Abstract
Current mathematical models of tumor growth are limited in their clinical application because they require input data that are nearly impossible to obtain with sufficient spatial resolution in patients even at a single time point—for example, extent of vascularization, immune infiltrate, ratio of tumor-to-normal cells, or extracellular matrix status. Here we propose the use of emerging, quantitative tumor imaging methods to initialize a new generation of predictive models. In the near future, these models could be able to forecast clinical outputs, such as overall response to treatment and time to progression, which will provide opportunities for guided intervention and improved patient care.
NEXT-GEN MODELS
At the turn of the 20th century, meteorology was in its infancy, and weather forecasts were based on historical trends, experience, intuition, and guesswork. It was not until the development of realistic mathematical models of atmospheric phenomena, as well as advances in computation, that accurate weather prediction became a reality. At the onset of the 21st century, our approach to “tumor-specific treatment” of cancer and personalized patient prognosis resembles the early days of weather forecasting. However, with proper deployment of noninvasive imaging and appropriate investment in the development and validation of mathematical models, momentous advances in forecasting an individual patient’s treatment outcomes are within reach. To make this vision a reality requires realistic mathematical models of tumor growth initialized and constrained by patient-specific data.
Although much progress has been made building mathematical models of tumor growth, they have not been centered on clinical data. Consequently, these models have had limited impact on clinical practice. It is not enough to test the effect of various assumptions mathematically (in silico); if they are to be of clinical value, these models must make predictions based on data that can be readily measured in people and that can be readily tested (falsified) in the clinic.
In this Perspective, we argue that the development of clinically relevant mathematical models of tumor growth and treatment response should meet a fundamental prerequisite: the ability to incorporate quantitative, spatiotemporal data from the individual patient. Advances in noninvasive imaging technologies make this possible. Such data integration into models will catalyze acceptance of tumor-model–based forecasting into clinical practice.
A MODEL IMPASSE
There is an extensive literature on the mathematical modeling of tumor growth and treatment response. Proposed models range in complexity from exponential growth of an avascular tumor to complex equations describing molecules that promote invasion and angiogenesis. Although these approaches have provided insights into tumor biology (1–4), progress in this field has been somewhat overlooked by cancer biologists and clinical practitioners (5). Furthermore, modelers are frequently not involved in experimental design or data interpretation (6). In short, the measure-model feedback loop that is vital for model refinement and traction has not been closed.
We suspect that a reason for the lack of interactions between modelers and experimentalists is the way that models are constructed, studied, and presented. Briefly, papers reporting on mathematical modeling of tumors contain a theoretical section in which the model is described. Immediately after this exposition, there is frequently a table listing the model parameters (sometimes dozens) and how parameter values are assigned. With these values assigned, the papers then illustrate how model simulations evolve in time and how various perturbations to the parameters lead to distinct outcomes. In principle, the structure of these papers is correct and should produce powerful insights into tumor biology. In practice, there are (at least) two translational weaknesses: (i) model parameter values are constrained from published literature, often from different biological systems; and (ii) model predictions depend sensitively on these parameters, yet they are frequently unable to be measured in an actual patient. These limitations dampen enthusiasm from both the cancer biology and oncology communities.
The impasse in the application of mathematical modeling to cancer therapy may be overcome by alternative frameworks. For example, certain patient-specific, imaging-based measurements obtained before and during therapy could be used to initialize, constrain, and update models of tumor growth. The model predictions could then be directly compared with clinical outcomes iteratively and the patient-specific model refined and adjusted. Rigorous testing and subsequent adoption of this approach will require greater communication between imaging scientists, modelers, cancer biologists, and oncologists and will be hastened by the recent advances in both the quality and quantity of data available from emerging medical imaging techniques.
QUANTITATIVE IMAGING
There are several quantitative, noninvasive imaging methods that are now capable of reporting on biologically relevant, complementary tumor variables or parameters. Magnetic resonance imaging (MRI) and positron emission tomography (PET) have matured to the point where they offer patient-specific measures of tumor status at the physiological, cellular, and molecular levels. The methods described below have independently proven clinical utility. It is therefore natural to explore their integration into computational models.
Tumor-cell density
The microscopic, thermally induced behavior of molecules moving in a random pattern is referred to as self-diffusion or Brownian motion. The rate of diffusion in cellular tissues is described by means of an apparent diffusion coefficient (ADC), which largely depends on the number and separation of barriers that a diffusing water molecule encounters. Diffusion-weighted MRI (DW-MRI) methods have been developed to map the ADC and have been shown to correlate inversely with tissue cellularity (7, 8). ADC measurements can report on the early effects of cytotoxic therapies and have therefore been widely deployed in clinical trials as a surrogate biomarker of treatment response.
Tumor vascular characteristics
In dynamic contrast-enhanced MRI (DCE-MRI), images are collected before, during, and after a contrast agent is injected into a peripheral vein. The signal can be analyzed over time with a pharmacokinetic model so as to estimate physiological parameters related to vessel perfusion and permeability, the extravascular volume fractions, and the plasma volume. DCE-MRI methods are routinely used in clinical trials exploring the effects of antiangiogenic therapies (9).
Tumor glucose metabolism
The PET radiotracer most frequently used in clinical practice is fluorodeoxyglucose (FDG). As a glucose analog, FDG is taken up by tumor cells and phosphorylated to FDG-6-phosphate. However, FDG-6-phosphate is not metabolized further and therefore remains trapped intracellularly. PET quantification of FDG accumulation in tumors is a method of assessing and, in some cases, predicting therapeutic response by providing a surrogate for the metabolic activity of the tumor (10).
Tumor oxygen status
It is well known that hypoxia can induce changes in gene expression in tumor cells that lead to a more aggressive phenotype, including stimulation of angiogenesis, inhibition of apoptosis, and cell invasion. Clinical detection of hypoxia became available with the introduction of radiotracers, such as fluoromisonidazole and copper diacetyl-bis(N4-methylthiosemicarbazone), as well as blood oxygen level–dependent MRI methods (11). To date, the most common uses for hypoxia imaging are for selecting patients who may benefit from therapies designed to overcome hypoxia and for longitudinally assessing reduction in hypoxia (12).
These are but a few examples of quantitative, dynamic tumor variables or parameters that are currently obtained in the clinic with noninvasive imaging. As multimodal imaging of tumors continues to develop and combined PET/MRI scanners come online in the clinic, availability of tumor data from patients is bound to expand at a fast pace. Yet, harnessing the predictive power of these data remains a challenge. The paradigm described in this Perspective represents a realistic approach to optimizing the use of imaging data for predicting the response of tumors on an individual patient basis.
INTEGRATING IMAGING AND MATHEMATICAL MODELS
There is preliminary evidence that mathematical models of tumor growth, if conceived stringently with translational application as a goal, can be driven by data from noninvasive, patient-specific imaging studies.
Step 1: Cell number
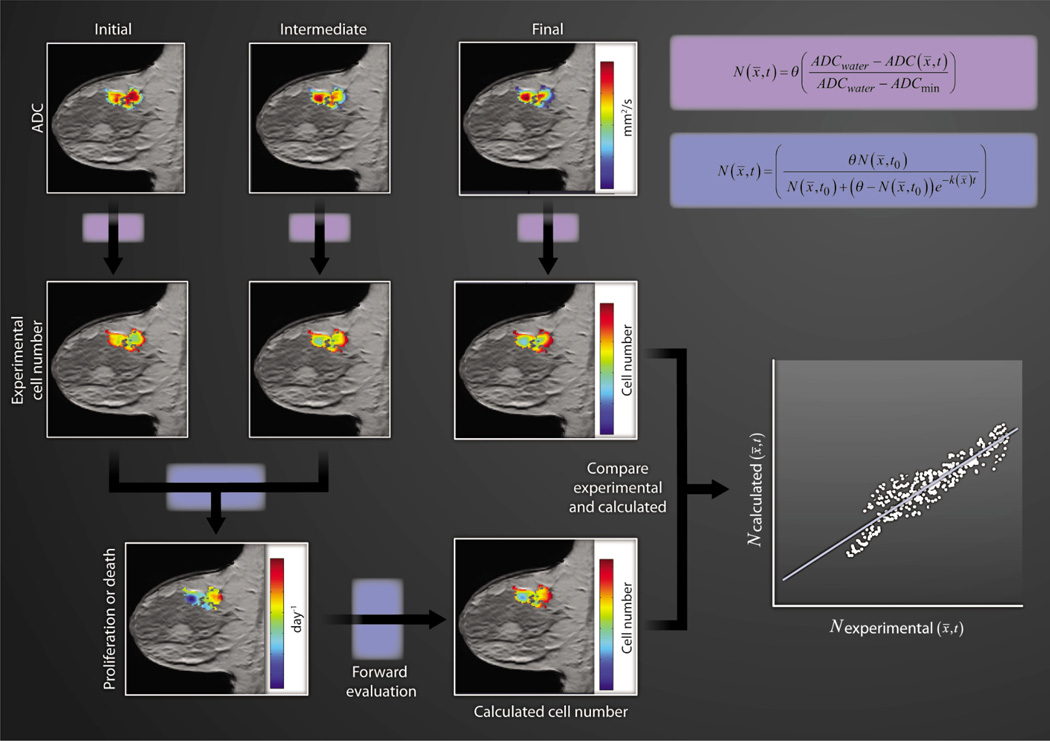
Figure 1 provides an example of how clinically acquired, quantitative MRI data of an invasive ductal carcinoma can be incorporated into a simple mathematical model of tumor growth. The top row of three images depicts ADC maps of a tumor locus superimposed on anatomical T1-weighted images obtained at three time points: before, after one cycle, and at the conclusion of chemotherapy. These data can be transformed to an estimation of cell number on a voxel-by-voxel basis. Then, the change in tumor cellularity from the pretreatment to the post-one-cycle treatment is used to calculate proliferation or death rate (depending on whether this value is positive or negative, respectively) for each voxel via the logistic model of tumor growth. In both equations in Fig. 1, the carrying capacity θ is the total number of tumor cells that fit within a given section of tissue. In imaging, the relevant length scale is the voxel, which has a well-defined size; thus, assuming a reasonable (measurable) mean tumor cell size fixes θ. Using the tumor cellularity (N) measured after one therapeutic time point in conjunction with the proliferation/death rate (k), one can predict cellularity at a future time point—and this predicted map can then be directly compared with experimental data (13, 14).
Fig. 1. Modeling cell number.
ADC maps of a tumor can be superimposed on anatomical T1-weighted MR images obtained at three time points during neoadjuvant chemotherapy. These data are first transformed to an estimation of cell number. So transformed, these data enable the calculation of the associated proliferation (k > 0) or death (k < 0) rate. The carrying capacity θ is estimated as the total number of tumor cells that fit within a voxel. Then by using the tumor cellularity measured at the second time point, the proliferation/death rate, and the logistic model of growth, one can predict future cellularity, which enables explicit comparison with experimental data (scatter plot).
Step 2: Incorporating cell motility
To improve the previous example of imaging correlating with tumor cell number, one could incorporate the movement of tumor cells. A natural extension would include the random diffusion of tumor cells as a function of both space and time—an approach studied by Wang et al. (15) in human glioma. They used a reaction-diffusion partial differential equation that described the rate of change in density of glioma cancer cells as the sum of a motility term (random diffusion) and a proliferation term (similar to the logistic model of Fig. 1). By analyzing serial MRI scans, the net proliferation and invasion rates can be estimated, and these rates can then be used to grow in silico an “untreated virtual control,” which can be compared directly with patient data. The authors showed that only patients with a low velocity of radial expansion or a low net proliferation rate survive longer than the median prognosis. Extending this approach for low-grade gliomas, Jbabdi et al. used diffusion tensor imaging (DTI) to allow for anisotropic movement of tumor cells along fiber tracts, which yielded improved shape and kinetic evolution of the brain tumors (16).
Step 3: Adding in mechanics
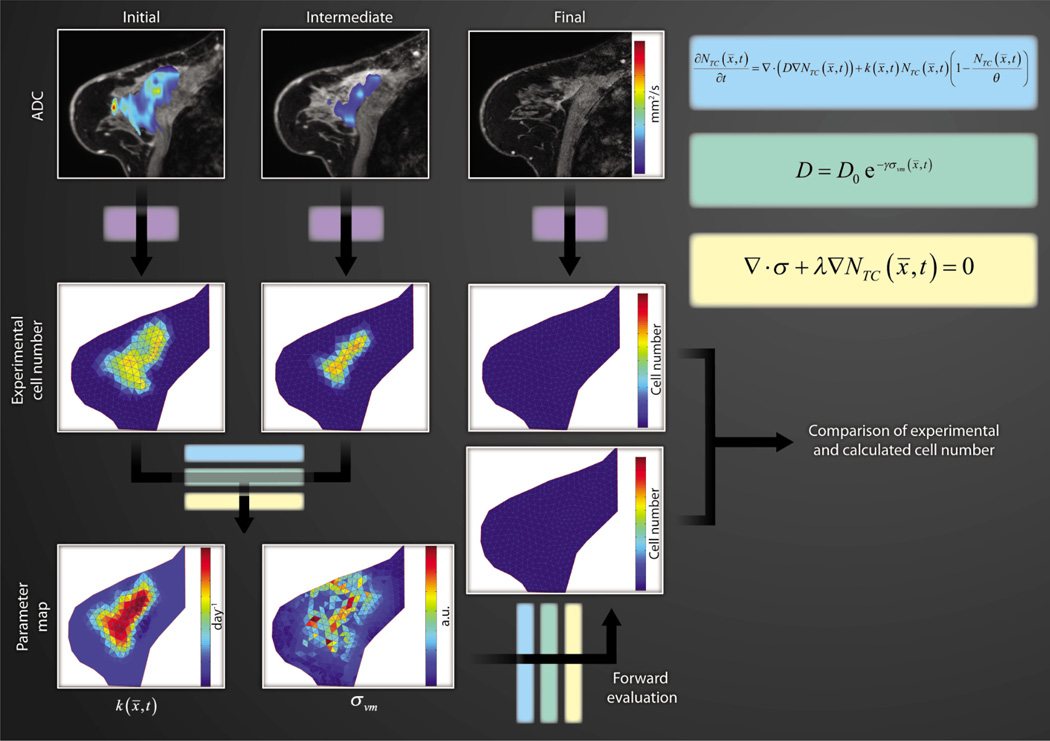
Tumor cell movement can be mechanically coupled to the surrounding tissue structure (17). As shown in Fig. 2, we express the rate of change of tumor cell number (NTC) at a particular location (x) and time (t) as the sum of random cell diffusion and logistic growth. Cell diffusion (D) is also linked to the nascent mechanical stress environment as described by the von Mises stress (σvm) and to an empirical coupling constant (γ). The mechanical stress tensor (σ) in Fig. 2 is influenced by an expansive force originating from mass changes associated with the proliferation of tumor cells and an empirical coupling constant (λ). Prediction of tumor cellularity at the conclusion of therapy is analogous to that presented in Fig. 1. Namely, the ADC maps obtained at three time points are transformed to estimates of cell number. Using cellularity data from the first two time points and a model, the proliferation parameter (k) can be estimated. As in Fig. 1, these initial cell number maps, the model, and the extracted proliferation parameter are then used to predict the cell number and distribution found in a human breast cancer patient at the conclusion of therapy and compared with experimental data. A similar approach was offered by Clatz et al. (18), who simulated the mechanical interaction of an invading glioblastoma into healthy brain parenchyma, albeit without considering the mechanotransductive effects on tumor growth mentioned here.
Fig. 2. Modeling cell number and biomechanics.
Similar to Fig. 1, the top row of three images depicts color-coded ADC maps of a tumor superimposed on (fat-saturated) T1-weighted MR images obtained at three time points during chemotherapy. Again, the initial and intermediate data are transformed to an estimation of cell number that, in conjunction with a biomechanical model (set of equations), is used to estimate the proliferation k. Using the estimated k and the biomechanical model, one can make predictions related to future cellularity, which can then be compared directly with experimental data (in this case, the acquired third time point acts as a comparator).
MULTISCALE IMAGING = MULTISCALE MODELING
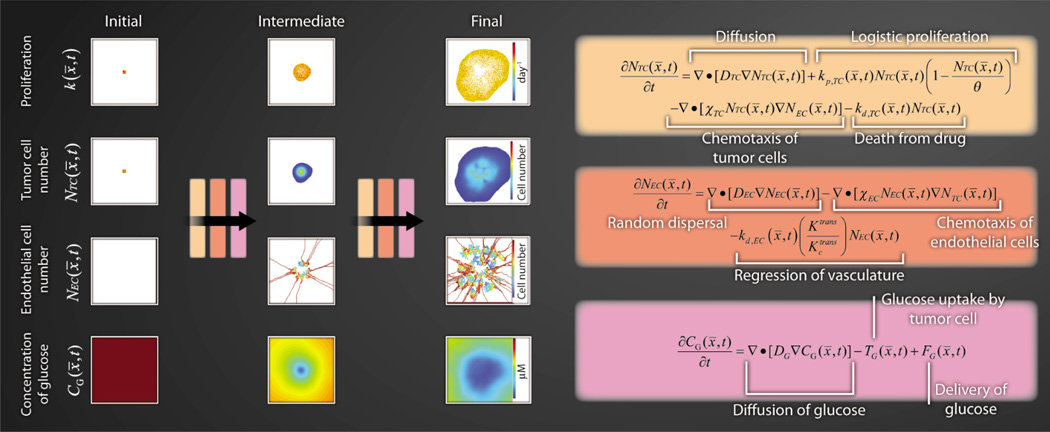
Multiscale data from combined PET-MRI could be used to initialize mathematical models incorporating many realistic tumor properties. A simulation of one such example is presented in Fig. 3: a coupled set of partial differential equations describing tumor cell proliferation, angiogenesis, and glucose consumption. T is system, although currently untested, closely resembles previous efforts (19–21) and has been recast to accept data from DCE-MRI, DW-MRI, and FDG-PET studies to initialize the model and lead to patient-realistic outcomes. Equations describing the rate of change of tumor cells, endothelial cells, and glucose at point x and time t take values of the above imaging data to initialize the model in order to predict distributions of these same parameters at future time points, which can then be directly compared with experimental data. The diffusion of tumor cells (DTC) can be estimated by using sequential anatomical MRI data (15). Similarly, sequential blood volume maps available from DCE-MRI can potentially inform the diffusion of endothelial cells (DEC). Once an estimate is obtained for DEC, chemotaxis of ECs (χEC) can be estimated as the value that results in a local blood volume increase equal to what is seen in vivo. MRI data can provide an estimate of NTC and θ. By adding a third imaging scan, the equation describing rate of change of tumor cells (Fig. 3) can be fit to sequential DW-MRI data in order to estimate kp,TC and kd,TC, which are the proliferation and death rates of the tumor cells, respectively. Chemotaxis of tumor cells (χTC) can be estimated to provide an equal tumor cell migratory/chemotaxic response to blood volume gradients as compared with literature studies of migratory/chemotaxic response to oxygen gradients.
Fig. 3. Multimodal outputs to describe tumor and vessel dynamics.
Multimodal (PET and MRI), multiparametric imaging data (estimates of glucose utilization, tumor cellularity, vascularity, perfusion, and tissue volume fractions) could be used to initialize and constrain a coupled set of partial-differential equations describing tumor cell proliferation, angiogenesis, and glucose utilization. The far left column represents the type of data that would be available from routine DW-MRI (k, NTC), DCE-MRI (NEC), and FDG-PET imaging (CG). These data can then be input into the equations provided to predict distributions of these parameters at future time points (intermediate and final outcomes). As with Figs. 1 and 2, the model outcomes can be directly compared with experimental data.
The diffusion value of glucose (DG) is a physical constant and can be safely assumed from literature values. Glucose uptake by tumor cells (TG) can be estimated by the product of the number of tumor cells and the glucose consumption rate per tumor cell. Although the number of tumor cells can be estimated from ADC data (Fig. 1), glucose consumption is more difficult to capture and may have to be assigned an empirical value. The delivery of glucose to tumor cells (FG) can be written as the product of glucose delivery to a given voxel, glucose in the blood, and the extraction fraction of glucose. Delivery is estimated from DCE-MRI data, blood glucose concentration (CG) is readily accessible by a blood draw, and the extraction fraction can be estimated by the difference between the concentration of FDG in the voxel and blood glucose levels.
LINKING THE SCALES
If we take the macroscopic-scale structure of the tumor to be its volume, surface area, and gross morphological features, then the mesoscopic-scale structure can be taken to be tumor cellularity (or cell density) and vascularity (Fig. 1). At finer scales, we encounter cellular properties such as glucose metabolism, proliferation, motility, or stress response (Figs. 2 and 3). Current, clinically available medical imaging methods report on all of these scales. [The Cancer Imaging Archive (www.cancerimagingarchive.net) provides a range of imaging data types available.] Although routinely available in vivo medical imaging approaches do not offer adequate spatial or temporal resolution at finer scales (such as the gene scale) to completely link all scales, it may be possible to link data accessible from imaging to the genomic and histopathology data available from, for example, biopsies (22). In short, every effort should be made to incorporate all available patient-specific data to more completely constrain a predictive model.
The models presented above are informed by recent work in cancer cell biology, but efforts are required to ensure measurements and associated predictions are consistent with the growing understanding at the microscopic level. We do not yet have a thorough understanding of how the biological properties not measured through imaging affect tumor modeling. Indeed, there are many ongoing efforts to understand the physical and biological principles at the microscopic scales, perhaps most notably by the members of the National Cancer Institute–National Science Foundation Physical Sciences in Oncology program (23). However, phenomenological, macroscopic models have been successful in other disciplines in both managing practical applications and providing lampposts for microscopic theories. The methodology we suggest could bring mathematical modeling to immediate clinical relevance through the use of phenomenological models that predict patient outcome from currently available patient measurables and thus can be used to improve patient care.
Acknowledgments
We thank M. Kardar (Massachusetts Institute of Technology) for several illuminating discussions.
REFERENCES AND NOTES
- 1.Norton L, Simon R. Growth curve of an experimental solid tumor following radiotherapy. J. Natl. Cancer Inst. 1977;58:1735–1741. doi: 10.1093/jnci/58.6.1735. [DOI] [PubMed] [Google Scholar]
- 2.Chaplain M, Anderson A. Mathematical modelling of tumour-induced angiogenesis: Network growth and structure. Cancer Treat. Res. 2004;117:51–75. doi: 10.1007/978-1-4419-8871-3_3. [DOI] [PubMed] [Google Scholar]
- 3.Anderson AR, Hassanein M, Branch KM, Lu J, Lobdell NA, Maier J, Basanta D, Weidow B, Narasanna A, Arteaga CL, Reynolds AB, Quaranta V, Estrada L, Weaver AM. Microenvironmental independence associated with tumor progression. Cancer Res. 2009;69:8797–8806. doi: 10.1158/0008-5472.CAN-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, Wang L, Amato KR, Arcila M, Sos ML, Socci ND, Viale A, de Stanchina E, Ginsberg MS, Thomas RK, Kris MG, Inoue A, Ladanyi M, Miller VA, Michor F, Pao W. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3002356. 90ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson ARA, Quaranta V. Integrative mathematical oncology. Nat. Rev. Cancer. 2008;8:227–234. doi: 10.1038/nrc2329. [DOI] [PubMed] [Google Scholar]
- 6.Byrne HM. Dissecting cancer through mathematics: From the cell to the animal model. Nat. Rev. Cancer. 2010;10:221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 7.Anderson AW, Xie J, Pizzonia J, Bronen RA, Spencer DD, Gore JC. Effects of cell volume fraction changes on apparent diffusion in human cells. Magn. Reson. Imaging. 2000;18:689–695. doi: 10.1016/s0730-725x(00)00147-8. [DOI] [PubMed] [Google Scholar]
- 8.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA, Rustin GJ, Taouli B, Choyke PL. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: Consensus and recommendations. Neoplasia. 2009;11:102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connor JP, Jackson A, Parker GJ, Roberts C, Jayson GC. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nat. Rev. Clin. Oncol. 2012;9:167–177. doi: 10.1038/nrclinonc.2012.2. [DOI] [PubMed] [Google Scholar]
- 10.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: Evolving considerations for PET response criteria in solid tumors. J. Nucl. Med. 2009;50(suppl. 1):122S–150S. doi: 10.2967/jnumed.108.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padhani AR, Krohn KA, Lewis JS, Alber M. Imaging oxygenation of human tumours. Eur. Radiol. 2007;17:861–872. doi: 10.1007/s00330-006-0431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krohn KA, Link JM, Mason RP. Molecular imaging of hypoxia. J. Nucl. Med. 2008;49(suppl. 2):129S–148S. doi: 10.2967/jnumed.107.045914. [DOI] [PubMed] [Google Scholar]
- 13.Atuegwu NC, Colvin DC, Loveless ME, Xu L, Gore JC, Yankeelov TE. Incorporation of diffusion-weighted magnetic resonance imaging data into a simple mathematical model of tumor growth. Phys. Med. Biol. 2012;57:225–240. doi: 10.1088/0031-9155/57/1/225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellingson BM, LaViolette PS, Rand SD, Malkin MG, Connelly JM, Mueller WM, Prost RW, Schmainda KM. Spatially quantifying microscopic tumor invasion and proliferation using a voxel-wise solution to a glioma growth model and serial diffusion MRI. Magn. Reson. Med. 2011;65:1131–1143. doi: 10.1002/mrm.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang CH, Rockhill JK, Mrugala M, Peacock DL, Lai A, Jusenius K, Wardlaw JM, Cloughesy T, Spence AM, Rockne R, Alvord EC, Jr, Swanson KR. Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel biomathematical model. Cancer Res. 2009;69:9133–9140. doi: 10.1158/0008-5472.CAN-08-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jbabdi S, Mandonnet E, Duffau H, Capelle L, Swanson KR, Pélégrini-Issac M, Guillevin R, Benali H. Simulation of anisotropic growth of low-grade gliomas using diffusion tensor imaging. Magn. Reson. Med. 2005;54:616–624. doi: 10.1002/mrm.20625. [DOI] [PubMed] [Google Scholar]
- 17.Garg I, Miga MI. Preliminary investigation of the inhibitory effects of mechanical stress in tumor growth, SPIE Medical Imaging 2008: Visualization, Image-Guided Procedures, and Modeling Conference. 2008;6918:69182L1–69182L11. [Google Scholar]
- 18.Clatz O, Sermesant M, Bondiau P-Y, Delingette H, Warfield SK, Malandain G, Ayache N. Realistic simulation of the 3D growth of brain tumors in MR Images coupling diffusion with biomechanical deformation. IEEE Trans. Med. Imaging. 2005;24:1334–1346. doi: 10.1109/TMI.2005.857217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, Xu SX, Wu J, Long Q. Coupled modelling of tumour angiogenesis, tumour growth and blood perfusion. J. Theor. Biol. 2011;279:90–101. doi: 10.1016/j.jtbi.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Gerlee P, Anderson ARA. A hybrid cellular automaton model of clonal evolution in cancer: The emergence of the glycolytic phenotype. J. Theor. Biol. 2008;250:705–722. doi: 10.1016/j.jtbi.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson ARA, Chaplain MAJ. Continuous and discrete mathematical models of tumor-induced angiogenesis. Bull. Math. Biol. 1998;60:857–899. doi: 10.1006/bulm.1998.0042. [DOI] [PubMed] [Google Scholar]
- 22.Cheng W-Y, Ou Yang T-H, Anastassiou D. Development of a prognostic model for breast cancer survival in an open challenge environment. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005974. 181ra50. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn NZ, Nagahara LA. Integrating physical sciences perspectives in cancer research. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005804. 183fs14. [DOI] [PubMed] [Google Scholar]