Abstract
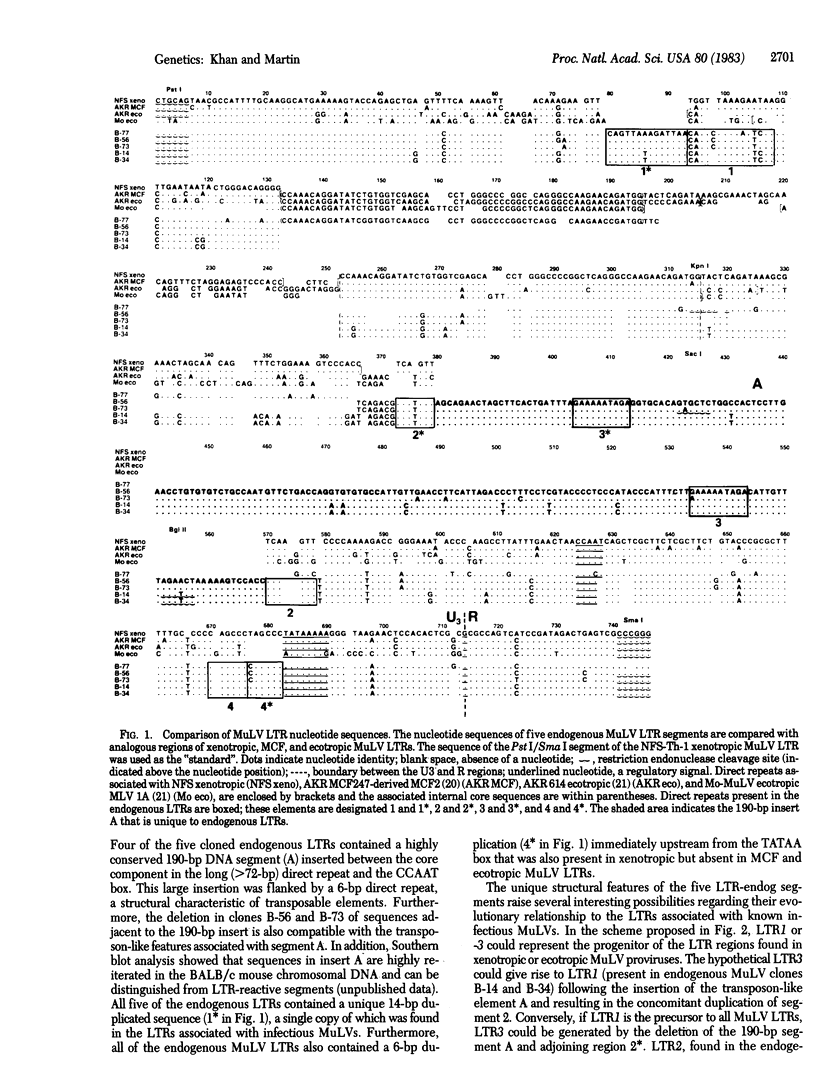
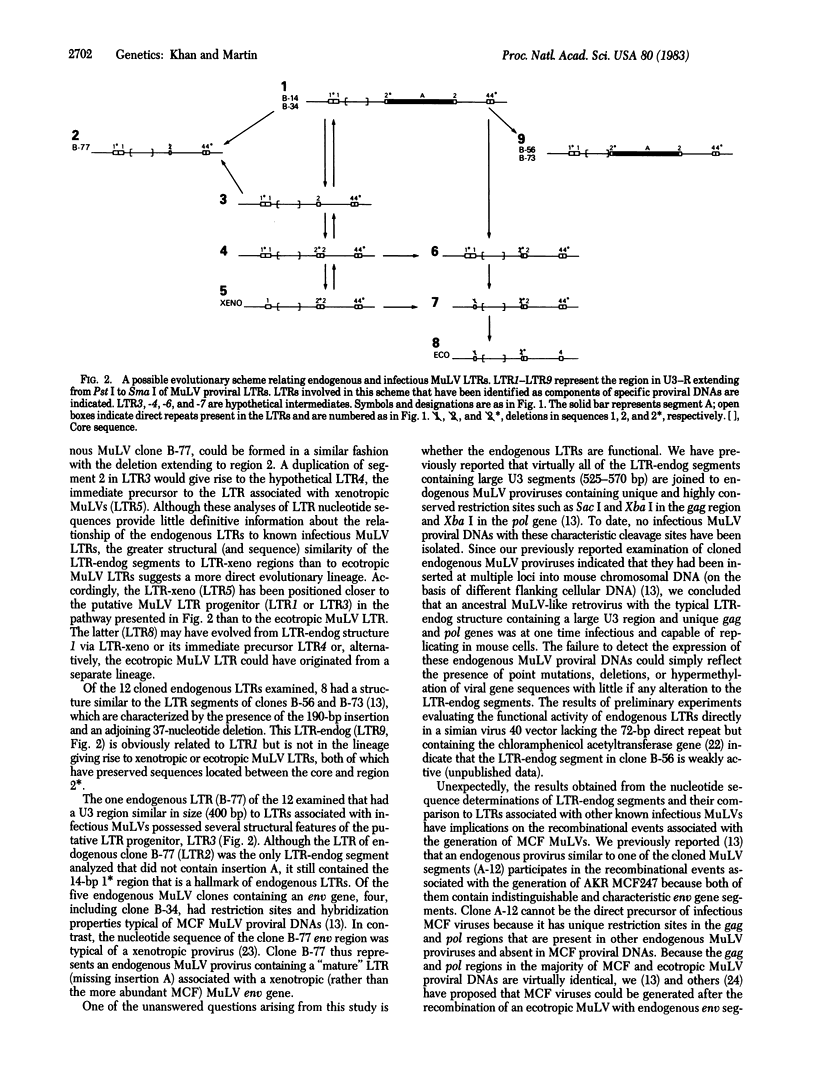
We have determined the nucleotide sequence in the U3-R regions of the long terminal repeat (LTR) associated with NFS-Th-1 xenotropic murine leukemia virus (MuLV) DNA and the LTR components of five endogenous proviruses cloned from BALB/c mouse chromosomal DNA. The five endogenous MuLV LTRs contained the regulatory signals thought to be important in viral transcription, such as "TATAA" and CCAAT-like boxes. A unique feature in four of the endogenous LTRs was the presence of a highly conserved 190-base-pair (bp) insert bounded by 6-bp direct repeats located 48 bp upstream from the C-C-A-A-T sequence. This segment was absent from LTRs associated with ecotropic, xenotropic, or mink cell focus-forming (MCF) MuLV proviruses. All five endogenous LTR segments also contained a 14-bp duplication of a sequence located near the 5' end of the first component of the long (greater than 72-bp) direct repeat of ecotropic and MCF MuLV LTRs. An evolutionary scheme relating LTRs associated with endogenous MuLV proviral DNAs to those found in ecotropic or xenotropic proviruses is presented. Nucleotide sequence analysis also suggested that the U3 region of the MCF247 MuLV LTR is derived from an NFS xenotropic related MuLV.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buckler C. E., Hoggan M. D., Chan H. W., Sears J. F., Khan A. S., Moore J. L., Hartley J. W., Rowe W. P., Martin M. A. Cloning and characterization of an envelope-specific probe from xenotropic murine leukemia proviral DNA. J Virol. 1982 Jan;41(1):228–236. doi: 10.1128/jvi.41.1.228-236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Cloyd M. W., Linemeyer D. L., Lander M. R., Rands E., Lowy D. R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature. 1982 Jan 7;295(5844):25–31. doi: 10.1038/295025a0. [DOI] [PubMed] [Google Scholar]
- Coffin J. M., Hageman T. C., Maxam A. M., Haseltine W. A. Structure of the genome of Moloney murine leukemia virus: a terminally redundant sequence. Cell. 1978 Apr;13(4):761–773. doi: 10.1016/0092-8674(78)90226-x. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishinuma F., DeBona P. J., Astrin S., Skalka A. M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981 Jan;23(1):155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Vanderryn D. F., Meltzer M. L., Martin M. A. Characterization of polyoma viral DNA sequences in polyoma-induced hamster tumor cell lines. J Biol Chem. 1980 Apr 25;255(8):3798–3805. [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Kelly M., Holland C. A., Lung M. L., Chattopadhyay S. K., Lowy D. R., Hopkins N. H. Nucleotide sequence of the 3' end of MCF 247 murine leukemia virus. J Virol. 1983 Jan;45(1):291–298. doi: 10.1128/jvi.45.1.291-298.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Repaske R., Garon C. F., Chan H. W., Rowe W. P., Martin M. A. Characterization of proviruses cloned from mink cell focus-forming virus-infected cellular DNA. J Virol. 1982 Feb;41(2):435–448. doi: 10.1128/jvi.41.2.435-448.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Rowe W. P., Martin M. A. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J mice: identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J Virol. 1982 Nov;44(2):625–636. doi: 10.1128/jvi.44.2.625-636.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung M. L., Hering C., Hartley J. W., Rowe W. P., Hopkins N. Analysis of the genomes of mink cell focus-inducing murine type-C viruses: a progress report. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1269–1274. doi: 10.1101/sqb.1980.044.01.138. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Martin M. A., Bryan T., McCutchan T. F., Chan H. W. Detection and cloning of murine leukemia virus-related sequences from African green monkey liver DNA. J Virol. 1981 Sep;39(3):835–844. doi: 10.1128/jvi.39.3.835-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy E. P., Smith M. J., Canaani E., Robbins K. C., Tronick S. R., Zain S., Aaronson S. A. Nucleotide sequence analysis of the transforming region and large terminal redundancies of Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5234–5238. doi: 10.1073/pnas.77.9.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repaske R., O'Neill R. R., Khan A. S., Martin M. A. Nucleotide sequence of the env-specific segment of NFS-Th-1 xenotropic murine leukemia virus. J Virol. 1983 Apr;46(1):204–211. doi: 10.1128/jvi.46.1.204-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Thomas C. Y., Coffin J. M. Genetic alterations of RNA leukemia viruses associated with the development of spontaneous thymic leukemia in AKR/J mice. J Virol. 1982 Aug;43(2):416–426. doi: 10.1128/jvi.43.2.416-426.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Goddard J. G., Berns A., Verma I. M. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]



