HIV and simian immunodeficiency virus (SIV) establish latent infection in long-lived cells that form a reservoir of virus that persists in infected individuals even after years of treatment with highly active antiretroviral therapy (HAART). We present here an innovative technique that can be used to detect latent and active infected cells in tissues. The HIV/SIV reservoir is not fully characterized and to cure HIV we need to determine where the virus is hiding either latently integrated into the host DNA or replicating at very low levels producing viral RNA. The SIV infection model currently used in the Tulane National Primate Research Center (TNPRC) is suitable to study the latency and emergence of the virus in the presence of treatment and/or after lifting the pressure imposed by HAART to remain latent.
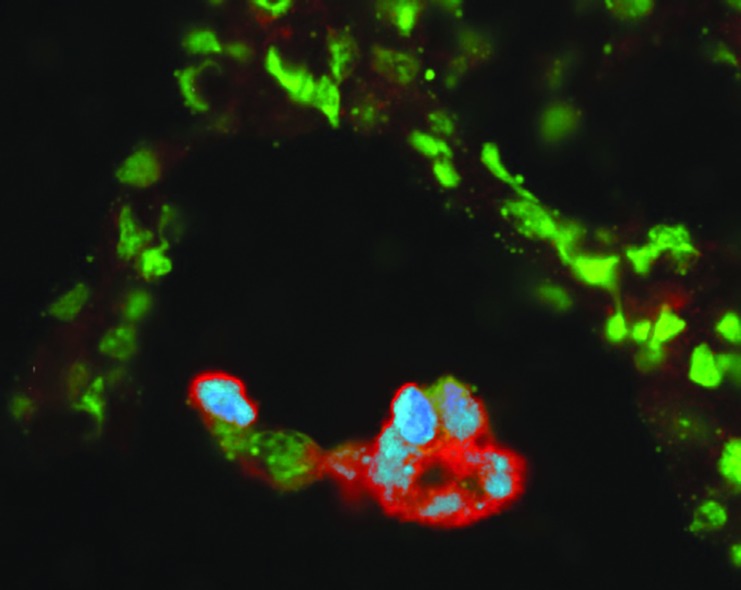
Figure 1 demonstrates simultaneous detection and colocalization of SIV RNA (red) and DNA (blue) in an infected multinuclear giant cell in the lung of an SIV-infected macaque. The image reveals that not all the nuclei have integrated the SIV. This method demonstrates that SIV-infected cells are relatively rare in vivo. The technique consists of double in situ hybridization: RNA hybridization and color development and after an RNase step a DNA hybridization developed with other color. The procedure combines previously described elements.1–3 This technique could be used to detect the virus integrated in cells that are not actively producing virus. This would be revealed by cells that are DNA positive, but RNA negative. Approaches identifying the expression of viral proteins or RNA will not work to identify latently infected cells. Only viral DNA detection can allow the identification of latently infected cells because they are defined by a lack of viral gene expression. This approach shows great promise to identify the location of cells, latently infected with SIV, and provide critical information to advance the efforts to develop a cure for HIV infection.
FIG. 1.
The image shows the identification of simian immunodeficiency virus (SIV) nucleic acids in the lung of an SIV-infected macaque. In situ hybridization of SIV RNA (red) and DNA (blue) identifies individual infected cells within tissue. All nuclei in the tissue are revealed by green staining with Bopro1.
Acknowledgments
This work was supported by the National Institutes of Health, grant OD011104 to Tulane National Primate Research Center, and grants AI097059, AI087302, and AI091501 to M.J.K.
References
- 1.Borda JT, Alvarez X, Kondova I, Aye P, Simon MA, Desrosiers RC,and Lackner AA: Cell tropism of simian immunodeficiency virus in culture is not predictive of in vivo tropism or pathogenesis. Am J Pathol 2004;165(6):2111–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jilbert AR: Probes. In: Methods in Molecular Biology, Vol. 123: In Situ Hybridization Protocols (Darby IA, Ed.). Humana Press Inc., Totowa, NJ: 2000 [DOI] [PubMed] [Google Scholar]
- 3.Hagiwara T, Hattori J, and Kaneda T: PNA–in situ hybridization method for detection of HIV-1 DNA in virus-infected cells and subsequent detection of cellular and viral proteins. In: Methods in Molecular Biology, Vol. 326: In Situ Hybridization Protocols, 3rd ed. (Darby IA. and Hewitson TD, Eds.). Humana Press Inc., Totowa, NJ, 2006 [DOI] [PubMed] [Google Scholar]