A group of plant-specific protein phosphatases has a rich evolutionary history and performs essential roles in development.
Abstract
Protein phosphatases with Kelch-like domains (PPKL) are members of the phosphoprotein phosphatases family present only in plants and alveolates. PPKL have been described as positive effectors of brassinosteroid (BR) signaling in plants. Most of the evidence supporting this role has been gathered using one of the four homologs in Arabidopsis (Arabidopsis thaliana), BRASSINOSTEROID-INSENSITIVE1 SUPPRESSOR (BSU1). We reappraised the roles of the other three members of the family, BSL1, BSL2, and BSL3, through phylogenetic, functional, and genetic analyses. We show that BSL1 and BSL2/BSL3 belong to two ancient evolutionary clades that have been highly conserved in land plants. In contrast, BSU1-type genes are exclusively found in the Brassicaceae and display a remarkable sequence divergence, even among closely related species. Simultaneous loss of function of the close paralogs BSL2 and BSL3 brings about a peculiar array of phenotypic alterations, but with marginal effects on BR signaling; loss of function of BSL1 is, in turn, phenotypically silent. Still, the products of these three genes account for the bulk of PPKL-related activity in Arabidopsis and together have an essential role in the early stages of development that BSU1 is unable to supplement. Our results underline the functional relevance of BSL phosphatases in plants and suggest that BSL2/BSL3 and BSU1 may have contrasting effects on BR signaling. Given that BSU1-type genes have likely undergone a functional shift and are phylogenetically restricted, we caution that inferences based on these genes to the whole family or to other species may be misleading.
The polyhydroxylated sterols collectively known as brassinosteroids (BRs) mediate several growth and developmental processes in plants. BRs induce strong elongation responses in bioassays, and plants unable to produce or perceive them are severely dwarfed, with small, dark green leaves caused by concerted defects in cell division and elongation (Zhiponova et al., 2013). Signaling by BRs is one of the more thoroughly explored transduction pathways in flowering plants (for review, see Zhu et al., 2013). The perception of brassinolide (BL; the most active of BRs) by the extracellular domain of BRASSINOSTEROID-INSENSITIVE1 (BRI1)-type receptor kinases results in the formation of a signaling complex that includes coreceptors of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE family. A series of dual-specificity auto- and trans-phosphorylation reactions in the intracellular kinase domains trigger a signaling cascade that, through shifting phosphorylation states, ultimately reaches the transcription factors BRI1-ETHYL METHANESULFONATE-SUPPRESSOR1 (BES1) and BRASSINAZOLE-RESISTANT1 (BZR1). BES1 and BZR1 belong to a plant-specific group of basic helix-loop-helix proteins with a C-terminal regulatory domain rife with Pro-associated Ser/Thr residues, substrates of a group of GLYCOGEN-SYNTHASE KINASE3 (GSK3) kinases whose most conspicuous member is BRASSINOSTEROID-INSENSITIVE2 (BIN2). In the absence of BL, BIN2-mediated phosphorylation prevents these transcription factors from binding to DNA, promotes their export from the nucleus mediated by 14-3-3 proteins, and possibly targets them for proteolytic turnover. The perception of BL inactivates BIN2 and leads to the accumulation of the dephosphorylated, active forms of BES1 and BZR1 in the nucleus.
Protein phosphatases have also found a role in this pathway rich in phosphorylation events. Protein Phosphatase2A (PP2A) holoenzymes have been shown to participate in at least two steps. On the one hand, the activity and/or localization of PP2A presumably leads to the dephosphorylation of activated BRI1, accelerating its turnover and shutting off the response (Wu et al., 2011). On the other hand, regulatory subunits of PP2A associate with BZR1; upon inactivation of BIN2, dephosphorylation of BZR1 allows its rapid accumulation in the nucleus and the consequent transcriptional effects (Tang et al., 2011).
The other phosphatases that have been involved in the pathway are also members of the Phosphoprotein phosphatase (PPP) family of Ser/Thr phosphatases, but they belong to a subfamily more closely related to PP1 characterized by a C-terminal catalytic domain with diagnostic sequence features linked to an N-terminal Kelch-repeat β-propeller domain. For this reason, they have been called protein phosphatases with Kelch-like domains (PPKL; Kutuzov and Andreeva, 2002; Moorhead et al., 2009; Oh et al., 2011; Uhrig et al., 2013). Arabidopsis (Arabidopsis thaliana) BSU1 (for BRI1-SUPPRESSOR1) was the first PPKL to be functionally characterized as a positive effector of BR signaling (Mora-García et al., 2004). Overexpression of BSU1 in plants suppressed the phenotype of a weak bri1 mutant and led to the accumulation of dephosphorylated BES1/BZR1 (Mora-García et al., 2004; Kim et al., 2009); also, the expression of BSU1 in protoplasts increased the levels of the nuclear pool of BES1/BZR1, overcoming the effects of coexpressed BIN2 (Ryu et al., 2007, 2010a, 2010b; Kim et al., 2009). BSU1 has been shown to interact with members of the membrane-bound receptor-like cytoplasmic kinase (RLCK) families XII (BR Signaling kinase [BSK]; Kim et al., 2009) and VIIc (Constitutive differential growth [CDG1]; Kim et al., 2011) that are phosphorylated by activated BRI1 (Tang et al., 2008; Kim et al., 2011; Sreeramulu et al., 2013). BSU1 interacts with and inactivates BIN2 through the dephosphorylation of a GSK3-specific autophosphorylated Tyr necessary for full activity (Kim et al., 2009) but is unable to dephosphorylate a variant carrying the dominant-negative bin2-1 mutation, explaining why this mutation is epistatic over the overexpression of BSU1 (Kim et al., 2009). Taken together, these results led to the current working model, whereby, upon BL perception, BSU1 is activated by the membrane-bound signaling complex and, in turn, inactivates BIN2.
BSU1 is one of a four-member gene family in Arabidopsis (throughout this work, we also call them BSL, for BSU1-Like; Mora-García et al., 2004). The remaining three homologs, BSL1 to BSL3, have received comparatively less attention and are assumed to perform overlapping roles in the pathway (Mora-García et al., 2004; Kim et al., 2009, 2011). In fact, a double knockout for BSU1 and its closest paralog, BSL1, combined with a double knockdown for BSL2 and BSL3 (called bsu-q) produced dwarfed plants apparently defective in BR signaling (Kim et al., 2009, 2012). However, evidence for contrasting activities between the members of the family was recently obtained in rice (Oryza sativa), where an amino acid change in the Kelch domain of a BSL2 homolog (GL3.1) was identified as a quantitative trait locus that impacts on grain length (Hu et al., 2012; Qi et al., 2012; Zhang et al., 2012). Grain length increased in loss-of-function mutant plants for Os03g44500 or its closest paralog but decreased in plants lacking the remaining homolog, related to BSL1 (Zhang et al., 2012). Divergent evolutionary paths suggestive of neofunctionalization have also been noted between Arabidopsis BSU1 and BSL1 (Liu et al., 2011).
In this work, using phylogenetic, functional, and genetic evidence, we reappraised the roles of the BSL genes in Arabidopsis, with special emphasis on BSL1, BSL2, and BSL3. We show that these three genes have been highly conserved in all land plants; in contrast, BSU1-type genes are only found in the Brassicaceae and have anomalously divergent sequences that set them apart in an otherwise conservative family. Moreover, we found that, early in development, the three most conserved members of the family together have a critical role that BSU1 cannot substitute. Loss of function of the two closest paralogs, BSL2 and BSL3, causes a peculiar set of phenotypic alterations that are unrelated to the typical BR-deficient syndrome. In fact, the responses to BRs in bsl loss-of-function mutants are only marginally affected. BSL1, BSL2, and BSL3 interact with intermediates of the BR signaling pathway, and strong overexpression of BSL2 has a stimulatory effect on the BR pathway, but in all cases in a quite different manner from what has been reported for BSU1. Therefore, our results show that BSL phosphatases play an essential function in plants and, at the same time, that their role in BR signaling should be reassessed. In addition, we suggest that drawing mechanistic inferences based on the divergent and phylogenetically restricted BSU1 may be misleading.
RESULTS
Phylogenetic Analysis of BSL Phosphatases in Land Plants
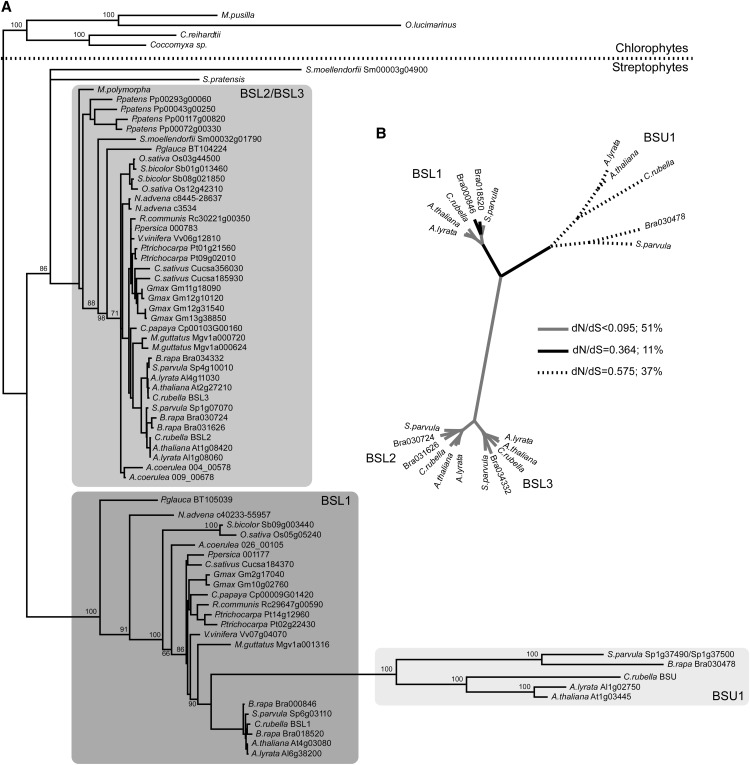
In order to clarify the phylogenetic relationships of PPKL phosphatases in land plants, we performed a search and curation of related sequences in fully sequenced genomes and EST collections from green organisms (Supplemental Data Set S1). Figure 1A shows the phylogenetic tree recovered using full-length amino acid sequences and some EST sequences that were long enough to allow for a robust assignment. We also built a tree using the highly conserved C-terminal phosphatase domain, to both maximize the stringency of the comparisons and to include some relevant taxonomic groups for which only 3′ end sequences were available (Supplemental Fig. S1). The general topology of both trees agrees and reflects accepted phylogenetic affinities (Soltis et al., 2008; Leliaert et al., 2011). PPKL phosphatases are found in all Viridiplantae, from the tiniest unicellular algae (Micromonas and Ostreococcus spp.; Vaulot et al., 2008) to multicellular land plants. In the latter, these genes can be grouped into two clearly defined clades: the BSL2/BSL3 clade, with representatives from liverworts to angiosperms, and the BSL1 clade, where bona fide sequences are first detected in Cycas rumphii, a member of the most ancient extant group of gymnosperms (Brenner et al., 2003). Sampling gaps make it difficult to confirm the presence of BSL1-type sequences in intermediate groups. The genome of the clubmoss Selaginella moellendorfii codes for two BSL genes: one of them (Sm32g01790) clearly belongs in the BSL2/BSL3 group, whereas the other (Sm3g04900) shows an unstable position but tends to cluster within the BSL1 clade. The available ESTs from the fern Pteridium aquilinum (Der et al., 2011; not included in the trees but shown in Supplemental Data Set S1) revealed the presence of BSL2/BSL3-type sequences (contig_13341 and contig_12782), but we were unable to unequivocally assign any to the BSL1 clade. Charophyte algae (Timme et al., 2012) appear as a sister group to all land plants in the tree built with the C-terminal sequences (Supplemental Fig. S1); however, the longest charophyte EST available (from Spirogyra pratensis) clusters with the BSL2/BSL3 clade in the tree built using full-length sequences (Fig. 1A). Overall, both clades are characterized by very short branches indicative of a robust sequence conservation.
Figure 1.
Evolutionary relationships of BSL genes. A, Maximum likelihood phylogenetic tree built with aligned amino acid sequences of BSL genes from Viridiplantae; only complete sequences derived from fully sequenced genomes or the longest EST clones available were used. Bootstrap values over 50% are shown; values over 50% in tightly grouped nodes were excluded for clarity. B, Distribution of model-averaged dN/dS values among the branches of a maximum likelihood tree built using 22 BSL genes in the Brassicaceae. GA-Branch inferred six different categories of dN/dS (range, 0.575–0.000), which we grouped into three for convenience: 0.575 (dotted lines), 0.364 (black lines), and 0.094 and lower (gray lines). The percentage values represent the proportion of total tree length measured in expected substitutions per site per unit of time evolving under the corresponding value of dN/dS.
In contrast, BSU1-type sequences stand out as divergent paralogs of the BSL1 subfamily, with very long branches that suggest a faster evolutionary pace. BSU1-type genes are found exclusively in the Brassicaceae. In Arabidopsis, BSL2/BSL3 and BSL1/BSU1 are two pairs of paralogs located at the α2 and α1 syntenic regions, respectively (Bowers et al., 2003), derived from the At-α whole-genome duplication that took place before the radiation of the Brassicaceae (Barker et al., 2009; Beilstein et al., 2010). To better characterize the evolution of BSL genes in this family, we first looked at the evolutionary rates in nucleotide sequences in the 22 genes shown in Figure 1A with a test of the molecular clock: the null hypothesis of equal evolutionary rates throughout the tree was soundly rejected (Supplemental Table S1). Unequal evolutionary rates after duplication events can be assessed by computing the ratio between synonymous and nonsynonymous nucleotide substitutions (dN/dS; ω). In a simplified scenario, one of the duplicated copies retains the ancestral function and remains under the same selective regime (ω < 1). The other copy, if not under pressure for keeping the ancestral role, will either lose or change its function. If the original function is lost, no selective pressure will prevent the fixation of random substitutions, resulting in a scenario of relaxed selection (ω ∼ 1) that will ultimately lead to pseudogenization unless a new selective regime is attained. In the process of adjusting to a new function, the gene will undergo a period of selection favoring an excess of nonsynonymous substitutions (ω > 1; Hahn, 2009). We inferred dN/dS values in all branches and computed the probability of ω > 1 at each branch. Low ω values were distributed across most of the tree, indicating that strong purifying selection is acting on BSL1-, BSL2-, and BSL3-type genes. In contrast, the highest values for model-averaged dN/dS were assigned to the branch separating the BSL1 and BSU1 clades and, significantly, to all the branches in the BSU1 clade (Fig. 1B). High ω values indicate that BSU1-type genes have diverged from the rest of their homologs under a relaxed selective regime.
Sequence divergence is not the only unusual feature of BSU1-type genes. Splice patterns appear to be very conservative in many functionally relevant land plant genes (Teich et al., 2007). Indeed, the positions and phases of introns have been strictly conserved in PPKL from liverworts to angiosperms, the only exception being BSU1-type genes: not only has the splice pattern preceding the catalytic domain been rearranged, but this pattern differs between species in the Brassicaceae (Supplemental Fig. S2).
Land plant PPKL phosphatases thus belong to two ancient and highly conserved clades. Brassicaceae BSU1-type sequences are, in contrast, characterized by an unusually high degree of sequence divergence, even between closely related species. In the next sections, we will explore some functional inferences derived from these observations.
Contrasting Expression Patterns and Subcellular Localization of BSL Phosphatases
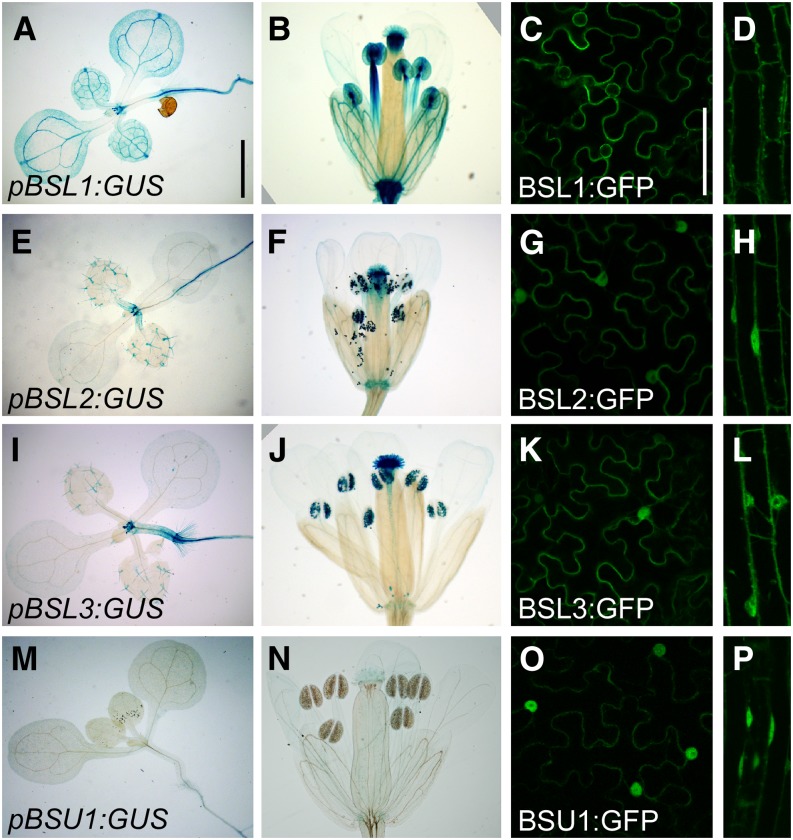
Considering their evolutionary history, BSL-type phosphatases may differ in distribution and/or function. We evaluated the expression patterns of the four BSL genes in Arabidopsis using fusions of their promoter regions to GUS (Fig. 2). We found two contrasting patterns. BSL2 and BSL3 showed identical expression patterns; expression was particularly strong in pollen but weak in vegetative tissues, where it mostly localized to the periphery of the apical meristem and to the root vasculature (Fig. 2, E, F, I, and J). BSL1 expression, in contrast, was considerably stronger in vegetative organs, concentrating in vascular strands and at the tip of cotyledons and leaves, in stamen filaments, and in anthers, but excluded from pollen (Fig. 2, A and B; for a closeup view of mature anthers, see Supplemental Fig. S3A). We could not detect a consistent expression of the BSU1 promoter after screening several independent lines with different staining conditions (Fig. 2, M and N). BSU1 promoter activity was previously detected in several tissues and organs (Mora-García et al., 2004), but the construct used was carried on a vector backbone prone to produce ectopic expression patterns due to a double 35S promoter. Our results, instead, are in good agreement with data from transcriptomic studies, both for expression levels and patterns (Zimmermann et al., 2004; Schmid et al., 2005; Winter et al., 2007; Liu et al., 2011).
Figure 2.
Expression patterns of Arabidopsis BSL genes. GUS expression is shown in representative 10-d-old T3 seedlings and mature flowers of ProBSL1:GUS (A and B), ProBSL2:GUS (E and F), ProBSL3:GUS (I and J), and ProBSU1:GUS (M and N). Bar = 2 mm. Localization of GFP-tagged versions of BSL1 (C and D), BSL2 (G and H), BSL3 (K and L), and BSU1 (O and P) is shown, expressed transiently in N. benthamiana leaves (C, G, K, and O) or stably in 4-d-old etiolated T2 Arabidopsis seedlings (D, H, L, and P). Bar = 100 µm. [See online article for color version of this figure.]
We also evaluated the subcellular localization of the four GFP-tagged proteins expressed (as p35S:complementary DNA [cDNA]) in Nicotiana benthamiana leaves (Fig. 2, C, G, K, and O) and in stably transformed Arabidopsis seedlings (Fig. 2, D, H, L, and P). Both systems showed identical results. BSL2 and BSL3 were found in the cytoplasm and nuclei; an identical pattern was obtained using a functional GFP-tagged BSL2 expressed from a genomic construct (see below). BSL1, in contrast, was localized only in the cytoplasm and excluded from nuclei, whereas BSU1 showed a strong nuclear and weak cytoplasmic localization. Similar patterns have been reported for Arabidopsis BSU1 and BSL1 (Mora-García et al., 2004; Kim et al., 2009), for a rice member of the BSL2/BSL3 clade (Qi et al., 2012), and for tomato (Solanum lycopersicum) BSL1 expressed in N. benthamiana leaves (Saunders et al., 2012).
The contrasting expression and subcellular localization patterns of BSL2/BSL3 and BSL1 suggest that these genes perform a similar function in different cell types or compartments or have different roles. We next studied the phenotypic effects caused by the loss of function of these genes.
Phenotypic Effects in bsl Loss-of-Function Mutants
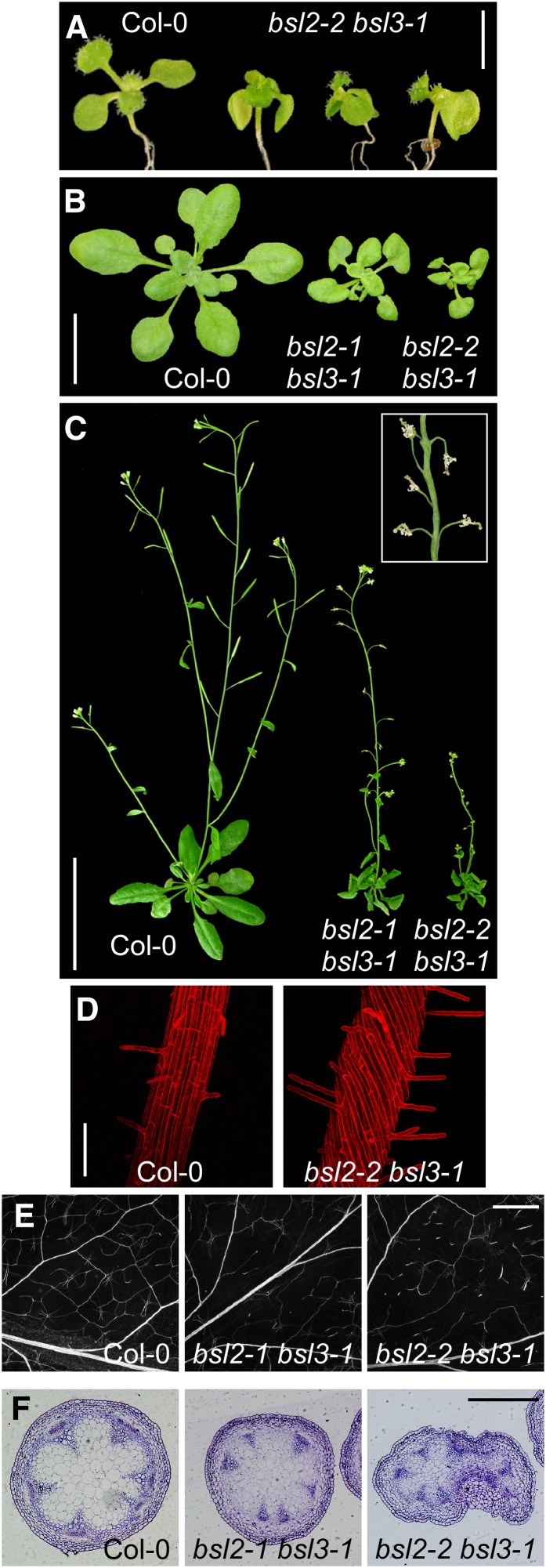
We identified several transferred DNA (T-DNA) insertion mutants in BSL1, BSL2, and BSL3 (Supplemental Fig. S4A). All but one appeared to produce complete loss-of-function alleles. Individual insertions failed to produce noticeable alterations; BSU1 knockout mutants have also been shown to be phenotypically silent (Mora-García et al., 2004; Kim et al., 2009). The fact that BSL2 and BSL3 are close homologs with identical expression patterns suggested that they may have a significant functional overlap. Indeed, all combinations between bsl2 and bsl3 alleles produced the same and characteristic set of phenotypic effects. The bsl2-1 allele was caused by a T-DNA inserted at the 5′ end of the coding region; whenever this allele was present, the phenotypic effects were milder. bsl2-1 plants probably produce low amounts of a truncated protein: we detected transcripts extending to the 3′ untranslated region (UTR) by reverse transcription-PCR (Supplemental Fig. S4B) and shorter mRNA species by northern blot, but no differential protein bands on western blots (data not shown).
bsl2 bsl3 double mutant seedlings were small, had epinastic cotyledons, and displayed different degrees of symmetry breakdown and cotyledon fusion (Fig. 3A). The proportion of laterally fused cotyledons correlated with the severity of the allele: five of 80 (6%) in bsl2-1 bsl3-1 but 19 of 73 (19%) in bsl2-2 bsl3-1. Although light-grown seedlings were shorter than the wild type (Fig. 3B), cell elongation was not intrinsically affected, as shown by the ability of dark-grown hypocotyls to elongate as much as those of the wild type (Supplemental Fig. S3B). In contrast, roots were significantly shorter than in the wild type and produced fewer lateral roots (Supplemental Fig. S3, C and D). Hypocotyls and primary roots in the double mutants were remarkably twisted around their axes (Fig. 3D; Supplemental Fig. S3E). Double mutant adult plants remained small, with twisted leaves (Fig. 3C), but their developmental transitions were not unlike those of the wild type. Inflorescence stems in bsl2 bsl3 mutants carrying strong-effect alleles were fasciated, with extensive fusions between petioles and the main stem (Fig. 3C). In leaves and derived organs (petals and ovaries), the vascular pattern was noticeably disrupted and the proportion of stray higher order vascular bundles correlated with the severity of the allelic combination (Fig. 3E; Supplemental Fig. S3F). In the inflorescence stems, vascular bundles were less developed and lacked interfascicular fibers (Fig. 3F).
Figure 3.
Phenotypic syndrome of bsl2 bsl3 double mutants. A, Twelve-day-old bsl2-2 bsl3-1 seedlings showing lateralized cotyledons. Bar = 2.5 mm. B and C, Three- and 5-week-old Columbia-0 (Col-0), bsl2-1 bsl3-1, and bsl2-2 bsl3-1 plants. Bars = 1 cm (B) and 5 cm (C). The inset in C shows the detail of a bsl2-2 bsl3-1 inflorescence. D, Confocal microscopy images of 5-d-old Col-0 and bsl2-2 bsl3-1 primary roots stained with propidium iodide. Bar = 1 mm. E, Vasculature in cleared leaves of Col-0, bsl2-1 bsl3-1, and bsl2-2 bsl3-1. Bar = 1 mm. F, Cross sections of young inflorescence stems of the indicated genotypes. Bar = 0.5 mm. [See online article for color version of this figure.]
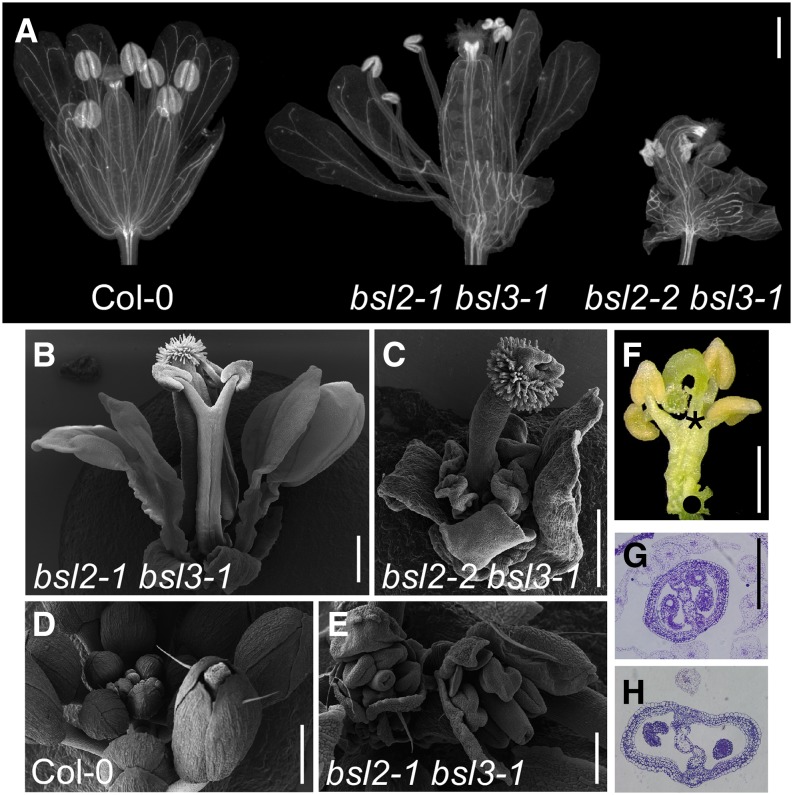
Flowers were particularly affected in the double mutants. Flower organs were mostly normal in shape and number in bsl2-1 combinations but failed to properly develop in bsl2-2 combinations (Fig. 4, A–C). Stamen filaments showed different degrees of fusion and were altogether unable to elongate in bsl2-2 mutants (Fig. 4, B and C). Buds were never closed, due to outwardly opening sepals (Fig. 4, D and E). Ovaries stood on top of gynophore-like structures, away from the insertion of sepals and petals; in bsl2-2 mutants, ovaries ranged from having two to no carpels at all (Fig. 4F). All bsl2 bls3 double mutants were male and female sterile; mutant ovules, when present, had a collapsed structure (Fig. 4, G and H). Despite their strong expression in pollen, loss of function of BSL2 and BSL3 did not significantly impair the production of mutant progeny, as long as the sporophyte carried at least one wild-type allele. However, a segregation bias in the proportion of double bsl2-2 bsl3-1 mutant siblings (73 of 360 observed versus 90 of 360 expected) suggests an effect on the development of gametes or embryos (see last section).
Figure 4.
Flower architecture in bsl2 bsl3 double mutants. A, Col-0, bsl2-1 bsl3-1, and bsl2-2 bsl3-1 cleared mature flowers. Bar = 500 µm. B, Scanning electron microscopy of a bsl2-1 bsl3-1 mature flower showing two stamens fused along their filaments. Bar = 500 µm. C, Scanning electron microscopy of a bsl2-2 bsl3-1 mature flower with no developed carpels. Bar = 500 µm. D and E, Scanning electron microscopy of Col-0 (D) and bsl2-1 bsl3-1 (E) flower buds. Bars = 500 µm for the wild type and 200 µm for the mutant. F, A gynophore-like structure subtends the ovary in a bsl2-2 bsl3-1 mutant. Dots mark the insertion of sepals and petals, and the asterisk indicates the seat of the gynoecium. Bar = 500 µm. G and H, Cross sections of Col-0 (G) and bsl2-1 bsl3-1 (H) ovaries. Bar = 250 µm. [See online article for color version of this figure.]
The phenotype of bsl2 bsl3 double mutants could be rescued by a genomic fragment comprising the full BSL2 coding sequence. This fragment was engineered to make translational fusions to GFP or hemagglutinin (HA); only C-terminal fusions complemented the mutant phenotype (Supplemental Fig. S6B).
BSL phosphatases have been linked to BR signaling (Mora-García et al., 2004; Kim et al., 2009, 2011; Ryu et al., 2010b). Despite their reduced size and sterility, the phenotype of the bsl2 bsl3 mutants did not match that of BR-related mutants. Therefore, we analyzed in detail the state of BR signaling in plants with either reduced or increased expression of BSL proteins.
BR Responses in Plants with Altered Levels of BSL Proteins
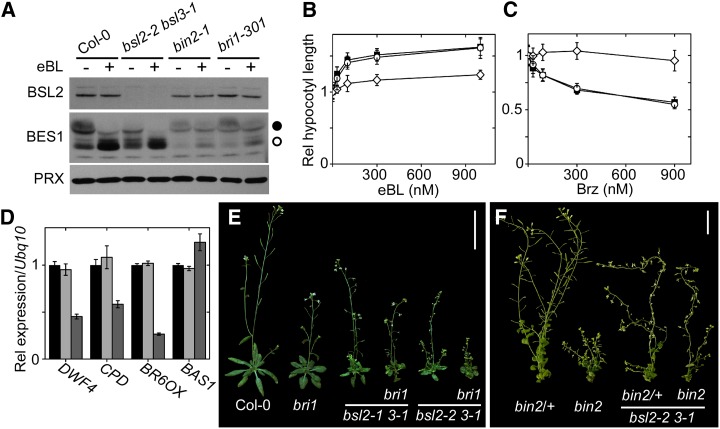
We first surveyed several outputs of BR signaling in bsl1, bsl2, bsl3, and bsl2 bsl3 mutants, using the strong bsl2-2 allele to better reveal possible effects. The phosphorylation state of BES1/BZR1 is a sensitive readout of the signal intensity through the pathway. Whereas the state of BES1 in bsl1 mutants was identical to that in the wild type, the abundance of fully phosphorylated form(s) in bsl2 and bsl3 single mutants decreased; at the same time, the levels of forms migrating between the fully phosphorylated and dephosphorylated species increased (Supplemental Fig. S5A). In the bsl2 bsl3 mutants, the amount of fully dephosphorylated BES1 was reduced compared with the wild type and correlated with the severity of the bsl2 allele (Fig. 5A; Supplemental Fig. S5B). Nonetheless, in response to the BL analog 24-epibrassinolide (eBL), BES1 was as readily dephosphorylated in the double mutants as in the wild type or in the BL-deficient de-etiolated2 mutant; this response was clearly different in BR-insensitive mutants (Fig. 5A; Supplemental Fig. S5B). The signaling pathway thus appears to be fully functional. In fact, the responses of the bsl2 bsl3 mutants to treatments with eBL (Fig. 5B) or with the BR biosynthesis inhibitor brassinazole (Brz; Fig. 5C; Supplemental Fig. S5C) were identical to those of the wild type. Also, the expression of target genes under tight feedback regulation by BR remained unchanged (Fig. 5D). We next tested the effects of the double mutation in sensitized genetic backgrounds, such as in bri1-301, a kinase-dead BRI1 mutant with a mild phenotype (Xu et al., 2008), and in bin2-1, which carries a gain-of-function mutation that disrupts the BSU1-dependent regulation but does not block BSU1 binding (Kim et al., 2009). The bsl2 bsl3 combination had additive, but not synergistic, effects in homozygous bri1-301 or in heterozygous bin2-1 mutants (Fig. 5, E and F). Surprisingly, the double mutation significantly relieved, rather than enhanced, the stuntedness of homozygous bin2-1 inflorescences (Fig. 5F).
Figure 5.
BR responses in bsl2 bsl3 mutants. A, Change in BES1 phosphorylation in response to treatment with eBL. Twelve-day-old seedlings of the indicated genotypes were treated with 1 μm eBL or with the equivalent amount of solvent for 2 h in the light. Western blots were processed as described in “Materials and Methods.” Black and white dots indicate fully phosphorylated and dephosphorylated BES1, respectively. PRX, Peroxiredoxin. B, Response to eBL in bsl2-2 bsl3-1 mutants. Four-day-old Col-0 (black squares), bsl2-2 bsl3-1 (white circles), and bri1-301 (white diamonds) seedlings were grown in dim white light in the presence of the indicated amounts of eBL. Data were normalized to the values with no eBL. Data are averages ± se of three independent experiments, with n = 15 seedlings. C, Responses to Brz in bsl2-2 bsl3-1 mutants. Four-day-old etiolated Col-0 (black squares), bsl2-2 bsl3-1 (white circles), and bes1-D (white diamonds) seedlings were grown with the indicated amounts of Brz. Data were normalized to the values with no Brz. Data are averages ± se of three independent experiments, with n = 15 seedlings. D, Expression levels of BL-regulated genes in 12-d-old, light-grown Col-0 (black bars), bsl2-2 bsl3-1 (light gray bars), and bzr1-d (dark gray bars) seedlings. No significant differences were detected between Col-0 and bsl2-2 bsl3-1. Data were collected in triplicate ± se. E, The bsl2-1 bsl3-1 mutant combination has an additive, not synergistic, effect on a bri1-301 mutant background; 40-d-old plants are shown. Bar = 5 cm. bri1 indicates homozygous bri1-301. F, The bsl2-2 bsl3-1 combination has additive, but not synergistic, effects on bin2-1 heterozygous plants but partly suppressive on homozygous bin2-1 plants. bin2-1/+ denotes heterozygous plants and bin2-1 indicates homozygous plants; 55-d-old plants are shown. Bar = 2 cm. [See online article for color version of this figure.]
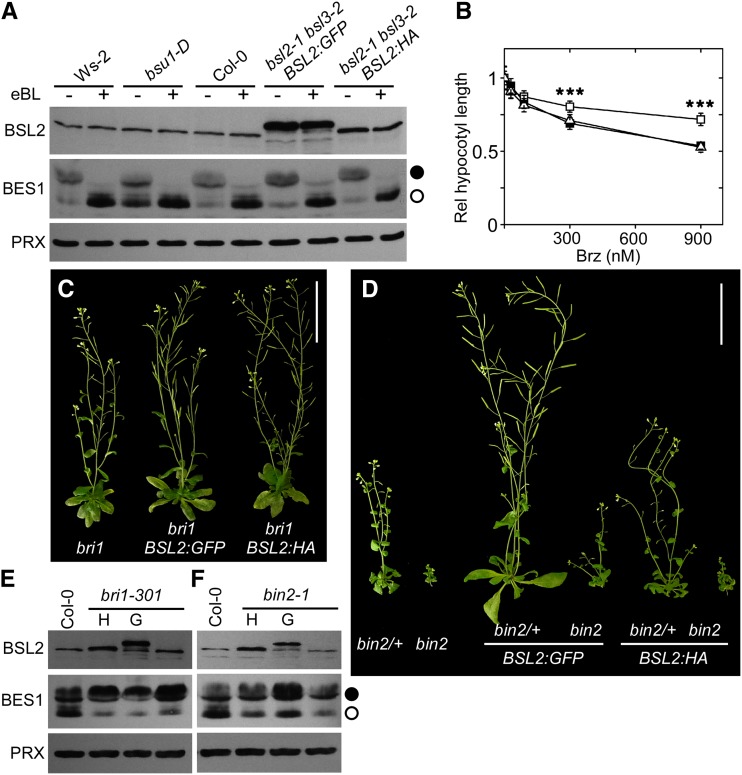
BSU1 was first identified as a gene that, when overexpressed, partially suppressed the phenotype of weak bri1 mutants (Mora-García et al., 2004). The inability of overexpressed BSU1 to suppress the phenotype of homozygous bin2-1 mutants placed this gene, and by extension the whole family, upstream of BIN2 in the signaling pathway (Kim et al., 2009). When analyzed, the GFP- and HA-tagged BSL2 lines complemented the bsl2 bsl3 phenotype (see previous section). In both cases, the genomic sequences were placed under the influence of multimerized 35S enhancers (Weigel et al., 2000). The BSL2:HA line expressed protein levels slightly over those of the wild type, whereas the BSL2:GFP line accumulated significantly higher amounts of protein (Supplemental Fig. S6A; as an example of the specificity of the anti-BSL antiserum, see Supplemental Fig. S7A). Rosette leaves in these lines were longer and more erect than in the wild type (Supplemental Fig. S6B), resembling weak versions of BRI1- or DWARF4-overexpressing lines (Mora-García et al., 2004). However, we could not detect an enrichment in dephosphorylated BES1 in these lines, as happens in bsu1-D plants (Fig. 6A; Supplemental Fig. S6A), even if the line with the highest expression levels showed reduced sensitivity to Brz (Fig. 6B). When introduced into the bri1-301 background, enhanced BSL2 expression mildly alleviated the mutant phenotype (Fig. 6C; Supplemental Fig. S6C), but the levels and phosphorylation of BES1 remained unchanged (Fig. 6E). Remarkably, overexpression of BSL2 suppressed, in a dose-dependent manner, the phenotype of both heterozygous and homozygous bin2-1 mutants (Fig. 6D; Supplemental Fig. S6D). In bin2-1 plants with higher levels of BSL2, the overall amount of BES1 increased, but this was barely noticeable in the moderate overexpressors. In any case, the ratio between the phosphorylated and dephosphorylated forms remained unchanged and resembled that of bin2-1 rather than the wild type (Fig. 6F).
Figure 6.
Phenotypic effects of BSL2 overexpression. A, BES1 phosphorylation in 12-d-old seedlings of the indicated genotypes treated with 1 μm eBL or with the equivalent amount of solvent for 2 h in the light. bsu1-D (in a BRI1 background) is compared with its wild type (Wassilewskija-2 [Ws-2]). PRX, Peroxiredoxin. B, Response of 4-d-old etiolated Col-0 (black squares), BSL2:HA (white triangles), and BSL2:GFP (white squares) seedlings to Brz treatment. Data were compared at each concentration by one-way ANOVA; where significant differences were found, a post hoc Bonferroni test was performed. Asterisks indicate significant differences (***P < 0.001). Data are averages ± se of three independent experiments, with n > 15 seedlings. C, Effect of BSL2 overexpression on 45-d-old adult plants of the indicated genotypes. Bar = 5 cm. D, Effect of BSL2 overexpression on 45-d-old adult plants of the indicated genotypes. Bar = 5 cm. E, BES1 phosphorylation in the bri1-301 combinations shown in C. F, BES1 phosphorylation in the bin2-1 combinations shown in D. Western blots were processed as described in “Materials and Methods.” In E and F, G denotes BSL2:GFP and H denotes BSL2:HA. [See online article for color version of this figure.]
The BR signaling pathway thus appears to be mostly intact in bsl1 single and bsl2 bsl3 double mutants and is only stimulated in lines with greatly enhanced levels of BSL2. To further clarify the involvement of these phosphatases in BR signaling, we assayed their interaction with known actors in the pathway.
Interaction of BSL Phosphatases with Members of the BR Signaling Pathway
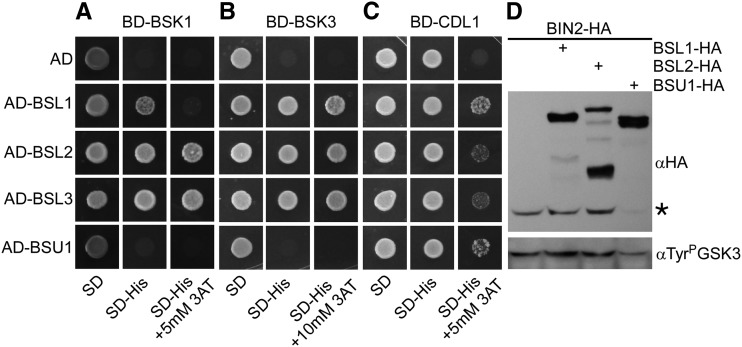
We tested the ability of the four BSL proteins to interact with some of the RLCKs identified as partners of BSU1. We chose BSK1 because it was shown to be phosphorylated by activated BRI1 in vivo (Tang et al., 2008), BSK3 because its absence produces a mild BR-deficient phenotype (Tang et al., 2008; Sreeramulu et al., 2013), and CDL1 because its expression levels are significantly higher than those of CDG1 and was reported to interact with both BSU1 and BSL1 (Kim et al., 2011). We found that BSL2 and BSL3 were able to interact with BSK1 and BSK3, whereas BSL1 interacted with BSK3 but only weakly with BSK1 (Fig. 7, A and B). Surprisingly, BSU1 did not interact with any BSK in this system (all proteins were similarly expressed; Supplemental Fig. S7B). The interaction of CDL1 with BSLs was obscured by a strong background activation of the reporter when CDL1 was used as bait. Still, BSL1 and BSU1 appeared to weakly interact with CDL1, whereas the interaction with BSL2 and BSL3 was not significantly different from the background (Fig. 7C).
Figure 7.
Interaction between BSL and BR signaling intermediates. A to C, The interaction between the BSL phosphatases and BSK1 (A), BSK3 (B), and CDL1 (C) was assessed in Saccharomyces cerevisiae two-hybrid assays. Representative colonies were spotted on selective medium with increasing stringency. AD, Activation domain; 3AT, 3-amino-1,2,4-triazole; BD, DNA-binding domain; SD, synthetic dextrose. D, BSU1, but not BSL1 or BSL2, accelerates the turnover of BIN2 when coexpressed in N. benthamiana leaves. The asterisk marks BIN2:HA. The top gel was probed with anti-HA antiserum, and the bottom gel was probed with anti-TyrP-GSK3 monoclonal antibody.
BR-activated BSU1 inactivates BIN2, dephosphorylating a conserved Tyr residue (Tyr-200 in BIN2); as a result, BIN2 becomes unstable (Kim et al., 2009). We thus assayed the effect of BSL phosphatases on the stability and Tyr-200 phosphorylation of BIN2 in a transient expression assay (Kim et al., 2009). The expression of BSU1 did significantly reduce the abundance of coexpressed BIN2. However, the level of phosphorylated Tyr-200 correlated, in this steady-state experimental setting, with the amount of protein, precluding any conclusion on the relevance or kinetics of the dephosphorylating reaction (Fig. 7D). Strikingly, no changes in BIN2 abundance or phosphorylation could be evoked by BSL1, BSL2 (Fig. 7D), and BSL3 (data not shown).
Taken together, these results confirm and extend previous evidence for the interaction of BSL phosphatases with intermediates of the BR signaling pathway (Kim et al., 2009, 2011), but at the same time they show that the isoforms display different, and even contrasting, reactivities and/or affinities toward these partners.
BSL1, BSL2, and BSL3 Together Play an Essential Function in Arabidopsis
We surmised that knocking out the function of the three most conserved genes in Arabidopsis might better uncover the role of this protein family in BR signaling and at the same time clarify the possible function(s) of the divergent, Brassicaceae-specific BSU1.
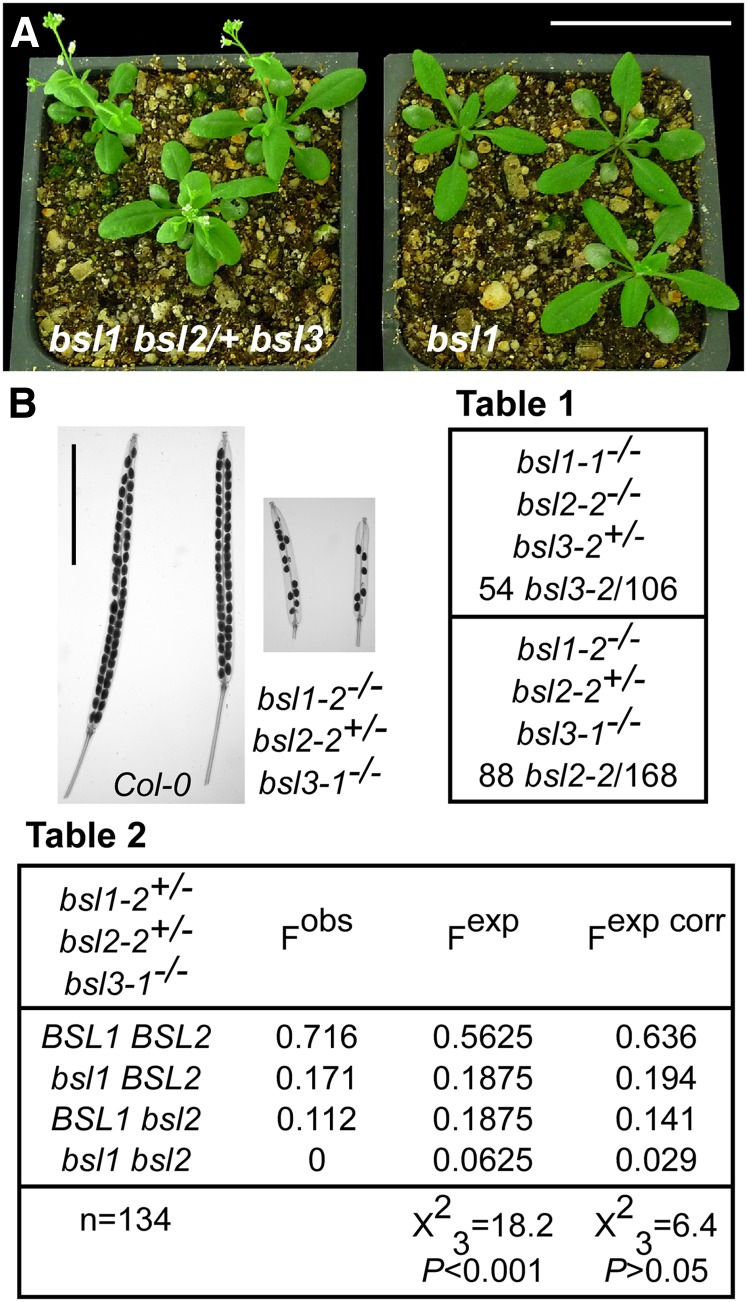
In sensitized genetic backgrounds (bsl2−/− bsl3+/− or bsl2+/− bsl3−/−), loss of function of BSL1 caused mild phenotypic perturbations: mutant plants carrying only one functional copy of either BSL2 or BSL3 were somewhat smaller, flowered earlier, and produced more ramified inflorescences and fewer seeds than the wild type (Fig. 8; Supplemental Fig. S8). However, in none of these features did they recall BR-related mutants. The ratio of T-DNA-positive versus T-DNA-negative plants in the progeny of bsl1-1−/− bsl2-2−/− bsl3-2+/− or bsl1-2−/− bsl2-2+/− bsl3-1−/− plants was close to 1:1, rather that the expected 3:1, and triple mutants could never be recovered (Fig. 8, Table 1). We did not observe seedling lethality but instead found conspicuous gaps in the siliques of selfed bsl2-2+/− bsl3-1−/− bsl1-2−/− plants (Fig. 8B). Even if the three genes are on different chromosomes, significant departures from a Mendelian segregation were evident in the progeny of bsl1-2+/− bsl2-2+/− bsl3-1−/− plants (Fig. 8, Table 2). Segregation distortions could be due to the inviability of one of the gametes or to early embryo lethality. Both pollen and ovules carrying the three mutant alleles were able to produce progeny in reciprocal crosses between bsl2-2+/− bsl3-1−/− bsl1-2−/− and wild-type plants. However, whereas the transmission of the bsl2-2 allele through pollen was not statistically different from that expected for completely viable male gametophytes, we recovered only one-third of the expected descendants of ovules carrying the bsl2-2 allele, suggesting that female gametophytes suffer from incompletely penetrant lethality. Correcting the proportions of viable ovules carrying different allele combinations by the observed ratios (see above), we obtained a reasonable fit to the data (Fig. 8, Table 2).
Figure 8.
Effects of the simultaneous loss of function of BSL1, BSL2, and BSL3. A, Comparison between 3-week-old bsl1-2−/− bsl2-2+/− bsl3-1−/− (left) and bsl1-2−/− (right) plants. Bar = 5 cm. B, Cleared mature siliques of the indicated genotypes. Bar = 5 mm. Table 1, The progeny of the indicated genotypes carrying the segregating mutant allele significantly departed from the expected 3:1 ratio. Observed frequencies are as follows: 51% for bsl1-1−/− bsl2-2−/− bsl3-2+/− and 52% for bsl1-2−/− bsl2-2+/− bsl3-1−/−. Table 2, The progeny of the indicated genotypes significantly departed from the expected 9:3:3:1 ratio for two unlinked loci and fully viable gametes (Fobs versus Fexp). When the proportion of female gametes is corrected as 0.33 (BSL1 BSL2):0.33 (bsl1 BSL2):0.225 (BSL1 bsl2):0.115 (bsl1 bsl2), while the proportion of male gametes is kept at 0.25 for each genotype, the frequency distributions are not significantly different (Fobs versus Fexp corr). bsl1 and bsl2 indicate the presence of the T-DNA insertions. Data were compared with a χ2 test. [See online article for color version of this figure.]
The complete absence of triple mutants thus appears to be the result of both partial female gametophyte malfunction and outright early embryonic lethality. Together with the indication of strong purifying selection, these results show that, in Arabidopsis, the conserved BSL1 and BSL2/BSL3 together perform an essential role that BSU1 is unable to replace.
DISCUSSION
PPKL are protein phosphatases of still poorly characterized roles found only in plants and alveolates. We set out to explore the relevance of these proteins in Arabidopsis, using the BR signaling pathway as a testing system. We found that BSLs perform an essential function in plants and characterized the alterations caused by the loss of two close paralogs. Our results also indicate that the evolutionary history of this small gene family must be taken into account to precisely gauge the contribution of each isoform. This applies to their role in BR signaling, which we suggest should be reconsidered.
Essential Role of BSLs
Loss of BSL1, BSL2, and BSL3 causes early developmental arrest. Evidence from other species supports a crucial role of PPKL. In the liverwort Marchantia polymorpha, the dominant generation is a dioecious haploid gametophyte with a chromosomal sex-determination mechanism. A PPKL gene that groups within the BSL2/BSL3 clade is codified in the pseudoautosomal regions of the X and Y chromosomes. Both male and female alleles seem to have reached a saturation of synonymous changes, with a low proportion of nonsynonymous substitutions. Recombination or gene conversion being apparently suppressed between pseudoautosomal regions, these features suggest that the alleles have been isolated for a long time but have been subject to a strong purifying selection that prevented their sequences from drifting apart, because they perform the same and essential function in both sexes (Yamato et al., 2007). In the apicomplexan Plasmodium falciparum, the single-copy PPKL is preferentially expressed during the sexual and diploid stages. Deletion of this gene compromises the proper differentiation and motility of the zygote to the point that it can no longer invade host cells (Clouse, 2011; Guttery et al., 2012).
BSLs join the ranks of essential plant protein PPP phosphatases. PP1 and PP2A are the most prominent members of the family, responsible for the bulk of Ser/Thr dephosphorylation in eukaryotic cells (Shi, 2009). Accordingly, perturbations in the function of these enzymes have important consequences. Phenotypic screens have not identified mutations in any of the nine PP1 catalytic subunits in Arabidopsis (Uhrig et al., 2013), but lack of a shared regulatory subunit, Inhibitor3, causes embryonic lethality (Takemiya et al., 2009). The lack of several PP2A subunits brings about a plethora of related perturbations, ranging from agravitropism and root meristem collapse to early developmental arrest (Zhou et al., 2004; Michniewicz et al., 2007; Ballesteros et al., 2013). In these cases, mislocalization of auxin efflux transporters and, as a result, alterations in auxin distribution are to blame. Double bsl2 bsl3 mutants show a strong twisting in primary roots and hypocotyls, without a fixed handedness. This feature, together with defects in root growth, organ separation, and vascular patterning, have all been linked to anomalies in the perception or transport of auxin (Petrásek and Friml, 2009). However, our attempts to connect the mutant phenotype to auxin have thus far proved inconclusive (S. Mora-García, unpublished data). One of the BSL2 homologs in rice has been shown to interact with Cyclin T1 (Qi et al., 2012), suggesting that these phosphatases modulate cell cycle progression. Although we could not find a phenotypic correlate to the loss of BSL1, this gene, together with BSL2 and BSL3, modulates some essential process(es). Remarkably, the presence of a functional BSU1 failed to rescue the absence of its homologs, despite the fact that this gene is specifically expressed in pollen (Borges et al., 2008; Liu et al., 2011) and during early endosperm development (Wolff et al., 2011). Analyzing only the couple BSL1/BSU1 in Arabidopsis, Liu et al. (2011) suggested that BSU1 must have undergone a process of neofunctionalization. Our analyses showed that BSU1-type genes transit an ongoing process of sequence divergence under a more relaxed evolutionary regime than their highly constrained paralogs. The analysis of the role of BSLs in BR signaling provided further support for functional splits inside the family.
Functional Divergence of BSL Phosphatases
The link between BSLs and BR signaling has been built mostly on BSU1, the first member of the family identified (Mora-García et al., 2004; Ryu et al., 2007, 2010b; Kim et al., 2009, 2011). We found that BSL phosphatases appear to modulate BR signaling to some extent, but in a way that calls the current model into question.
Neither bsl1 nor bsl2 bsl3 mutants showed altered responses to BRs. Cotyledon epinasty and reduced growth notwithstanding, bsl2 bsl3 double mutants did not resemble BR-insensitive plants, nor did the loss of BSL2 and BSL3 synergistically enhance the phenotype of BR-insensitive mutants. Intriguingly, the shape of bsl2-2 bsl3-1 leaves and the widespread fusion processes in inflorescences and flowers are phenotypes associated with runaway BR responses, as in cdg-1D mutants (Muto et al., 2004) or in plants overexpressing the hypermorphic bzr1-D allele (Wang et al., 2002). Loss of function of BSL2, BSL3, or both had a dose-dependent but silent effect on the phosphorylation state of BES1/BZR1, altering the ratio between electrophoretic variants. To the best of our knowledge, this is the first example of a lack of tight coupling between BES1 phosphorylation states and phenotypic outputs. Phosphorylation shifts in BES1/BZR1 have been considered an on-off process, but these proteins bear many putative phosphorylation sites, and whether different phosphorylation states exist and have specific properties is an unresolved issue. Overexpression of BSU1 leads to the accumulation of dephosphorylated BES1 but is unable to suppress the phenotype of homozygous bin2-1 mutants (Mora-García et al., 2004; Kim et al., 2009). In contrast, overexpression of BSL2 suppressed the bin2-1 phenotype, but the relative levels of phosphorylated and unphosphorylated BES1 remained unchanged; at most, both forms increased in the bin2-1 background. A similar effect has been observed in plants overexpressing the regulatory subunits PP2AB α or β, involved in the direct dephosphorylation of BZR1 downstream of BIN2 (Tang et al., 2011).
BRs signal through a membrane-based receptor complex that includes SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE coreceptors and RLCKs. We found that, in yeasts (Saccharomyces cerevisiae), BSL1, BSL2, and BSL3 associated with different apparent affinities to BSK1 and BSK3 but, strikingly, BSU1 did not interact with any. BSU1 has been shown to associate with BSK1 in vitro by membrane overlays or in vivo by bimolecular fluorescence complementation (Kim et al., 2009). It is possible that the interaction between BSU1 and BSK1 fails to activate the transcription of the reporters in yeasts but can be detected with sufficiently concentrated proteins on a membrane or when fixed by the irreversible reconstitution of GFP. Still, when expression patterns, subcellular localization, and interactions are considered, BSL1, BSL2, and BSL3 appear to be the most likely partners of BSKs and, therefore, should transduce the signal to BIN2. However, only BSU1 was able to affect the stability of BIN2.
Our results imply that BSU1 and BSL2/BSL3 have seemingly opposite activities in the BR transduction pathway. Given that BSL2/BSL3 genes have been highly conserved in all land plants but BSU1 genes are found exclusively in Brassicaceae, some aspects of our understanding of the BR pathway, gleaned in Arabidopsis, may not apply to other species. A closer look at the evolutionary patterns inside the family may help contextualize these findings.
Evolutionary Relationships and Functional Correlates
Land plants carry two clearly defined types of BSL genes; in each clade, distinct evolutionary trends are evident. On the one hand, pairs of paralogs are found in the BSL2/BSL3 clade, a feature apparent in the best sampled families, Poaceae, Fabaceae, and Brassicaceae, where duplications are an ancestral character predating speciation. On the other hand, a rapid pruning is apparent in the BSL1 branch, where only one gene per genome is the rule. Exceptions are found in relatively recent polyploids like soybean (Glycine max) and Chinese cabbage (Brassica campestris; Severin et al., 2011; Wang et al., 2011) or slowly evolving polyploids like poplar (Populus spp.; Tang et al., 2008). In the long run, the basic angiosperm BSL set appears to be formed by only one of each BSL1 and BSL2/BSL3 genes. Such is the configuration found in papaya (Carica papaya), grapevine (Vitis vinifera), and castor bean (Ricinus communis), unrelated species that have not undergone further genome multiplications after the basal eudicot hexaploidization approximately 140 million years ago (Proost et al., 2011). Brassicaceae stands out among all other land plants for the presence of the anomalous BSU1 genes that arose as paralogs of BSL1. In fact, no other angiosperm family thus far surveyed bears such a divergent group of BSL sequences. If BSL1 genes tend to be preserved as single copies, BSU1-type genes have probably escaped elimination through divergence.
No matter the ploidy, in every species the ratio BSL1:BSL2/BSL3 ranges from 1:2 to 1:1, which suggests that a certain balance between both isoforms has been selected. The described functional and evolutionary patterns can be accounted for if we consider that, in all likelihood, BSL phosphatases act in the context of multiprotein complexes that are sensitive to the stoichiometry of its components. This situation is dealt with by two complementary hypotheses: duplication resistance (De Smet et al., 2013) and gene dosage balance (Birchler and Veitia, 2012).
The first one states that certain classes of genes that perform essential functions tend to be highly conserved and codified by single copies, probably because mutations in duplicates can act in a dominant-negative manner and interfere with the wild-type function. There is an indication that the coexistence of functional BSL variants may indeed have an effect. In rice, a conservative mutation in the Kelch domain of a BSL2/BSL3 homolog (Os03g44500) was mapped as a recessive trait with a positive effect on grain length. However, when the mutation was introduced into other cultivars, F1 hybrids also produced longer grains (Zhang et al., 2012). This could be due to a higher dose sensitivity of the BSL2/BSL3 complement in rice (even in the presence of the remaining BSL2/BSL3 paralog, Os12g42310) or to a semidominant effect of the mutant protein.
Defect or excess of certain subunits can also perturb the stoichiometry and function of multiprotein complexes (Birchler and Veitia, 2010). Both loss of bsl2 and bsl3 and overexpression of BSL2 altered the phenotype of homozygous bin2-1 mutants in the same direction, albeit to different extents. Overexpression of BSL2 produced nonlinear effects (e.g. only plants with the highest protein levels showed a detectable tolerance to Brz or increased levels of BES1 in a bin2-1 background), indicating that mass-action effects might be at work. BSLs appear to be part of a multiprotein assemblage that includes BSKs, proteins with scaffolding functions that form homodimers and heterodimers and associate with and are phosphorylated by BIN2 (Sreeramulu et al., 2013). Although the functional correlate of these observations is still unclear, overrepresentation or underrepresentation of BSL2/BSL3 could perturb the signaling complex, even more so if burdened by the presence of a dominant-negative form of BIN2.
It should be borne in mind that BSU1 was identified through an activation tagging strategy and that a great deal of the experimental support for its role in BR signaling has been gained through overexpression. As discussed before, it is possible that the interaction of BSU1 with the BR signaling complex is weak but can be detected in some conditions. If this divergent and phylogenetically restricted gene has changed function(s), we should ponder to what extent it interferes, rather than concurs, with its conserved and ubiquitous homologs when ectopically overexpressed. BSU1-type genes manifest, on close inspection, unusual features not observed in other BSLs. The full-length AtBSU1 mRNA coexists with an alternatively spliced form that retains the 11th intron, introducing a stop codon at the end of the Kelch domain. Our attempts to detect a truncated form of the protein or assign a function to an isolated Kelch domain have been unsuccessful (G.A. Maselli and S. Mora-García, unpublished data). In CrBSU, a nucleotide is missing at the ninth exon, altering the reading frame and forcing an anomalous splicing in the annotated protein sequence. We introduced the missing nucleotide to restore the consensus splicing and protein sequence, but whether this mutation is real or a sequencing error remains to be proved. In SpBSU, the genomic region between the Kelch and catalytic domains is rich in repetitive sequences, making the splicing pattern we suggest speculative. These peculiarities suggest that BSU1-type genes might be susceptible to nonsense-mediated RNA decay, explaining in part the low expression levels observed for AtBSU1. Added to the evidence for a relaxed selective regime, it might be worth considering whether we witness the birth of a new function, or the demise thereof. In fact, it has been suggested that genes in Arabidopsis with very low expression levels might be en route to pseudogenization (Yang et al., 2011); examples of decaying functions are not unheard of (Cui et al., 2011) and will probably become less of an oddity as more full gene complements are analyzed.
Besides their role in BR signaling, BSLs likely take part in other signaling processes that we have only begun to explore and are worth studying in detail. The green lineage, rich in model organisms and genome duplication events, probably offers the best tools to understand how ancient functions have been modified and coopted across different phylogenetic groups and levels of organization.
MATERIALS AND METHODS
Materials and General Plant Methods
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines and bacterial artificial chromosome (BAC) clones were obtained from the Arabidopsis Biological Resource Center (abrc.osu.edu). All T-DNA lines were back-crossed twice into the Columbia-0 (Col-0) background. Arabidopsis full-length cDNA clones for BSL1 (RAFL09-11-J01), BSL2 (RAFL16-56-H16), and BSL3 (RAFL16-39-G23) were obtained from RIKEN (www.brc.riken.jp/lab/epd/catalog/cdnaclone.html). cDNA clones for BSL1 and BSL3 were edited before use: the BSL1 clone carries a mutation in a highly conserved residue of the phosphatase domain (Q794P), and the BLS3 clone is a misspliced form that retains part of the 13th intron, introducing a premature stop codon. In both cases, the sequences around these positions were replaced by their respective wild-type cassettes amplified from Col-0 retrotranscribed RNA. The full-length cDNA for BSU1 was amplified from retrotranscribed RNA derived from bsu1-D plants (Mora-García et al., 2004). Full-length cDNAs for BIN2, BES1, BSK1, BSK3, and CDL1 were amplified from Col-0 retrotranscribed RNA.
Arabidopsis seeds were surface sterilized with 70% (v/v) ethanol and 0.05% (v/v) Triton X-100, sown on 0.5× Murashige and Skoog (MS), 0.8% (w/v) agar plates, stratified for 3 d at 4°C, and induced to germinate with a 1-h white-light pulse. Ten-day-old seedlings were transferred to pots containing a 1:1:1 mixture of vermiculite, perlite, and peat and grown under long-day conditions (16 h of light/8 h of darkness) at 22°C. Transformation was carried out by the floral dip method (Clough and Bent, 1998).
Nicotiana benthamiana plants were grown on pots containing a 1:1:1 mixture of vermiculite, perlite, and peat under long-day conditions at 24°C with weekly fertilization.
Molecular Techniques
Oligonucleotides used are listed in Supplemental Table S2.
For promoter analyses, we used a derivative of the pBI101.3 plasmid in which we replaced the Escherichia coli uidA gene with the GUSPlus gene from pCAMBIA1305.1 (CAMBIA; www.cambia.org) in order to enhance the detection limit. Promoter sequences, from the end of the annotated 3′ UTR of the upstream gene to the third coding codon, were amplified from BAC clones T4I9 (BSL1), T27G7 (BSL2), T22O13 (BSL3), and F21B7 (BSU1).
For genotyping, DNA was extracted from individual leaves according to Berendzen et al. (2005). The bri1-301 and bin2-1 mutations were scored by treating the amplification products with Sau3AI and MboI, respectively, that cleave the wild-type sequence.
For expression analyses, RNA was extracted from the indicated genotypes using the RNeasy Plant Mini Kit (Qiagen; www.qiagen.com) and retrotranscribed with an oligo(dT) primer and SuperScript III (Invitrogen; www.invitrogen.com). For real-time PCR, we used the FastStart Universal SYBR Green master mix (Roche; www.roche-applied-science.com) and an Mx3005P cycler (Agilent; www.home.agilent.com). Primer pairs yielded single peaks in melting curves.
For the complementing GFP- and HA-tagged versions of BSL2, a SacI fragment of BAC T27G7 comprising the complete promoter, coding sequence, and 3′ UTR was inserted into pMN19 downstream of the 35S enhancers. A KpnI fragment including the 3′ region of the coding sequence was subcloned and mutagenized to introduce an MluI cleavage site replacing the penultimate codon. GFP and HA tags were introduced in frame at the MluI site, and the whole cassette was reintroduced in the context of the genomic clone. bsl2-1−/−bsl3-2+/− plants were transformed and their progeny screened for bsl3-2 homozygotes.
For 35S-driven GFP- and HA-tagged constructs, the full-length cDNA clones flanked by attachment sites were recombined using the Gateway system (Invitrogen) into pEARLEY103 (Earley et al., 2006) and into pK7m34GW, in combination with pEN-R2-3xHA-L3 and pDONRP4-P1_p35S (Karimi et al., 2007), respectively. Agrobacterium tumefaciens GV3101 carrying the corresponding constructs was infiltrated into 4- to 5-week-old N. benthamiana leaves. Briefly, each strain was grown overnight in rich medium with the appropriate selection, diluted at an optical density at 600 nm of 0.5 in infiltration medium (50 mm MES, 200 µm acetosyringone, and 5 mg mL−1 Suc, pH 5.6), and incubated for another 1 h before being mixed and used for infiltration.
Phylogenetic Analysis
Sequences were retrieved using TBLASTN (Altschul et al., 1997) from a variety of sources, using the amino acid sequences of the Arabidopsis or rice (Oryza sativa) BSL genes as query. Whenever possible, BLAST results were used as starting points to retrieve the full genomic sequences, which were manually spliced. Curated nucleotide sequences, translated to their corresponding amino acid sequences (Geneious 5.6.5; Biomatters; www.biomatters.com), and the list of databases used are shown in Supplemental Data Set S1.
Amino acid sequences were aligned using the MAFFT algorithm implemented in Geneious (Katoh et al., 2002). The aligned sequences were manually inspected and edited with eBioX version 1.5.1 (ebioinformatics.org) in order to refine the alignment and exclude hypervariable, ambiguously aligned regions. We generated a full-length data set and an alignment containing the C-terminal region only (starting at position 451 of the Arabidopsis BSU1 protein). For each of the data sets, we conducted a phylogenetic analysis using the RAxML version 2.2.3 method (Stamatakis, 2006) implemented in the graphical interface TOPALi 2.5 (Milne et al., 2004). Amino acid sequence evolution was modeled using the empirical model WAG (Whelan and Goldman, 2001). The parameter proportion of invariant sites and across-site variation (γ) were estimated for each data set and used in the analysis. Branch support was assessed with 300 bootstrap pseudoreplicates. To assess dN/dS values in the Brassicaceae BSL genes, we applied the Genetic Algorithm-Branch method (Pond and Frost, 2005).
Histological and Microscopical Techniques
For GUS staining, tissues were collected in cold 90% (v/v) acetone, rinsed with infiltration buffer (50 mm sodium phosphate buffer, pH 7.5, 0.1% (v/v) Triton X-100, 2 mm sodium ferricyanide, and 2 mm sodium ferrocyanide), and vacuum infiltrated with infiltration buffer plus 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (cyclohexylammonium salt; www.x-gluc.com). Staining was performed at 37°C and stopped after 5 h for ProBSL1:GUS and after 14 h for ProBSL2:GUS and ProBSL3:GUS lines; for ProBSU1:GUS lines, several staining conditions were assayed. After staining, samples were dehydrated through an ethanol series, fixed with 50% (v/v) ethanol, 10% (v/v) acetic acid, and 3.7% (v/v) formaldehyde, and mounted in chloral hydrate solution.
For the analysis of vascular patterns, mature flowers and 3-week-old leaves were fixed in 6:1 ethanol:acetic acid, dehydrated through an ethanol series, and mounted in chloral hydrate solution. Images were taken under dark-field illumination with a Zeiss Axioscope microscope.
For sectioning, samples were fixed in buffered 4% (v/v) glutaraldehyde, dehydrated through an ethanol series, and included in HistoResin (Leica; www.leica-microsystems.com). Five-micrometer sections were stained with 0.1% (w/v) toluidine blue. Stem sections were obtained from the second internode of young inflorescences; regular internodes are absent in bsl2-2 bsl3-1 inflorescences.
For scanning electron microscopy, specimens were fixed in 50% (v/v) ethanol, 10% (v/v) acetic acid, and 3.7% (v/v) formaldehyde and dehydrated through an ethanol series, flash dried under liquid CO2, mounted on tin disks with graphite glue, and coated with gold/palladium in a Thermo VG Scientific SC 7620 sputter coater. Images were captured with a Philips XL30 TMP scanning electron microscope.
Physiological Experiments
For treatments with Brz (Brz220; dissolved in dimethyl sulfoxide), seeds were germinated in the dark at 22°C; for treatments with eBL (Sigma-Aldrich; www.sigmaaldrich.com; dissolved in 80% (v/v) ethanol), seeds were germinated under continuous dim (3 µmol m−2 s−1) white light. After 4 d, all seedlings were laid onto fresh 0.5× MS, 0.8% (v/v) agar plates and scanned; double mutants were identified with a dissecting Leica EZ4D microscope by their twisted hypocotyls and roots, and hypocotyls were measured from the scanned images with NIH ImageJ (rsbweb.nih.gov/ij). Assays were repeated at least three times with 15 or more seedlings each time.
For measurements of root length and lateral root density, seeds from Col-0 and bsl2-2−/− bsl3-1+/− plants were sown on square 0.5× MS, 0.8% (v/v) agar plates and grown vertically under long-day conditions at 22°C. After 5 d, the plates were thinned for excess wild-type-looking siblings and grown for another 5 d. Emerged lateral roots were counted with a dissecting microscope, and root length was measured from scanned images as above.
Western Blots
The antiserum against BSL2 was raised in mice using as antigen a recombinant His-tagged protein comprising the region between the end of the Kelch domain and the beginning of the phosphatase domain (215 amino acids). As shown in Supplemental Figure S7A, this antiserum recognizes BSL2 and BSL3, only weakly reacts against BSL1, and reacts not at all against BSU1.
Twelve-day-old seedlings grown on 0.5× MS, 0.8% (v/v) agar plates under long-day conditions at 22°C were used for protein extraction. For eBL treatments, seedlings were soaked in liquid 0.5× MS medium with (1 μm) or without (equivalent amounts of solvent) the hormone for 2 h in the light, weighed, and frozen in liquid N2. Proteins were extracted with 2 volumes per weight of 2× Laemmli sample buffer, separated by 10% Tris-Gly SDS-PAGE, and transferred onto polyvinylidene difluoride (Bio-Rad; www.bio-rad.com). Usually, the top part of the blot (proteins with an apparent molecular mass greater than 80 kD) was probed with a 1:2,000 dilution of the anti-BSL2 antiserum. The middle part of the blot (proteins ranging from 30 to 80 kD) was probed with an immunopurified antiserum (1:300) raised in rabbits against recombinant BES1. The bottom part of the blot (proteins of less than 30 kD) was probed with a crude antiserum (1:2,000) raised in rabbits against rapeseed (Brassica napus) chloroplast 2-Cys peroxiredoxin as a loading control.
Yeast Two-Hybrid Assays
The corresponding cDNA clones were recombined using the Gateway system into pDEST22 or pDEST32 (Invitrogen) and introduced into the Saccharomyces cerevisiae AH109 strain (Clontech; www.clontech.com). Transformants were first assayed for β-Gal activity and then for the ability to grow on selective dropout medium (−Leu−Trp−His with the addition of different concentrations of 3-amino-1,2,4-triazole; Sigma-Aldrich). Exponentially growing yeast cultures (optical density at 600 nm ≤ 0.5) were used for protein extraction, following the Yeast Protocols Handbook (Clontech). Protein extracts were blotted as above and probed with anti-Gal4 DNA-binding or activation domain monoclonal antibodies (Santa Cruz Biotechnology; www.scbt.com).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Evolutionary relationships of BSL genes.
Supplemental Figure S2. Genomic organization of BSL genes.
Supplemental Figure S3. Phenotypic characterization of bsl2 bsl3 mutants.
Supplemental Figure S4. BSL1, BSL2, and BSL3 insertion alleles used in this study.
Supplemental Figure S5. Responses to BRs in single and double bsl mutants.
Supplemental Figure S6. Protein expression controls.
Supplemental Figure S7. Phenotypic effects of BSL2 overexpression.
Supplemental Figure S8. Phenotypes of bsl1-2 and bsl1-2 BSL2/bsl2-2 bsl3-1 plants.
Supplemental Table S1. Test of the molecular clock in BSL sequences from the Brassicaceae.
Supplemental Table S2. List of oligonucleotides used.
Supplemental Data Set S1. Curated BSL sequences used in this work.
Acknowledgments
We thank Fabián Tricárico (Museo Argentino de Ciencias Naturales “Bernardino Rivadavia”) for his assistance with scanning electron microscopy, Isabel Farías (Universidad de Buenos Aires) for her assistance with histological sections, Ruth Timme (National Center for Biotechnology Information), Charles Delwiche (University of Maryland), and Stefan Henz and Detlef Weigel (Max Planck Institute for Developmental Biology) for sharing their data from streptophyte algae and Capsella rubella prior to publication, and Marcelo Yanovsky, Pablo Cerdán, and Ricardo Wolosiuk (Fundación Instituto Leloir) for helpful comments and support. Tadao Asami (RIKEN) provided the Brz220, Yanhai Yin (Iowa State University) the anti-BES1 antibody, Ricardo Wolosiuk (Fundación Instituto Leloir) the anti-2-Cys peroxiredoxin antiserum, Miguel Blázquez (Instituto de Biología Molecular y Celular de Plantas) the anti-Gal4 activation and DNA-binding domain antibodies, and Eugenia Russinova (Department of Plant Systems Biology) the Gateway-compatible plant vectors.
Glossary
- BR
brassinosteroid
- BL
brassinolide
- PPKL
protein phosphatases with Kelch-like domains
- RLCK
receptor-like cytoplasmic kinase
- dN/dS
ratio between synonymous and nonsynonymous nucleotide substitutions
- cDNA
complementary DNA
- T-DNA
transferred DNA
- UTR
untranslated region
- HA
hemagglutinin
- eBL
24-epibrassinolide
- Brz
brassinazole
- BAC
bacterial artificial chromosome
- Col-0
Columbia-0
- MS
Murashige and Skoog
Footnotes
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (grant no. PICT–38012) and the Consejo Nacional de Investigaciones Científicas y Técnicas (grant no. PIP–414) to S.M.-G.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros I, Domínguez T, Sauer M, Paredes P, Duprat A, Rojo E, Sanmartín M, Sánchez-Serrano JJ. (2013) Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. Plant J 73: 862–872 [DOI] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME. (2009) Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol 1: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen K, Searle I, Ravenscroft D, Koncz C, Batschauer A, Coupland G, Somssich IE, Ulker B. (2005) A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. (2010) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. (2012) Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA 109: 14746–14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Brenner ED, Stevenson DW, McCombie RW, Katari MS, Rudd SA, Mayer KF, Palenchar PM, Runko SJ, Twigg RW, Dai G, et al. (2003) Expressed sequence tag analysis in Cycas, the most primitive living seed plant. Genome Biol 4: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Pan YH, Zhang Y, Jones G, Zhang S. (2011) Progressive pseudogenization: vitamin C synthesis and its loss in bats. Mol Biol Evol 28: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Der JP, Barker MS, Wickett NJ, dePamphilis CW, Wolf PG. (2011) De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics 12: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MC, Maere S, Van de Peer Y. (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci USA 110: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Guttery DS, Poulin B, Ferguson DJ, Szöőr B, Wickstead B, Carroll PL, Ramakrishnan C, Brady D, Patzewitz EM, Straschil U, et al. (2012) A unique protein phosphatase with kelch-like domains (PPKL) in Plasmodium modulates ookinete differentiation, motility and invasion. PLoS Pathog 8: e1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. (2009) Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100: 605–617 [DOI] [PubMed] [Google Scholar]
- Hu Z, He H, Zhang S, Sun F, Xin X, Wang W, Qian X, Yang J, Luo X. (2012) A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J Integr Plant Biol 54: 979–990 [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY. (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY. (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutuzov MA, Andreeva AV. (2002) Protein Ser/Thr phosphatases with kelch-like repeat domains. Cell Signal 14: 745–750 [DOI] [PubMed] [Google Scholar]
- Leliaert F, Verbruggen H, Zechman FW. (2011) Into the deep: new discoveries at the base of the green plant phylogeny. Bioessays 33: 683–692 [DOI] [PubMed] [Google Scholar]
- Liu SL, Baute GJ, Adams KL. (2011) Organ and cell type-specific complementary expression patterns and regulatory neofunctionalization between duplicated genes in Arabidopsis thaliana. Genome Biol Evol 3: 1419–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Milne I, Wright F, Rowe G, Marshall DF, Husmeier D, McGuire G. (2004) TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 20: 1806–1807 [DOI] [PubMed] [Google Scholar]
- Moorhead GB, De Wever V, Templeton G, Kerk D. (2009) Evolution of protein phosphatases in plants and animals. Biochem J 417: 401–409 [DOI] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J. (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Yabe N, Asami T, Hasunuma K, Yamamoto KT. (2004) Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol 136: 3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Sun J, Oh DH, Zielinski RE, Clouse SD, Huber SC. (2011) Enhancing Arabidopsis leaf growth by engineering the BRASSINOSTEROID INSENSITIVE1 receptor kinase. Plant Physiol 157: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. (2005) A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol 22: 478–485 [DOI] [PubMed] [Google Scholar]
- Proost S, Pattyn P, Gerats T, Van de Peer Y. (2011) Journey through the past: 150 million years of plant genome evolution. Plant J 66: 58–65 [DOI] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX. (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22: 1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Cho H, Kim K, Hwang I. (2010a) Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol Cells 29: 283–290 [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Hwang I. (2010b) Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells 29: 291–296 [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DG, Breen S, Win J, Schornack S, Hein I, Bozkurt TO, Champouret N, Vleeshouwers VG, Birch PR, Gilroy EM, et al. (2012) Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24: 3420–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Severin AJ, Cannon SB, Graham MM, Grant D, Shoemaker RC. (2011) Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 23: 3129–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Bell CD, Kim S, Soltis PS. (2008) Origin and early evolution of angiosperms. Ann N Y Acad Sci 1133: 3–25 [DOI] [PubMed] [Google Scholar]
- Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, et al. (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74: 905–919 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Takemiya A, Ariyoshi C, Shimazaki K. (2009) Identification and functional characterization of inhibitor-3, a regulatory subunit of protein phosphatase 1 in plants. Plant Physiol 150: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teich R, Grauvogel C, Petersen J. (2007) Intron distribution in Plantae: 500 million years of stasis during land plant evolution. Gene 394: 96–104 [DOI] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. (2012) Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 7: e29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Labandera AM, Moorhead GB. (2013) Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci 18: 505–513 [DOI] [PubMed] [Google Scholar]
- Vaulot D, Eikrem W, Viprey M, Moreau H. (2008) The diversity of small eukaryotic phytoplankton (< or =3 microm) in marine ecosystems. FEMS Microbiol Rev 32: 795–820 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Köhler C. (2011) High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J. (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y. (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Yamato KT, Ishizaki K, Fujisawa M, Okada S, Nakayama S, Fujishita M, Bando H, Yodoya K, Hayashi K, Bando T, et al. (2007) Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc Natl Acad Sci USA 104: 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Takuno S, Waters ER, Gaut BS. (2011) Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol Biol Evol 28: 1193–1203 [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J, et al. (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109: 21534–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhiponova MK, Vanhoutte I, Boudolf V, Betti C, Dhondt S, Coppens F, Mylle E, Maes S, González-García MP, Caño-Delgado AI, et al. (2013) Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol 197: 490–502 [DOI] [PubMed] [Google Scholar]
- Zhou HW, Nussbaumer C, Chao Y, DeLong A. (2004) Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Sae-Seaw J, Wang ZY. (2013) Brassinosteroid signalling. Development 140: 1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]