: ***P < 0.00, *P < 0.05. E, Spray infection (inoculum, 107 colony-forming units mL−1) of 4-week-old plants with Pto DC3000 COR− as in C. Results are averages ± 2× se across three independent experiments. Two-way ANOVA/Holm-Sidak: a≠bP < 0.001. All experiments were repeated at least twice with similar results.
Given their homology and their analogous regulation by PAMP treatment, we tested if BEE2 or CIB1 overexpression could also inhibit immunity. To this end, we first generated a homozygous transgenic line expressing 35S:BEE2-YELLOW FLUORESCENT PROTEIN (YFP)-HA in the Col-0 background (BEE2-ox) and confirmed BEE2 overexpression in this line (Supplemental Fig. S3). The BEE2 overexpression line reproducibly exhibited a reduced seedling growth inhibition in response to elf18 treatment (Fig. 4B), although this effect was not necessarily statistically significant. However, the BEE2-ox line exhibited significant hypersusceptibility toward Pto DC3000 COR− compared with wild-type Col-0, equivalent to fls2c efr-1 plants (Fig. 4C), indicating that BEE2 overexpression also impairs immunity. To test if CIB1 has a similar effect in immunity, we used a previously published transgenic line overexpressing CIB1 (CIB1-ox; Liu et al., 2008; Supplemental Fig. S3). Notably, CIB1 overexpression also led to reduced elf18-triggered seedling growth inhibition (Fig. 4D) but did not appear to increase susceptibility to Pto DC3000 COR− (Fig. 4E).
These results suggest that the HBI1 closest homologs BEE2 and CIB1 can also negatively regulate immunity and may act partially redundantly with HBI1.
HBI1 Is a Positive Regulator of BR Responses
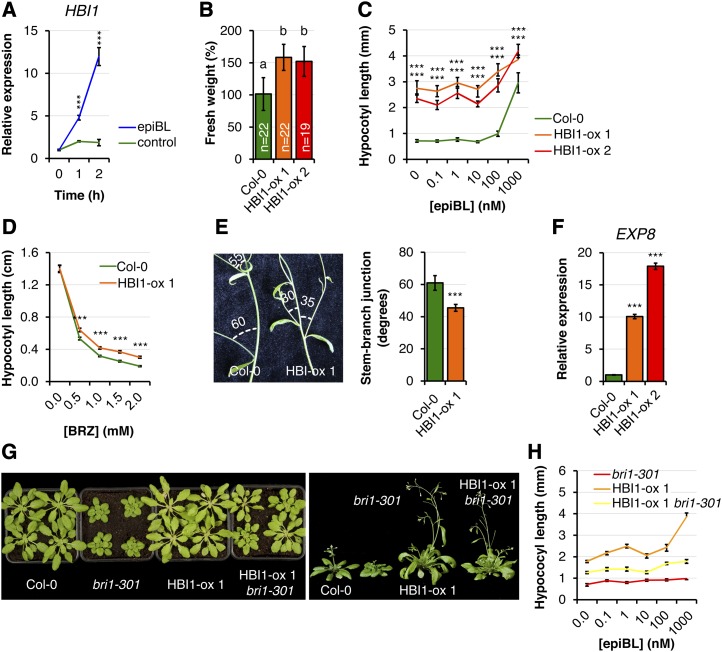
BEE2 was initially identified based on its homology with BEE1, whose expression is induced by brassinolide (BL) treatment in the BR biosynthetic mutant de-etiolatied2 (Friedrichsen et al., 2002). Northern-blot and promoter-luciferase fusion analyses confirmed that BEE1 and BEE2 were BL inducible (Friedrichsen et al., 2002). However, BL-induced expression of HBI1 was not observed (Friedrichsen et al., 2002). It has nevertheless been shown since that HBI1 is induced by BL in both seedlings and adult plants and that HBI1 is a direct target of the master transcriptional regulator BZR1 (Sun et al., 2010; Yu et al., 2011; Fig. 1A). Consistently, we confirmed by RT-qPCR that treatment of seedlings with epiBL increases the accumulation of HBI1 transcripts (Fig. 5A). These results indicate that HBI1 may also regulate BR responses.
Figure 5.
HBI1 overexpression increases BR responses. A, HBI1 expression kinetics after epiBL treatment monitored by RT-qPCR. Two-week-old Col-0 seedlings were treated with 1 µm epiBL for the indicated times. Transcript levels were normalized to U-box transcript levels and relative to time zero. Results are averages ± 2× se (n = 3). One-way ANOVA/Holm-Sidak: ***P < 0.001. B, Fresh weight of 2-week-old seedlings grown in liquid MS medium. Results are averages ± 2× se across four independent experiments. Two-way ANOVA/Holm-Sidak: a≠bP < 0.003. C, Hypocotyl lengths of 6-d-old seedlings grown under long-day conditions on one-half-strength MS medium supplemented with a concentration gradient of epiBL. Results are averages ± 2× se (n = 11–23). One-way ANOVA/Holm-Sidak: ***P < 0.00. D, Hypocotyl lengths of 6-d-old seedlings grown in the dark on one-half-strength MS medium supplemented with a concentration gradient of BRZ. Results are averages ± 2× se (n = 17–21). One-way ANOVA/Dunnett: ***P < 0.001. E, Stem-branch junction bending of long-day-grown 5-week-old plants. Results are averages ± 2× se (n = 20). One-way ANOVA/Holm-Sidak: ***P < 0.001. F, Steady-state expression of the BR marker gene EXP8 in 2-week-old seedlings measured by RT-qPCR. The transcript levels were normalized to U-box gene expression and relative to Col-0. Results are averages ± 2× se (n = 3). One-way ANOVA/Holm-Sidak: ***P < 0.00. G, Phenotypes of 4-week-old short-day-grown (left) and 5-week-old long-day-grown (right) plants. H, Hypocotyl lengths of 6-d-old seedlings grown under long-day conditions on one-half-strength MS medium supplemented with a concentration gradient of epiBL. Results are averages ± 2× se (n = 12–23). All genotypes are different from each other: P < 0.001. All experiments were repeated at least three times with similar results.
We observed that adult plants and seedlings of the two independent HBI1-ox lines are larger than wild-type Col-0 (Fig. 5B; Supplemental Fig. S1). Consistent with a potential role of HBI1 in BR responses, hypocotyls of light-grown HBI1-ox seedlings were longer than wild-type Col-0 (Fig. 5C). In addition, dark-grown HBI1-ox seedlings were less sensitive to the BR biosynthesis inhibitor brassinazole (BRZ) than wild-type Col-0 seedlings, as measured by hypocotyl length measurements (Fig. 5D). Another feature regulated by BR is the angle of stem-branch junctions (Vert and Chory, 2011). Accordingly, the angle of the stem-branch junctions was narrower in HBI1-ox than in wild-type Col-0 plants (Fig. 5E). Also, the expression of the BR marker gene EXPANSIN8 (EXP8), whose expression is induced by BR, was constitutively up-regulated in HBI1-ox seedlings (Fig. 5F). Lastly, HBI1 overexpression suppressed the semidwarf phenotype of the weak bri1 allele bri1-301 in both short-day and long-day conditions (Fig. 5G) and partially reverted the epiBL insensitivity of bri1-301 (Fig. 5H). These results indicate that HBI1 is a positive regulator of BR responses.
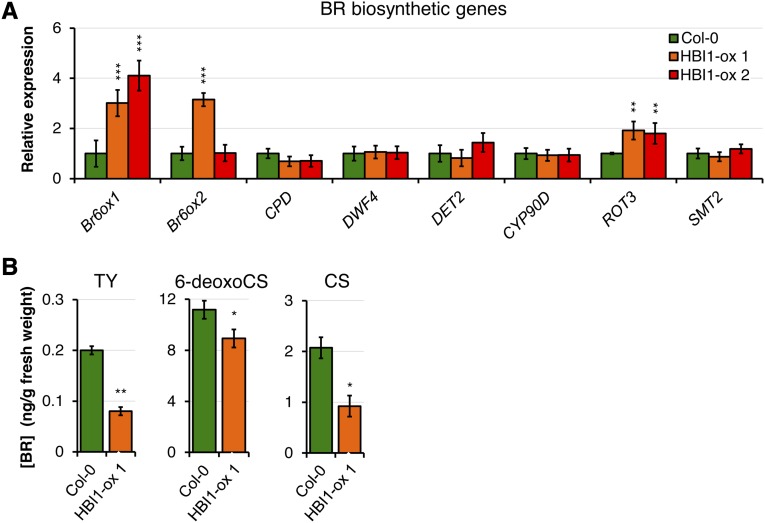
The positive effect of HBI1 overexpression on growth may be due to an augmented BR biosynthesis. Indeed, several Arabidopsis transcription factors were recently identified as directly regulating the expression of key BR biosynthetic enzyme genes (Guo et al., 2010; Poppenberger et al., 2011), including the bHLH transcription factor CESTA (CES), which is closely related to BEE1 and BEE3 (Fig. 1, A and C). While its expression is not affected by BR perception, CES plays a role in BR biosynthesis by regulating the expression of the BR biosynthetic gene Constitutive Photomorphogenesis and Dwarfism (CPD) (Poppenberger et al., 2011). Based on these previous observations, we tested if the increased BR responses of HBI1-ox plants could be due to enhanced BR biosynthesis by measuring the steady-state expression level of different BR biosynthetic genes. HBI1 overexpression had no significant impact on the expression of CPD but enhanced the expression of Brassinosteroid-6-Oxidase1/CYTOCHROME P450 85A1 (Br6ox1/CYP85A1), Br6ox1/CYP85A2, and ROTUNDIFOLIA3 (Fig. 6A).
Figure 6.
Impact of HBI1 overexpression on BR biosynthesis. A, Expression of BR biosynthetic genes in 2-week-old seedlings measured by RT-qPCR. The transcript levels were normalized to U-box gene expression and then relative to time zero. Results are averages ± 2× se (n = 3). Two-way ANOVA/Holm-Sidak: **P < 0.01, ***P < 0.001. B, Endogenous BR levels of 4-week-old short-day-grown plants: typhasterol (TY), 6-deoxoCS, and CS. Results are averages from three independent biological repeats ± 2× se (n = 3). Two-way ANOVA/Holm-Sidak: **P < 0.01, *P < 0.05. All experiments were repeated at least three times with similar results.
We next tested if the up-regulation of a subset of BR biosynthetic genes affects BR levels in HBI1-ox lines. Unexpectedly, we found that the steady-state levels of typhasterol, 6-deoxocastasterone (6-deoxoCS), and castasterone (CS) were significantly lower in HBI1-ox plants compared with wild-type Col-0 (Fig. 6B). Together with Br6ox2/CYP85A2, Br6ox1/CYP85A1 acts in the late C-6 oxidation pathway to mediate the conversion of 6-deoxoCS to CS (Kim et al., 2004, 2005). However, unlike Br6ox2/CYP85A2, Br6ox1/CYP85A1 does not catalyze the conversion of CS to BL (Kim et al., 2005). In addition, compared with Br6ox2/CYP85A2, the expression of Br6ox1/CYP85A1 is subsidiary and restricted to specific tissues, and its C-6-oxidase activity is lower (Shimada et al., 2003; Kim et al., 2005). Consistent with its apparent minor role in BR biosynthesis, Br6ox1/CYP85A1 loss of function or overexpression does not have any noticeable impact on BR biosynthesis and/or plant growth (Kim et al., 2005). Therefore, although we could observe that several BR biosynthesis genes are constitutively up-regulated in HBI1-ox lines (Fig. 6A), it is highly unlikely that the effect of HBI1 overexpression on growth and BR responses is caused by increased BR biosynthesis. Rather, our results (particularly the BR measurements and reduced sensitivity to BRZ) suggest that HBI1 is involved in BR signaling. Active BR signaling negatively regulates the expression of BR biosynthetic genes in a feedback-loop regulation (Bancoş et al., 2002; Tanaka et al., 2005). Notably, this regulation seems to be bypassed by HBI1 overexpression, as no significant down-regulation of BR biosynthetic genes could be observed in the HBI1-ox lines (Fig. 6A). Yet, constitutive BR signaling (as observed in bak1/elongated-D, bzr1-D, or bes1-D, for example) leads to a depletion of bioactive BRs and, therefore, to reduced levels of endogenous CS (Noguchi et al., 1999; Wang et al., 2002; Chung et al., 2012). A similar situation likely occurs in HBI1-ox lines, which exhibit increased BR signaling (Fig. 5), thereby using more CS and ultimately resulting in a reduction of endogenous CS levels (Fig. 6B).
Mutation of the Conserved Leu-214 Residue Has a Dominant-Negative Effect on HBI1 Function
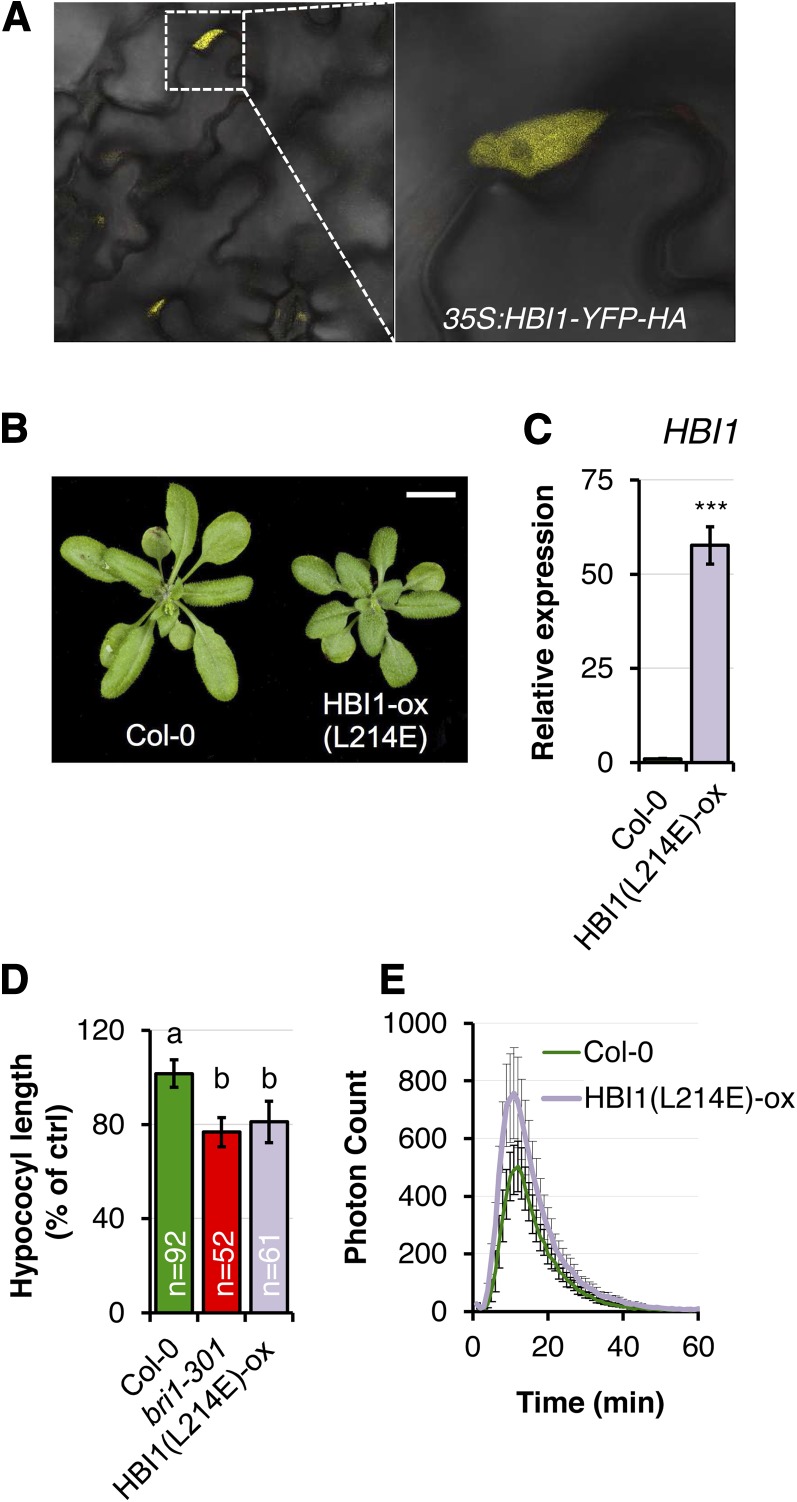
HBI1 encodes a hypothetical bHLH protein that contains the conserved His/Arg-9, Glu-13, and Arg-17 residues (corresponding to His-196, Glu-200 and Arg-204 in HBI1, respectively; indicated by red dots in Fig. 4A) in the basic motif of DNA-binding bHLH (Toledo-Ortiz et al., 2003; Carretero-Paulet et al., 2010). Consistent with a potential role as a DNA-binding bHLH transcription factor, we observed that HBI1-YFP-HA localizes to the nucleus in HBI1-ox lines (Fig. 7A).
Figure 7.
Mutation of the conserved residue Leu-214 impairs HBI1 function. A, Subcellular localization of HBI1-YFP in HBI1-ox 1 epidermal cells of 2-week-old seedlings. B, Phenotypes of long-day-grown 4-week-old Col-0 and a homozygous Arabidopsis line stably expressing 35S:HBI1(L214E)-YFP-HA [HBI1(L214E)-ox]. C, HBI1 expression levels in 2-week-old seedlings measured by RT-qPCR. The transcript levels were normalized to U-box gene expression and relative to Col-0. Results are averages ± 2× se (n = 3). One-way ANOVA/Holm-Sidak: ***P < 0.001. D, Hypocotyl lengths of 6-d-old seedlings grown in continuous light on one-half-strength MS medium. Results are averages ± 2× se across four independent experiments. Two-way ANOVA/Holm-Sidak: a≠bP < 0.001. E, Oxidative burst measured in leaf discs of 4-week-old plants in response to 100 nm elf18. Results are averages ± 2× se (n = 8). All experiments were repeated at least twice with similar results.
Our results based on HBI1 overexpression indicate that HBI1 is a negative regulator of immunity but a positive regulator of BR signaling. We next tested if HBI1 loss of function would support these conclusions. Single HBI1 insertional mutants are unlikely to give a noticeable phenotype, given the homology between HBI1, BEE2, and CIB1 (Fig. 4A) and the partially redundant functions of these three bHLHs in negatively regulating immunity (Fig. 4, B–E). Accordingly, the single bee2 mutant did not exhibit any obvious developmental phenotype (Friedrichsen et al., 2002). In addition, we could not identify true null HBI1 mutants among the SALK and SAIL transfer DNA lines for HBI1 currently available in the stock centers (SALK_090958, SALK_015771, SALKseq_46259, SALKseq_65819, SALKseq_79833, SAIL_540_E05, SAIL_77_D01, and SAIL_1263_G02). Therefore, we speculated that a mutant variant of HBI1 carrying a mutation in the conserved Leu-214 residue (L214E, indicated by the asterisk in Fig. 4A) within the bHLH domain (Brownlie et al., 1997; Roig-Villanova et al., 2007) could lead to a dominant-negative effect when overexpressed in wild-type Col-0. Similarly, it was recently shown that a mutation in the orthologous amino acid of the atypical bHLH PHYTOCHROME RAPIDLY REGULATED1, involved in shade avoidance, affects its homodimerization and compromises its signaling capacity (Roig-Villanova et al., 2007; Carretero-Paulet et al., 2010). We could not test whether the L214E mutation affects HBI1 homodimerization, as we found that HBI1 was always autoactive in yeast two-hybrid assays when fused to pLexA. Nevertheless, we found that soil-grown homozygous transgenic plants expressing 35S:HBI1(L214E)-YFP-HA in the Col-0 background [HBI1(L214E)-ox] were smaller than wild-type Col-0 under long-day conditions (Fig. 7, B and C). In addition, hypocotyls of light-grown HBI1(L214E)-ox seedlings were significantly shorter than hypocotyls of wild-type Col-0 seedlings and similar in length to bri1-301 hypocotyls (Fig. 7D). Therefore, overexpression of HBI1(L214E) led to a phenotype opposite to that of HBI1-ox plants, suggesting that HBI1(L214E) has a dominant-negative effect on endogenous HBI1 and potentially closely related proteins, such as BEE2 and CIB1.
HBI1 Interacts with the Inhibitory Helix-Loop-Helix Proteins ATBS1-INTERACTING FACTOR1-4 and INCREASED LAMINA INCLINATION1 BINDING BHLH PROTEIN1
bHLHs typically act as homodimers or heterodimers with other bHLHs or together with atypical non-DNA-binding bHLHs to regulate gene expression (Toledo-Ortiz et al., 2003). In particular, the Arabidopsis atypical bHLH PACLOBUTRAZOL RESISTANT1 (PRE1) interacts with the atypical bHLH INCREASED LAMINA INCLINATION1 BINDING BHLH PROTEIN1 (IBH1) to regulate BR signaling as positive and negative regulators, respectively (Zhang et al., 2009). In addition, the related atypical bHLH PRE3 (also called ATBS1, for ACTIVATION-TAGGED BRI1 SUPPRESSOR1) interacts with the atypical bHLHs ATBS1-INTERACTING FACTORS (AIFs; which are related to IBH1) to regulate BR signaling (Wang et al., 2009). Notably, interactions between BEE2 and AIF1, CIB1 and AIF2, and CIB1, BEE2, HBI1, and IBH1 were previously identified in yeast-two hybrid assays (Wang et al., 2009; http://interactome.dfci.harvard.edu/A_thaliana/). Given the homology of HBI1 with BEE2 and CIB1, we tested if HBI1 could also interact with AIF proteins. We found that HBI1 interacts with AIF1, AIF2, AIF3, and AIF4 in yeast two-hybrid assays (Supplemental Fig. S4A). The interaction between HBI1 and AIF2 was confirmed in planta by coimmunoprecipitation following transient expression of HBI1-GFP and AIF2-HA in Nicotiana benthamiana (Supplemental Fig. S4B). Next, we confirmed that HBI1 can interact with IBH1 in the yeast two-hybrid assay (Supplemental Fig. S4C) and extended these findings in planta by showing that HBI1 interacts with IBH1 in N. benthamiana upon transient expression as well as in stable transgenic Arabidopsis lines expressing both HBI1-GFP and IBH1-HA (Supplemental Fig. S4, D and E).
CONCLUSION
We initially hypothesized that negative regulators of PTI signaling might be under negative transcriptional regulation by different PAMPs. Among 15 genes commonly down-regulated by flg22, elf26, and chitin, we focused on the bHLH HBI1. We found that HBI1 overexpression leads to reduced PAMP-induced responses (such as ROS burst, seedling growth inhibition, and marker gene expression) and to enhanced disease susceptibility to Pto DC3000, indicating that HBI1 acts genetically as a negative regulator of PTI. HBI1 is phylogenetically closely related to BEE2, a bHLH previously shown to be involved in BR responsiveness (Friedrichsen et al., 2002). Accordingly, we found that BEE2 overexpression also leads to reduced elf18-triggered ROS burst and to increased susceptibility to Pto DC3000, suggesting that HBI1 and BEE2 can act in a partially redundant manner.
HBI1-overexpressing plants show phenotypes reminiscent of BR-hyperproducing or -hyperresponsive plants. Consequently, HBI1 overexpression leads to increased BR responsiveness, demonstrating that HBI1 is a positive regulator of BR signaling, similar to BEE2. Consistent with our findings, it was recently published, while this article was in preparation, that HBI1 is a positive regulator of cell elongation and of BR responses (Bai et al., 2012). HBI1 is a direct transcriptional target of the master regulator BZR1 and is induced in response to BL treatment (Sun et al., 2010; this study). In addition, HBI1 appears to be under the control of a complex regulatory protein network involving interactions with atypical helix-loop-helix proteins that are negative regulators of BR signaling (such as IBH1 and AIF1 to AIF4; Wang et al., 2009; Zhang et al., 2009; Bai et al., 2012; Ikeda et al., 2012; this study). While genome-wide studies are still required to identify HBI1 target genes, the expansin genes EXP1 and EXP8 are examples of BR-regulated genes that are controlled by HBI1 (Bai et al., 2012; this study).
Activation of the BR pathway has recently been shown to inhibit PTI signaling (Albrecht et al., 2012; Belkhadir et al., 2012). Therefore, the negative effect of HBI1 overexpression on PAMP-triggered responses and PTI is most likely caused by its primary positive role in BR signaling. Consistent with our previous findings (Albrecht et al., 2012), the increased BR responses caused by HBI1 overexpression lead to a reduced ROS burst and seedling growth inhibition in response to PAMP treatment, and this effect is not caused by a reduced PAMP-induced complex formation between PRRs and BAK1. Accordingly, HBI1 overexpression also leads to the inhibition of chitin-induced ROS burst, a response that is BAK1 independent. Notably, beyond confirming the previously reported antagonism between the BR and PTI pathways, this study uncovers a complex interlinked regulation of BR-mediated growth and PAMP-induced innate immunity. Consistent with HBI1 being a direct BZR1 transcriptional target (Sun et al., 2010; Bai et al., 2012), this study complements our recent findings that BZR1 is the key regulator of the BR-PTI antagonism (Lozano-Durán et al., 2013). Altogether, our results illustrate that BZR1 controls the expression of transcription factors (e.g. WRKY11, WRKY15, WRKY18, and HBI1; Lozano-Durán et al., 2013; this study), which themselves might control the expression of PTI components whose identities remain to be identified.
While activation of the BR pathway clearly inhibits PTI responses, one of the early transcriptional events resulting from PAMP perception is a dampening of this antagonism by repressing the expression of HBI1 that emerges as a novel key positive regulator of BR responses. It is tempting, therefore, to speculate that seedling growth inhibition triggered by long-term PAMP treatment may result from the inhibition of key transcription factors involved in BR-mediated growth, such as HBI1, ACTIVATORS FOR CELL ELONGATION, and other bHLHs (Bai et al., 2012; Ikeda et al., 2012; this study). It is now important to establish the mechanistic details of how the BR-PTI antagonism downstream of BAK1 is mediated and, conversely, how PAMP perception leads to the transcriptional repression of key components of the BR pathway.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The genetic background for the Arabidopsis (Arabidopsis thaliana) mutants and transgenic lines described in this study is Col-0, with the exception of 35S:Myc-CIB1 (CIB1-ox; Liu et al., 2008), which is in the Columbia-4 accession. The following lines were described previously: fls2c efr-1 (Nekrasov et al., 2009), efr-1/pEFR-EFR-eGFP-HA (Nekrasov et al., 2009), pBIK1:BIK1-HA (Zhang et al., 2010), 35S:Myc-CIB1 (Liu et al., 2008), bri1-301 (Xu et al., 2008), and 35S:myc-IBH1 (Zhang et al., 2009).
Arabidopsis plants were grown in soil at 22°C under a 10-h photoperiod and 75% humidity (short day), unless otherwise indicated. Long-day conditions were a 16-h photoperiod under 20°C and 80% humidity. Seedlings were grown axenically at 22°C under a 16-h photoperiod in Murashige and Skoog (MS) medium (Duchefa) with 1% (w/v) Suc with or without 0.8% (w/v) phytoagar (Duchefa).
Nicotiana benthamiana plants were grown in soil at 22°C under a 16-h photoperiod and 55% humidity.
Molecular Cloning and Generation of Transgenic Lines
To generate 35S:HBI1-YFP-HA and 35S:BEE2-YFP-HA, the complementary DNAs (cDNAs) for these genes were PCR amplified without stop codon and cloned into pENTR-/D-Topo (pENTR-HBI1; for a list of primers used for cloning, see Supplemental Table S2). Inserts from correct clones were then subcloned into pEarleyGate101 using Gateway LR Clonase II enzyme (Invitrogen). To generate HBI1(L214E), site-directed mutagenesis was performed on pENTR-HBI1 using the QuickChange XL site-directed mutagenesis kit (Stratagene) following the manufacturer’s instructions. The mutant HBI1 variant was then subcloned into pEarleyGate101. All resulting constructs were electroporated into Agrobacterium tumefaciens and then transformed into wild-type Col-0 Arabidopsis using the floral dipping method (Clough and Bent, 1998).
Similarly, the cDNAs of HBI1, IBH1, and AIF2 were PCR amplified without stop codon and cloned into pENTR-/D-Topo. Inserts from correct clones were then subcloned into pGWB5 (for HBI1) or pGWB14 (for IBH1 and AIF2) using Gateway LR Clonase II enzyme (Invitrogen). All resulting constructs were electroporated into A. tumefaciens and used for transient expression in N. benthamiana.
Chemicals
The flg22 and elf18 peptides were purchased from Peptron, and chitin was purchased from Yaizu Suisankagaku. epiBL was purchased from Xiamen Topusing Chemical, and BRZ was purchased from Sigma.
Phylogenetic Analysis
Amino acid sequences of the clade 18 bHLH transcription factors were downloaded from http://www.arabidopsis.org (Lamesch et al., 2012). Multiple sequence alignment was performed using the ClustalW program (Larkin et al., 2007), and the alignment was edited for the bHLH domain. The unrooted tree was constructed by the neighbor-joining algorithm using the bootstrapping option provided in ClustalW. Bootstrapping was performed 1,000 times.
PTI Assays
ROS burst and seedling growth inhibition assays were performed as described (Schwessinger et al., 2011).
RNA Isolation and RT-qPCR
Total RNA extraction and cDNA synthesis were performed as described by Albrecht et al. (2012). The relative expression values using U-box (At5g15400) as a reference were determined by quantitative PCR using SYBR Green JumpStart Taq ReadyMix (Sigma) and the Bio-Rad CFX96 Real-Time System C1000 Thermal Cycler. All measurements where done in triplicate. For a list of primers used for RT-qPCR, see Supplemental Table S2.
Bacterial Infection Assays
For bacterial infection assays, Pseudomonas syringae pv tomato DC3000 wild type, ∆avrPto/∆avrPtoB (Lin and Martin, 2005), and COR− (Brooks et al., 2004) were streaked from glycerol stocks onto solid Lysogeny medium (10g/L triptone [Merk], 5g/L yeast extract [Merk], 5g/L NaCl, 1g/L D-glucose, 1% [w/v] agar and grown for 2 d with appropriate selection, then restreaked on fresh Lysogeny plates and grown overnight at 28°C. The bacteria were then scraped off the plates, diluted in 10 mm MgCl2, and adjusted to an optical density at 600 nm (OD600) = 0.02, and 0.4 µL mL−1 Silwet L-77 was added to the suspension before spraying. The plants were kept at high humidity for 3 d after spraying.
Hypocotyl Length and Stem-Branch Junction Angle Assays
For hypocotyl length assays, seeds were sown on one-half-strength MS medium with 1% (w/v) Suc and 0.8% (w/v) phytoagar supplemented with BL or BRZ, stratified for 2 d in the dark at 4°C, and then grown at 22°C under the specified light conditions. Hypocotyl length was quantified with ImageJ software (http://rsbweb.nih.gov/ij/) after photography of 5-d-old seedlings. The stem-branch junction angles were measured with a protractor.
Confocal Microscopy
Analysis of the subcellular localization of HBI1-YFP-HA was done by confocal laser-scanning microscopy with a DM6000B/TCS SP5 confocal microscope (Leica Microsystems), as described by Nekrasov et al. (2009).
Protein Extraction, Coimmunoprecipitation, and Western Blotting
Coimmunoprecipitation assays were performed on extracts from either one N. benthamiana leaf or 48 2-week-old sterile-grown Arabidopsis seedlings. Extraction buffer (150 mm Tris, pH 7.5, 1 mm EDTA, pH 8, 150 mm NaCl, 10% [v/v] glycerol, 1% [v/v] IGEPAL CA630, 10 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail P9599 [Sigma-Aldrich]) was added to the liquid nitrogen-ground samples (1.5 mL g−1). The extracts were cleared by centrifugation (20 min, 13,000 rpm, 4°C) and filtered through Bio-Rad Poly-Prep chromatography columns. For immunoprecipitation, 1.4 mL of extract adjusted to the lowest concentration was incubated with 15 to 25 µL of immunoprecipitation bead slurry (2–4 h, 4°C). The beads were washed four to six times with 0.5% IGEPAL-Tris-buffered saline before boiling with 3× loading buffer (30% [v/v] glycerol, 3% [v/v] SDS, 100 mm Tris, pH 6.8, 0.05% bromphenol blue, and 50 mm dithiothreitol). The samples were loaded on 8% (w/v) to 15% (v/v) SDS-PAGE gels depending on the size of the protein of interest. SDS-PAGE and immunoblotting were done as described by Roux et al. (2011). BIK1-HA phosphorylation was assayed as described by Albrecht et al. (2012).
The following antibodies were diluted in 5% nonfat milk-Tris-buffered saline-Tween and used in the indicated dilutions: polyclonal anti-BAK1 from rabbit (1:1,000; Roux et al., 2011); polyclonal anti-FLS2 from rabbit (1:1,000; Chinchilla et al., 2007); anti-HA horseradish peroxidase (HRP) high affinity from rat IgG1 (1:1,000; Roche); anti-Myc antibody C-MYC (9E10) from mouse (Insight Biotechnology); anti-GFP GF-P [B-2] HRP (SC-9996; 1:5,000; Insight Biotechnology); anti-rabbit IgG anti-rabbit-HRP (1:10,000; Sigma-Aldrich); and TrueBlot anti-rabbit IgG HRP (1:10,000; eBIOSCIENCE). For the respective coimmunoprecipitations, GFPTrap-A beads (Chromotek), TrueBlot anti-rabbit Ig beads (eBIOSCIENCE), or anti-HA affinity matrix (Roche) was used as indicated by the manufacturer.
Transient Expression in N. benthamiana
A. tumefaciens strain GV3101 carrying 35S:HBI1-GFP, 35S:IBH1-HA, or 35S:AIF2-HA was streaked from glycerol stocks onto solid L medium supplemented with appropriate antibiotics and grown at 28°C. After 2 d, bacteria were restreaked on fresh plates and grown overnight at 28°C. The bacteria were scraped off the plates and resuspended in 10 mm MgCl2 to OD600 = 0.6. Bacterial suspensions were mixed 1:1 or with water to final OD600 = 0.3 per strain and syringe infiltrated into 3-week-old N. benthamiana leaves. Sampling in liquid nitrogen was done 2 d after inoculation.
Yeast Two-Hybrid Assays
Direct interactions were determined using the Matchmaker LexA Two-Hybrid System (Clontech). Auto activation (synthetic dextrose/−uracil/−Trp/−His) and interaction (synthetic defined/−uracil/−Trp/−His/−Leu) were tested between HBI1 in the pB42AD vector (activation domain) and IBH1 or AIFs in pLexA (DNA binding) following the manufacturer’s instructions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transgenic HBI1 over-expressing Arabidopsis lines.
Supplemental Figure S2. HBI1 over-expression affects flg22- and chitin induced ROS burst.
Supplemental Figure S3. Transgenic BEE2 and CIB1 over-expressing lines.
Supplemental Figure S4. HBI1 interacts with the atypical bHLHs AIFs and IBH1.
Supplemental Table S1. Genes down-regulated by elf26, flg22, and chitooctaose.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Zhi-Yong Wang, Jianming Li, Joanne Chory, Chentao Lin, and Jian-Min Zhou for providing published biomaterials. The excellent technical assistance of the the Sainsbury Laboratory Plant Transformation team and John Innes Centre Horticulture Services is much appreciated. We thank Rosa Lozano-Durán for commenting on the manuscript and all members of the Zipfel laboratory for useful discussions.
Glossary
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- PTI
PAMP-triggered immunity
- LRR-RK
leucine-rich repeat receptor kinase
- ROS
reactive oxygen species
- BR
brassinosteroid
- bHLH
basic helix-loop-helix
- RT-qPCR
quantitative real-time reverse transcription-PCR
- Col-0
Columbia-0
- Pto
Pseudomonas syringae pv tomato
- BL
brassinolide
- BRZ
brassinazole
- 6-deoxoCS
6-deoxocastasterone
- CS
castasterone
- MS
Murashige and Skoog
- OD600
optical density at 600 nm
- HRP
horseradish peroxidase
- cDNA
complementary DNA
Footnotes
This work was supported by the Gatsby Charitable Foundation (to C.Z.), by the United Kingdom Biotechnology and Biological Sciences Research Council (grant no. BB/E024874/1 ERA-PG [RLPRLKs] and grant no. BB/G024944/1 ERA-PG [Pathonet] to C.Z.), by the National Research Foundation of Korea (grant no. NRF.–2011–220–C00059 to S.-K.K.), and by the John Innes Centre and the Sainsbury Laboratory rotation program (to B.S.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY. (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancoş S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M. (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, Kunkel BN. (2004) Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Brownlie P, Ceska T, Lamers M, Romier C, Stier G, Teo H, Suck D. (1997) The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure 5: 509–520 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807–838 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2012) Benefits of brassinosteroid crosstalk. Trends Plant Sci 17: 594–605 [DOI] [PubMed] [Google Scholar]
- Chung Y, Choe V, Fujioka S, Takatsuto S, Han M, Jeon JS, Park YI, Lee KO, Choe S. (2012) Constitutive activation of brassinosteroid signaling in the Arabidopsis elongated-D/bak1 mutant. Plant Mol Biol 80: 489–501 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J. (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D, Tsuda K, Katagiri F. (2012) The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J 71: 194–204 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. (2012) A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24: 4483–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Chang SC, Lee JS, Hwang B, Takatsuto S, Yokota T, Kim SK. (2004) Cytochrome P450-catalyzed brassinosteroid pathway activation through synthesis of castasterone and brassinolide in Phaseolus vulgaris. Phytochemistry 65: 679–689 [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY. (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Hwang JY, Kim YS, Joo SH, Chang SC, Lee JS, Takatsuto S, Kim SK. (2005) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17: 2397–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lin NC, Martin GB. (2005) An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe Interact 18: 43–51 [DOI] [PubMed] [Google Scholar]
- Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L. (2013) Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA 110: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C. (2013) The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2: e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Mosher S, Seybold H, Rodriguez P, Stahl M, Davies KA, Dayaratne S, Morillo SA, Wierzba M, Favery B, Keller H, et al. (2013) The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J 73: 469–482 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. (2004) The transcriptional innate immune response to flg22: interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Rozhon W, Khan M, Husar S, Adam G, Luschnig C, Fujioka S, Sieberer T. (2011) CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J 30: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF. (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D. (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S. (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Kuo YC, Mishra S, Tsai CH, Chien CC, Chen CW, Desclos-Theveniau M, Chu PW, Schulze B, Chinchilla D, et al. (2012) The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24: 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho A, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. (2011) Crosstalk in cellular signaling: background noise or the real thing? Dev Cell 21: 985–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J. (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y. (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]