Abstract
Reports show that up to 30% of antiretroviral drug-naive patients in Johannesburg have CXCR4-utilizing HIV-1 subtype C. We assessed whether HIV-1 subtype C-infected individuals failing highly active antiretroviral therapy (HAART) have a higher proportion of CXCR4-utilizing viruses compared to antiretroviral drug-naive patients. The V3 loop was sequenced from plasma from 100 randomly selected HAART-failing patients, and tropism was established using predictive algorithms. All patients harbored HIV-1 subtype C with at least one antiretroviral drug resistance mutation. Viral tropism prediction in individuals failing HAART revealed similar proportions (29%) of X4-utilizing viruses compared to antiretroviral drug-naive patients (30%). Findings are in contrast to reports from Durban in which 60% of HAART-failing subjects harbored X4/dual/mixed-tropic viruses. Despite differences in proportions of X4-tropism within South Africa, the high proportion of thymidine analogue mutations (TAMs) and CXCR4-utilizing HIV-1 highlights the need for intensified monitoring of HAART patients and the predicament of diminishing drug options, including CCR5 antagonists, for patients failing therapy.
HIV-1 enters the host cell by sequentially binding its envelope glycoprotein to cell surface CD4, and a cellular chemokine coreceptor, either CCR5 and/or CXCR4,1–5 leading to fusion of the viral membrane with the target cell plasma membrane. CCR5 or R5-utilizing viruses are usually responsible for transmission/early infection, and are referred to as nonsyncytia-inducing (NSI) or macrophage-tropic (M-tropic).3,4 Viruses that use CXCR4 (X4-utilizing), also known as syncytia-inducing (SI) or T-cell-tropic (T-tropic), tend to be associated with accelerated CD4 decline and rapid disease progression.4,6,7 Some viruses can use both CCR5 and CXCR4 (dual tropic).8
Approximately 50% of individuals infected with HIV-1 subtype B switch from R5 to X4 or R5X4 tropism during disease progression to AIDS.9 By contrast, in HIV-1 subtype C-infected individuals R5 tropism occurs throughout all stages of disease with limited numbers of CXCR4-utilizing viruses being described.10–14 However, an increase in the incidence of X4 and R5X4 emerging at the late stages of infection in subtype C individuals has been reported.15 The introduction of entry inhibitors, such as the R5 antagonist maraviroc, as components of highly active antiretroviral therapy (HAART), has heightened interest of HIV-1 coreceptor usage because of the concern that preexisting X4 viruses may emerge in patients as a consequence of treatment, which necessitates tropism testing prior to treatment initiation.
This study aimed to assess whether South African HIV-1 subtype C-infected individuals failing HAART have a higher proportion of X4-utilizing viruses compared to antiretroviral drug-naive patients, and establish whether R5 antagonists can be used as alternative therapies for patients failing HAART or if they can be used as part of first-line regimens.
Samples from patients failing HAART and sent for routine genotyping between January and March 2013 to the Charlotte Maxeke Johannesburg Academic Hospital were available for the purposes of this study. Ethical clearance for the study was obtained for Research on Human Subjects (Medical) at the University of the Witwatersrand (clearance number M090688). One hundred patient samples with known antiretroviral drug resistance profiles were randomly selected. Of the patients failing treatment (median age of 27 years; median viral load of 4.81 log10), 51% had received two nucleoside reverse transcriptase inhibitors (NRTIs) plus one non-NRTI (NNRTI), 23% received two NRTIs plus one protease inhibitor (PI), 3% received three NRTIs, 7% received more than four antiretrovirals, while 16% were unknown. Antiretroviral drug treatment duration was unknown.
Viral RNA was extracted from 100 patient plasma samples using the automated NucliSENS easyMag system (bioMérieux) according to the manufacturer's instructions. The full length envelope glycoprotein was reverse transcriptase polymerase chain reaction (RT-PCR) amplified and sequenced (population based) on the ABI Prism 3730 (Applied Biosystems, Foster City, CA).15,16 Sequence data were edited using the Sequencing Analysis V5.3.1 program (Applied Biosystems), and the complete env sequences were assembled and manually edited using Sequencher V5.1 (Genecodes, Ann Arbor, MI). The V3 loop region was extracted, nucleotide sequences were converted to amino acid sequences, and coreceptor utilization and viral tropism were predicted manually by assessing sequences for typical features of X4 viruses15,16 and by using the two most widely used publicly available predictive algorithms for clinical samples, C-PSSMsinsi (http://indra.mullins.microbiol.washington.edu/webpssm/) and geno2pheno [coreceptor] (http://coreceptor.bioinf.mpi-inf.mpg.de/). The geno2pheno false-positive rate was set at 20% as per European guideline recommendations when a single PCR per patient is performed for analysis.17 Matched patient reverse transcriptase and protease inhibitor resistance profiles were extracted from the Viroscore database (Advanced Biological Laboratories, South Africa).
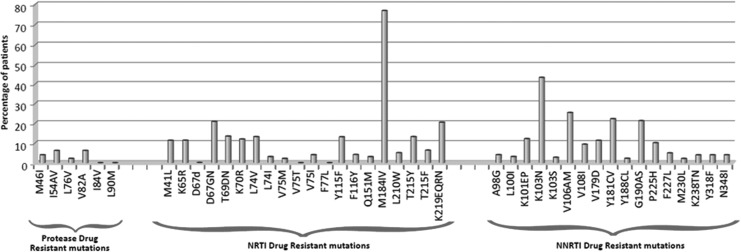
Drug resistance mutations and their frequencies are shown in Fig. 1. Mutations to all three major classes of drugs were noted. All 100 patients were infected with HIV-1 subtype C (www.hiv.lanl.gov) and harbored virus with at least one antiretroviral drug resistance mutation. Twenty-four percent had resistance mutations against only one class of drugs while 69% and 4% had at least one drug resistance mutation against two classes of drugs (NRTI and NNRTI or NNRTI and PI, respectively). A further 3% had mutations against all three classes of drugs (NRTI, NNRTI, and PI). The most common NRTI mutation was M184V (75%), with K103N (43%) and V106AM (26%) being the most common NNRTI mutations, similar to previously reported studies.18,19 K65R was present in 11% of patients, 9% of whom were on a tenofovir-containing regimen, consistent with recent findings.20 Thymidine analogue mutations (TAMs) were present in 35% of patient samples, with 10% possessing mutations associated with the TAM1 pathway, 19% with TAM2, and 6% with mutations common to both pathways. Thirteen percent had ≥3 TAMs. The high proportion of TAMs (35%) is comparable to previous South African studies that reported 32.2% TAMs in adult patients from KwaZulu-Natal.18 By contrast, TAMs were reported in 58.5% of HAART-failing children21 and 55% of HAART-failing adults19 from KwaZulu-Natal.
FIG. 1.
Frequency of selected antiretroviral drug resistance mutations in the 100 highly active antiretroviral therapy (HAART)-failing patients to protease inhibitors, nucleoside reverse transcriptase inhibitors (NRTIs), and non-NRTIs (NNRTIs).
The RT-PCR and PCR env amplification and sequencing were successful for 100% of patient samples. V3 loop analysis revealed that the overall positive amino acid charges ranged from +2 to +8, with overall lengths ranging from 33 to 37 amino acids. Signature sequence motifs within V3 such as the tetramer crown motif, overall positive charge, and positions 11 and 25, were strong predictors of coreceptor usage for each patient sample. Furthermore, V3 loop sequence-based predictive algorithms showed 40% and 42% of the HAART-failing patients had CXCR4-utilizing viruses using C-PSSMsinsi and geno2pheno, respectively (Table 1), but only 29 sequences gave concordant X4 prediction results between algorithms. Geno2pheno was shown to accurately predict X4/dual populations when compared to phenotyping.19,22 However, another study showed C-PSSMsinsi was most reliable at predicting X4 usage.21 Seclén et al.23 showed an overall concordance of 88% between PSSM and geno2pheno. Reanalysis of the Connell et al.15 data using the two algorithms showed 100% concordance of genotype with phenotype for PSSMsinsi and 95% with geno2pheno. Hence, based on concordant results from the two most widely used algorithms, a minimum of 29% of HAART-failing patients in our study had X4-utilizing viruses.
Table 1.
Amino Acid Sequence Alignment of V3 Loop Sequences of Highly Active Antiretroviral Therapy-Failing Patients Predicted as X4 Tropic by the C-PSSMsinsi and/or Geno2pheno [Coreceptor] (Using a 20% False-Positive Rate) Algorithms
| Patient ID | V3 loop sequence alignment | 11/25 aa | Net positive charge | C-PSSM predictiona | Geno2pheno predictionb |
|---|---|---|---|---|---|
| Cons C | CTRPNNNTRKSIRI––GPGQTFYATGDIIGDIRQAHC | SD | 4 | 0 | 83 |
| 13ZAIK001 | CTRPNNNTRKFIRIGPGGPGHAFYTNAVIGDIRKAHC | FA | 7 | 1 | 0.2 |
| 13ZAIK004 | CTRPGNNTRTRVGI––GPGRSFITTGQIIGDIRQAHC | RQ | 5 | 1 | 0.2 |
| 13ZAIK005 | CTRPHNNTSKRIKI––GPGRSFITTKSITGDIRQAHC | RS | 8 | 1 | 1.7 |
| 13ZAIK006 | CARPGNNTRSRVRVGIGRGQSVYATKAIIGDVRQAHC | RA | 7 | 1 | 0 |
| 13ZAIK007 | CTRPANNTRKSYRI––GPGQVFYTN-GVIGDIRQAHC | SG | 5 | 1 | 2.4 |
| 13ZAIK008 | CTRPSNNTRKKVRIGIGPGHTFYTTGNIIGRIREPHC | KN | 7 | 1 | 0.9 |
| 13ZAIK009 | CTRPNNNTRKSYRI––GPGQAFYTT-DIIGDIRQAHC | SD | 4 | 1 | 9.6 |
| 13ZAIK010 | CSRPNNNTRRRIHL––GLGRRFYTT-EIIGEIKQAYC | RE | 6 | 1 | 1.8 |
| 13ZAIK011 | CIRPGNNTRRSLRI––GPGQVFYTN-DIIGDIRKAHC | SD | 5 | 1 | 1.7 |
| 13ZAIK012 | CIRPGNNTRKSVRIGIGRGQVFYTN-SRIGDIRKAHC | SS | 8 | 1 | 0.2 |
| 13ZAIK013 | CTRPGNNTRRSIRI––GPGQVFYTN-NIVGGIRQAYC | SN | 5 | 1 | 8.2 |
| 13ZAIK014 | CTRPGNNTRRSIRI––GPGQVFYTN-PITGDIRKAYC | SP | 5 | 1 | 3.2 |
| 13ZAIK015 | CIRPGNNTRRSIRI––GPGHAFYAPRGIIGDIRKAYC | SG | 7 | 1 | 2.6 |
| 13ZAIK021 | CTRPGNNTRKSVPV––GTGRVIYATGAIIGDIRQAHC | SA | 5 | 1 | 5 |
| 13ZAIK025 | CTRPGNNTIKGIRI––GPGRQRFVAHKVIGDIRKAYC | GK | 8 | 1 | 1.1 |
| 13ZAIK039 | CTRPYKKIKRRVGI––GPGQAFRATEGITGDIRKAYC | RG | 7 | 1 | 0 |
| 13ZAIK050 | CVRPNQNTRRNIRI––GPGKVFYAT-DIKGSIREAHC | ND | 6 | 1 | 4 |
| 13ZAIK068 | CTRPGNNTRRSMGI––GPGRTFFATGDIIGDIRKAHC | SD | 5 | 1 | 5.3 |
| 13ZAIK069 | CTRPGNNTRKSVRI––GPGAAFYATRNMLGNIKKAHC | SN | 8 | 1 | 9.6 |
| 13ZAIK075 | CTRPNNNTRRGIGI––GPGRAVFATDKIIGNIRQAHC | GK | 6 | 1 | 1.7 |
| 13ZAIK076 | CIRPGNNTRRRIGI––GPGQAFHTHDRIIGDIRRAHC | RR | 8 | 1 | 0 |
| 13ZAIK078 | CMR-GNNTRKSIRI––GPGQAFYAHSNIIGD-RKAHC | SN | 7 | 1 | 1.7 |
| 13ZAIK080 | CTRPSNNTRKSVGI––GPGQVFYATEAVIGDIRQAHC | SA | 3 | 1 | 2.7 |
| 13ZAIK 83 | CTRPYKNTRQRVRI––GPGRTFVATSNIIGDIRTAYC | RN | 6 | 1 | 0.7 |
| 13ZAIK087 | CMRPGNKTRRRVGI––GPGQAFRATVGIIGNIRQAHC | RG | 8 | 1 | 0 |
| 13ZAIK091 | CTRPNNNTRKSVRIGPGPGQTFYAT-NIIGDIRQAHC | SN | 5 | 1 | 3.2 |
| 13ZAIK074 | CTRPNNNTRKSIGI––GPGQAFYANNNIIGDIRQAHC | SN | 4 | 1 | 13.7 |
| 13ZAIK002 | CTRPNNNTRKGIRI––GPGQVFYAN-EIIGDIREAHC | GE | 3 | 1 | 17.6 |
| 13ZAIK022 | CTRPGNNTRQSVGI––GPGQTIYATGAIIGDIRQAYC | SA | 2 | 1 | 19.1 |
| 13ZAIK094 | CVRPDNNTRKSVRI––GPGQTFYATESIIGDIRQAHC | SS | 3 | 1 | 41.3 |
| 13ZAIK098 | CIRPGNNTRQSIRI––GPGQTFYASKGIIGDIRQAHC | SG | 5 | 1 | 67.5 |
| 13ZAIK099 | CTRPNNNTRKSIRI––GPGQAFFANNNIIGDIRQAYC | SN | 4 | 1 | 58.7 |
| 13ZAIK003 | CSRPNNNTRKSIRI––GPGQAFYAN-DVIGDIRQAHC | SD | 4 | 1 | 32 |
| 13ZAIK032 | CTRTGNNTRQSVRI––GPGQTWYATGGIIGDIRKAYC | SG | 4 | 1 | 73.6 |
| 13ZAIK038 | CTRVANNTRRSVRI––GPGQAFYATGEVIGNIRQAHC | SE | 5 | 1 | 35.8 |
| 13ZAIK041 | CTRPNNNTSTGVRI––GPGQTFYATGRIIGDIRQAYC | GR | 3 | 1 | 33.7 |
| 13ZAIK053 | CIRPNNNTRKSVRI––GPGQAFFAPDDIIGDIRQAYC | SD | 2 | 1 | 52.1 |
| 13ZAIK061 | CTRPNNNTRKSIRI––GPGQALYTT-DIIGDIRKAYC | SD | 4 | 1 | 21.2 |
| 13ZAIK062 | CVRSNNNTRKSIRI––GPGQIFYAYGDIIGDIRQAYC | SD | 3 | 1 | 58.6 |
| 13ZAIK063 | CSRPNNNTRRSIHL––GLGRRFYTN-EIIGDIRQAYC | SE | 5 | 1 | 22.8 |
| 13ZAIK064 | CARPNNNTRKSVRI––GPGQVFYANNDIIGDIRQAHC | SD | 4 | 0 | 8.2 |
| 13ZAIK084 | CTRPGNNTRKSVRI––GPGQAFYATRDIIGDIRQAYC | SD | 4 | 0 | 6.9 |
| 13ZAIK023 | CMRPGNNTRKSIRI––GPGQTFYATGEIIGDIRQAHC | SE | 4 | 0 | 7.4 |
| 13ZAIK027 | CTRPGNNTRTSVRI––GPGQAFYATSDIIGDIRKAHC | SD | 4 | 0 | 9.3 |
| 13ZAIK035 | CTRPGNNTRKSIRI––GPGQTFYARGDIIGDIRKAHC | SD | 6 | 0 | 7.4 |
| 13ZAIK047 | CMRPNNNTRKSVRI––GPGQAFYATGEIIGNIRQAHC | SE | 5 | 0 | 6 |
| 13ZAIK051 | CARPNNNTRKSVRI––GPGSAFYATGDIIGDIREAHC | SD | 3 | 0 | 9.6 |
| 13ZAIK052 | CTRPNNNTRTSTRI––GPGQAFYATNDIIGDIRQAYC | SD | 2 | 0 | 7.4 |
| 13ZAIK054 | CTRPNNNTRKSIRI––GPGRAFSATGDIIGDIRQAYC | SD | 4 | 0 | 4.8 |
| 13ZAIK028 | CTRPNNNTRTSVRI––GPGQAFYATHDIIGDIRKAYC | SD | 6 | 0 | 16.9 |
| 13ZAIK093 | CIRPNNNTRKSIRI––GPGQAFYATNAIIGDIRQAYC | SA | 5 | 0 | 18.3 |
| 13ZAIK072 | CMRPNNNTRKGVRI––GPGQTFYATGEIIGNIRQAHC | GE | 6 | 0 | 10.8 |
| 13ZAIK058 | CTRPGNNTRKSVRI––GPGQVFYATNDIIGDIRQAHC | SD | 6 | 0 | 14.3 |
| Total X4 | 40 | 42 at 20% FPRc |
1, CXCR4 use predicted; 0, CCR5 use predicted.
All sequences<20% were predicted to use CXCR4.
False-positive rate.
The crown motif for each sequence is in bold and amino acids at positions 11 and 25 are in italics. Sequences predicted to be X4 by both algorithms are in bold and italics and discordant results are underlined.
Other tropism algorithms tested predicted different proportions of CXCR4-utilizing viruses for our sequences.24,25 For example, the CoRSeqv3-c algorithm25 predicted 27% CXCR4-utilizing viruses, thus its specificity and sensitivity must be evaluated further. Although improvements have been made to genotypic tropism prediction algorithms, the high proportion of discordant results implies that phenotypic characterization of viral tropism should be performed to resolve coreceptor usage.
Differences in prevalence of HAART-failing subjects having X4-tropic viruses in this study, as compared to 30% of HAART-naive subjects described in Connell et al.,15 are not statistically significant (p=1.0). Patients described in both studies were recruited from Gauteng province. Interestingly, our findings are in contrast to reports from KwaZulu-Natal that showed a significant difference between HAART-failing subjects with X4/dual//mixed-tropic viruses (60%) compared to HAART-naive subjects (p<0.02).19 Despite differences in proportions of X4-tropic viruses in HAART-failing subjects seen within South Africa, the high proportion of TAMs and X4-utilizing HIV-1 suggests that patients on HAART need to be monitored routinely for earlier detection of treatment failure and highlights the predicament of limited drug options, including R5 antagonists, for HAART-failing patients. As the South African epidemic evolves, patients failing second line treatment will increase, creating a need for the inclusion of alternative drugs such as maraviroc into third line regimens, and thus genotypic tropism testing must be refined to support this need.
Sequence Data
The V3 loop nucleotide and amino acid sequences were submitted to GenBank using Sequin V9.50 (www.ncbi.nlm.nih.gov/Sequin) and are available under accession numbers KF572487 to KF572586.
Acknowledgments
Research funding from the South African National Research Foundation (NRF), the Poliomyelitis Research Foundation (PRF), and the University of the Witwatersrand is gratefully acknowledged.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, et al. : Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996;381:661–666 [DOI] [PubMed] [Google Scholar]
- 2.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, et al.: HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 1996;381:667–673 [DOI] [PubMed] [Google Scholar]
- 3.Connor RI, Sheridan KE, Ceradini D, Choe S, and Landau NR: Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med 1997;184:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger EA, Murphy PM, and Farber JM: Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism and disease. Annu Rev Immunol 1999;17:657–700 [DOI] [PubMed] [Google Scholar]
- 5.Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, et al. : Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol 1997;71:7478–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho E, et al. : Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med 1993;118:681–688 [DOI] [PubMed] [Google Scholar]
- 7.Penn ML, Grivel JC, Schramm B, Goldsmith MA, and Margolis L: CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1 infected human lymphoid tissue. Proc Natl Acad Sci USA 1999;96:663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons G, Wilkinson D, Reeves JD, Dittmar MT, Beddows S, Weber J, et al. : Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol 1996;70:8355–8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, et al. : In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med 1997;3:1259–1265 [DOI] [PubMed] [Google Scholar]
- 10.Abebe A, Demissie D, Goudsmit J, Brouwer M, Kuiken CL, Pollakis G, et al. : HIV-1 subtype C syncytium and non-syncytium-inducing phenotype and coreceptor usage among Ethiopian patients with AIDS. AIDS 1999;13:1305–1311 [DOI] [PubMed] [Google Scholar]
- 11.Bjorndal A, Sonnerborg A, Tscherning C, Albert J, and Fenyo M: Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retroviruses 1999;15:647–653 [DOI] [PubMed] [Google Scholar]
- 12.Ping LH, Nelson JA, Hoffman IF, Schock J, Lamers SL, Goodman M, et al. : Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: Underrepresentation of X4 variants. J Virol 1999;73:6271–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecilia D, Kulkarni SS, Tripathy SP, Gangakhedkar RR, Paranjape RS, and Gadkari DA: Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 2000;271:253–258 [DOI] [PubMed] [Google Scholar]
- 14.Cilliers T, Nhlapo J, Coetzer M, Orlovic D, Ketas T, Olson WC, et al. : The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol 2003;77:4449–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connell BJ, Michler K, Capovilla A, Venter VD, Stevens WS, and Papathanasopoulos MA: Emergence of X4 usage among HIV-1 subtype C: Evidence for an evolving epidemic in South Africa. AIDS 2008;22:896–899 [DOI] [PubMed] [Google Scholar]
- 16.Michler K, Connell BJ, Venter VD, Stevens WS, Capovilla A, and Papathanasopoulos MA: Genotypic characterization and comparison of full-length envelope glycoproteins from South African HIV type 1 subtype C primary isolates that utilize CCR5 and/or CXCR4. AIDS Res Hum Retroviruses 2008;24:743–751 [DOI] [PubMed] [Google Scholar]
- 17.Vandekerckhove LPR, Wensing AMJ, Kaiser R, Brun-Vézinet F, Clotet B, De Luca A, et al. : European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 2011;11:394–407 [DOI] [PubMed] [Google Scholar]
- 18.Marconi VC, Sunpath H, Lu Z, Gordon M, Koranteng-Apeagyei K, Hampton J, et al. : Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis 2008;46:1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh A, Sunpath H, Green TN, Padayachi N, Hiramen K, Lie Y, et al. : Drug resistance and viral tropism in HIV-1 subtype C-infected patients in KwaZulu-Natal, South Africa: Implications for future treatment options. J Acquir Immune Defic Syndr 2011;58:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann CJ, Ledwaba J, Li JF, Johnston V, Hunt G, Fielding KL, et al. : Resistance to tenofovir-based regimens during treatment failure of subtype C HIV-1 in South Africa. Antivir Ther 2013;June10 (Epub ahead of print); DOI: 10.3851/IMP2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green TN, Archary M, Gordon ML, Padayachi N, Lie Y, Anton ED, et al. : Drug resistance and coreceptor usage in HIV type 1 subtype C-infected children initiating or failing highly active antiretroviral therapy in South Africa. AIDS Res Hum Retroviruses 2012;28:324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low AJ, Dong W, Chan D, Sing T, Swanstrom R, Jensen M, et al. : Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS 2007;21:F17–F24 [DOI] [PubMed] [Google Scholar]
- 23.Seclén E, Soriano V, Gonzalez MM, Gomez S, Thielen A, and Povedai E: High concordance between the position-specific scoring matrix and geno2pheno algorithms for genotypic interpretation of HIV-1 tropism: V3 length as the major cause of disagreement. J Clin Microbiol 2011;49:3380–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai S, Good B, Richman D, and Corbeil J: A new perspective on V3 phenotype prediction. AIDS Res Hum Retroviruses 2003;19:145–149 [DOI] [PubMed] [Google Scholar]
- 25.Cashin K, Gray LR, Jakobsen MR, Sterjovski J, Churchill MJ, and Gorry PR: CoRSeqv3-c: A novel HIV-1 subtype C specific V3 sequence based coreceptor usage prediction algorithm. Retrovirology 2013;10:24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]