Abstract
Eukaryotic chromatin is a hierarchical collection of nucleoprotein structures that package DNA to form chromosomes. The initial levels of packaging include folding of long strings of nucleosomes into secondary structures and array-array association into higher-order tertiary chromatin structures. The core histone tail domains are required for the assembly of higher-order structures and mediate short- and long-range intra- and inter-nucleosome interactions with both DNA and protein targets to direct their assembly. However, important details of these interactions remain unclear and are a subject of much interest and recent investigations. Here, we review work defining the interactions of the histone N-terminal tails with DNA and protein targets relevant to chromatin higher-order structures, with a specific emphasis on the contributions of H3 and H4 tails to oligonucleosome folding and stabilization. We evaluate both classic and recent experiments determining tail structures, affect of tail cleavage/loss, and posttranslational modifications of the tails on nucleosomes and nucleosome arrays, as well as internucleosomal and inter-array interactions of the H3 and H4 N-terminal tails.
Keywords: Chromatin histone tail domains, histone acetylation, intra-nucleosome, inter-nucleosome
Introduction
The human genome is comprised of several meters worth of DNA. In order to fit into the tiny volume of a human cell nucleus, the DNA is compacted ~10,000 fold via assembly into a hierarchical and multifaceted DNA-protein complex known as chromatin. Beyond orderly packaging of genomic DNA, chromatin structures play important roles in regulation of gene expression and in other DNA-dependent processes. A critical aspect of chromatin’s function in such processes involves transitions between highly compacted and condensed chromatin states, in which the DNA is inaccessible to trans-acting factors, and more accessible states. Indeed, such transitions provide for a level of regulation that is essential for genes in organisms from yeast to humans (Bai, Charvin, Siggia and Cross 2010; Han and Grunstein 1988). However, essential details regarding the formation and regulation of condensed chromatin structures are poorly understood. In this review, we focus on recent work defining inter-nucleosome interactions mediated by the core histone tail domains, essential to higher-order chromatin organization.
The Nucleosome
The initial level of chromatin structure is a basic repeating subunit known as the nucleosome. Approximately 200 base pair (bp) segments of genomic DNA are organized into each nucleosome repeat, which can be further subdivided into nucleosome core and linker DNA regions. The ~147 bp nucleosome core region is wrapped ~1.75 times in a left-handed superhelix around a protein spool comprised of an octamer of core histone proteins, to generate a 225 kDa nucleosome core structure (Arents, Burlingame, Wang, Love and Moudrianakis 1991; Luger, Mader, Richmond, Sargent and Richmond 1997). The core histone octamer contains two copies of each of the four core histones, H2A, H2B, H3 and H4, which are highly conserved in both length and amino acid sequence. H3/H4 and H2A/H2B form stable dimers in aqueous solution of physiological pH and ionic strength. Two H3/H4 dimers also form a stable tetramer under such conditions but the tetramer does not bind H2A/H2B dimers to form the complete octamer in the absence of DNA or high (≥2M) NaCl (Arents and Moudrianakis 1993; Luger, Mader, Richmond, Sargent and Richmond 1997).
In most species, an additional family of histone proteins is present in chromatin in approximately a 1:1 stoichiometry with nucleosomes. Linker histones (also referred to as H1s), are highly basic and acid soluble proteins but share no structural homology with the core histones (Allan, Hartman, Crane-Robinson and Aviles 1980; Syed, Goutte-Gattat, Becker, Meyer, Shukla, Hayes, Everaers, Angelov, Bednar and Dimitrov 2010). H1s bind to the exterior of nucleosomes and stabilize the wrapping of DNA within nucleosomes. H1s also contribute to the formation and stability of higher-order chromatin structures (see below) (Allan, Hartman, Crane-Robinson and Aviles 1980). Nucleosome-specific binding by H1s is due to an ~80 residue ‘globular’ domain that adopts a winged-helix structure, found in some sequence-specific DNA binding proteins (Ramakrishnan, Finch, Graziano, Lee and Sweet 1993) and recognizes a trio of juxtaposed DNA surfaces formed by the two exiting/entering linker DNA strands and the central superhelical wrap of DNA at the nucleosomal dyad (Syed, Goutte-Gattat, Becker, Meyer, Shukla, Hayes, Everaers, Angelov, Bednar and Dimitrov 2010). The chromatin-condensing function of H1 is primarily provided by the ~100 residue C-terminal ‘tail’ domain, which contains about 40 lysine residues and few, if any, acidic residues (Allan, Hartman, Crane-Robinson and Aviles 1980). Interestingly, H1 CTDs are disordered when the protein is free in solution but adopt a unique fold or ensemble of folds upon binding to nucleosomes (Caterino, Fang and Hayes 2011).
The Core Histone Tail Domains
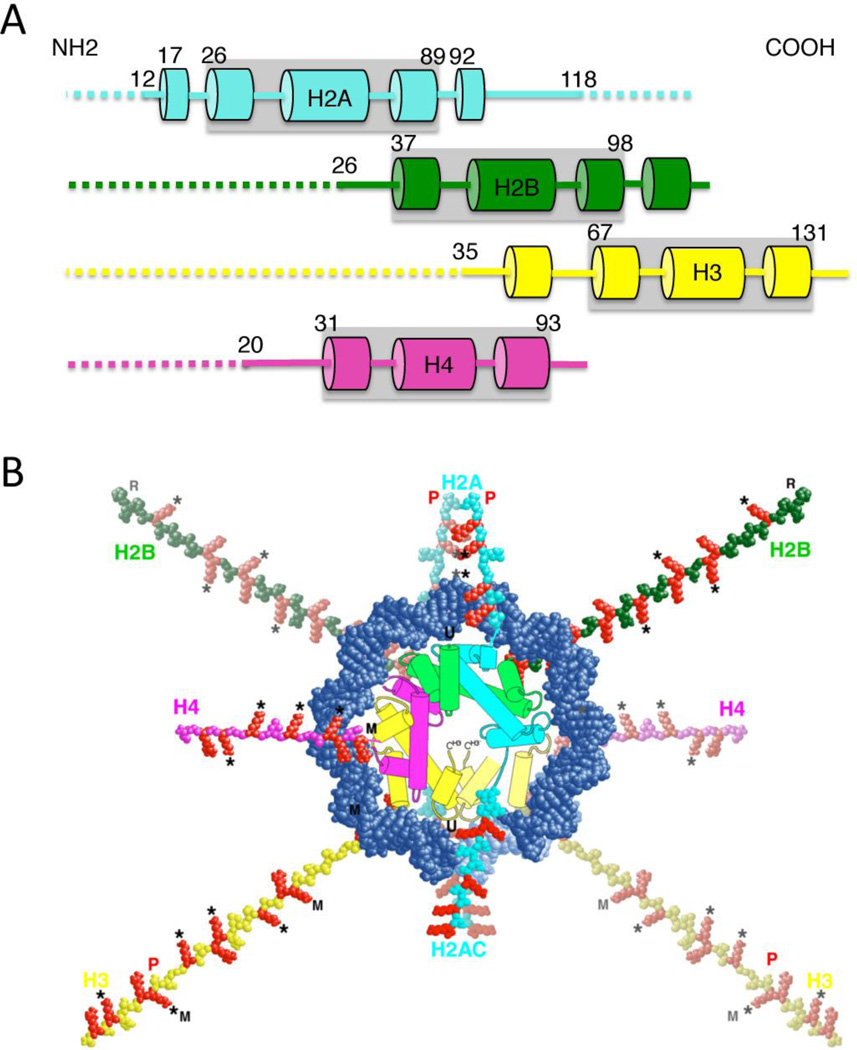
Each core histone contains a ~60 amino acid residue histone fold domain, which accounts for approximately 70–75% the mass of each protein (Arents and Moudrianakis 1993) and provides a highly structured and extensive dimerization interface between the H3/H4 and H2A/H2B pairs (Fig. 1A). Each histone also has an N-terminal tail domain that extends to the exterior of the nucleosome, while H2A contains an additional C-terminal tail domain. Thus, ten tails in total project from the nucleosome, exiting either between the superhelical gyres of DNA (N-H2B, N-H3) or over/under them (N-H2A, N-H4) (Fig. 1B) (Davey, Sargent, Luger, Maeder and Richmond 2002; Luger, Mader, Richmond, Sargent and Richmond 1997). The histone tails bind with low affinity to sites within nucleosome cores (Wang and Hayes 2007). Upon equilibration to unbound states or in the absence of DNA, the tails adopt unstructured random coil conformations (Smith and Rill 1989; Walker 1984) and thus are highly sensitive to proteolytic cleavage compared to the histone fold domains (Bohm and Crane-Robinson 1984; Grunstein 1997). The tail domains have a preponderance of lysines and arginines. For example, the ~35 residue H3 tail and the ~20 residue H4 tail contain 13 and 9 positively charged residues, respectively. Due to this abundance of positive charge, the tail domains contribute to the thermal stability of mononucleosomes (Ausio, Dong and van Holde 1989), while removal of the tail domains increases unwrapping of DNA from the nucleosome surface and accessibility of DNA-binding factors to nucleosome DNA (Lee, Hayes, Pruss and Wolffe 1993; Polach, Lowary and Widom 2000; Vettese-Dadey, Walter, Chen, Juan and Workman 1994; Vitolo, Yang, Basavappa and Hayes 2004). Note, however, that the effect of tail removal on nucleosome stability and DNA wrapping is relatively small compared to the stimulation of transcription factor binding and the tails central role in regulating transcription in vivo, indicating the tails play additional roles in these processes (see below) (Polach, 2000; Vitolo et al., 2004). Due to their intrinsic mobility, the tail domains are not well-ordered in most crystallographic studies of nucleosome cores (Luger, Mader, Richmond, Sargent and Richmond 1997), however, see (Davey, Sargent, Luger, Maeder and Richmond 2002).
Figure 1. The histone fold domains and N-terminal tails.
A. Cartoon of the histone fold domains (grey boxes) and tail domains (dashed lines) within a linear map of the core histones. α-helicies are indicated by columns. The positions of residues flanking the histone fold in each protein are indicated. The approximate residue at the end of the tail domain nearest the histone fold is also indicated. B. Cartoon of the nucleosome showing histone fold domains (colors correspond to A) and N-terminal tail domains extending away from the nucleosome core. Note that H2A also has a C-terminal tail. Basic residues (lysines and arginines) in the tail domains are colored red. Sites of lysine acetylation are indicated by a star. Figure adapted from (Wolffe and Hayes 1999).
Nucleosome Arrays and Higher-Order Chromatin Structure
In native chromatin, long chains of nucleosomes (hereafter referred to as nucleosome arrays) are folded and condensed into multiple higher-order structures. In vitro studies have shown that the conformation of nucleosome arrays is dependent on many factors, including but not limited to monovalent and multivalent counter-ion concentrations as well as the presence and particular subtype composition of histone H1. In low ionic strength solutions, nucleosome arrays exist as expanded “beads on a string” structures with extended linker DNA between successive nucleosome cores, as observed by both electron microscopy and atomic force microscopic techniques (Daban 2011; Olins and Olins 1974). Extended/unfolded nucleosome arrays [defined as primary chromatin structure (Woodcock and Dimitrov 2001)] spontaneously fold and condense into secondary structures, such as the so-called 30 nm diameter chromatin fiber, in solutions containing divalent cations such as Mg2+ (Thoma, Koller and Klug 1979; Widom 1986) (see below). Note that other secondary structures are possible and are often associated with the binding of non-histone ‘architectural’ factors such as HMGNs or PcG proteins (Luger and Hansen 2005). In addition to secondary chromatin structures, under physiological conditions nucleosomes arrays assemble into higher-order tertiary chromatin structures that may resemble the 100–130 nm and larger diameter ‘chromonema’ fibers originally observed by light microscopy (Belmont and Bruce 1994; Hansen 2002). Chromatin condensation is largely an electrostatic process, requiring counter-ion screening of DNA charge to allow close-packing of nucleosomes in condensed structures (Clark and Kimura 1990; Widom 1986).
It is interesting to note that the arrangement of nucleosomes and linker DNA within the 30 nm chromatin fiber still has not been definitively resolved. Current models posit either a solenoid model or a crossed-linker model. The solenoid model suggests that the 30 nm fiber folds through interactions between consecutive nucleosomes, in conjunction with bending of linker DNA, creating a continuous solenoid of condensed nucleosomes repeated every ~6 nucleosomes. Alternatively, a crossed linker model proposes that the linker DNA between nucleosomes within an array remains relatively straight during folding and crosses through the center of the 30 nm fiber, such that nucleosomes are arranged in a zigzag formation where the first nucleosome in the chain, “N1,” is closer in space to nucleosome “N3,” than nucleosome “N2”. (Additionally, a variation on this model proposes the linker DNA runs parallel to the fiber axis). Interestingly, compelling evidence has recently been presented where short-range order between nucleosomes thought to be associated with secondary structure is absent from metaphase chromosomes (Nishino, Eltsov, Joti, Ito, Takata, Takahashi, Hihara, Frangakis, Imamoto, Ishikawa and Maeshima 2012). A much fuller accounting of fiber models can be found in recent reviews (Grigoryev and Woodcock 2012; Luger, Dechassa and Tremethick 2012; Tremethick 2007).
Structural Studies of Native and Reconstituted Chromatin
Initial studies of chromatin folding employed native chromatin isolated from various nuclei and typically involved analysis by numerous physical techniques including electron microscopy, X-ray scattering, circular dichroism, and analytical centrifugation (van Holde 1989). Much information was obtained from these studies, including the relationship between counter-ions and overall condensation (Bates, Butler, Pearson and Thomas 1981; Clark and Kimura 1990; Thoma, Koller and Klug 1979; Widom, Finch and Thomas 1985), details of 30 nm diameter chromatin fiber structure (Graziano, Gerchman and Ramakrishnan 1988; Williams and Langmore 1991), effects of acetylation on higher-order structures (Annunziato and Hansen 2000) and interactions of the core histones with DNA (Mirzabekov, Bavykin, Karpov, Preobrazhenskaya, Ebralidze, Tuneev, Melnikova, Goguadze, Chenchick and Beabealashvili 1983). However, the heterogeneity present in native oligonucleosome samples and the inability to easily introduce modified or mutated histone proteins made detailed studies of tail function in chromatin structure difficult.
Although the ability to reconstitute nucleosomes has been available since the mid 1970’s (Camerini-Otero, Sollner-Webb and Felsenfeld 1976), oligonucleosome structures were not able to be prepared on long DNA templates due to the propensity of nucleosome cores to bind in a non-physiologically spaced, close-packed manner. This problem was resolved by the introduction of unique templates for reconstitution of oligonucleosome arrays by Simpson and colleagues (Simpson, Thoma and Brubaker 1985). These DNA templates contained tandemly arranged repeats of a DNA fragment from a Lytechinus variegatus 5 S rRNA gene, which possess nucleosome positioning properties, and allowed in vitro reconstitution of oligonucleosome arrays containing physiologically spaced nucleosomes with limited translational positions (Simpson, Thoma and Brubaker 1985) (Fig. 2). Recent development of the synthetic 601 family of nucleosome positioning elements (Lowary and Widom 1998), has lead to construction of arrays with extremely well-defined nucleosome positions (Dorigo, Schalch, Kulangara, Duda, Schroeder and Richmond 2004).
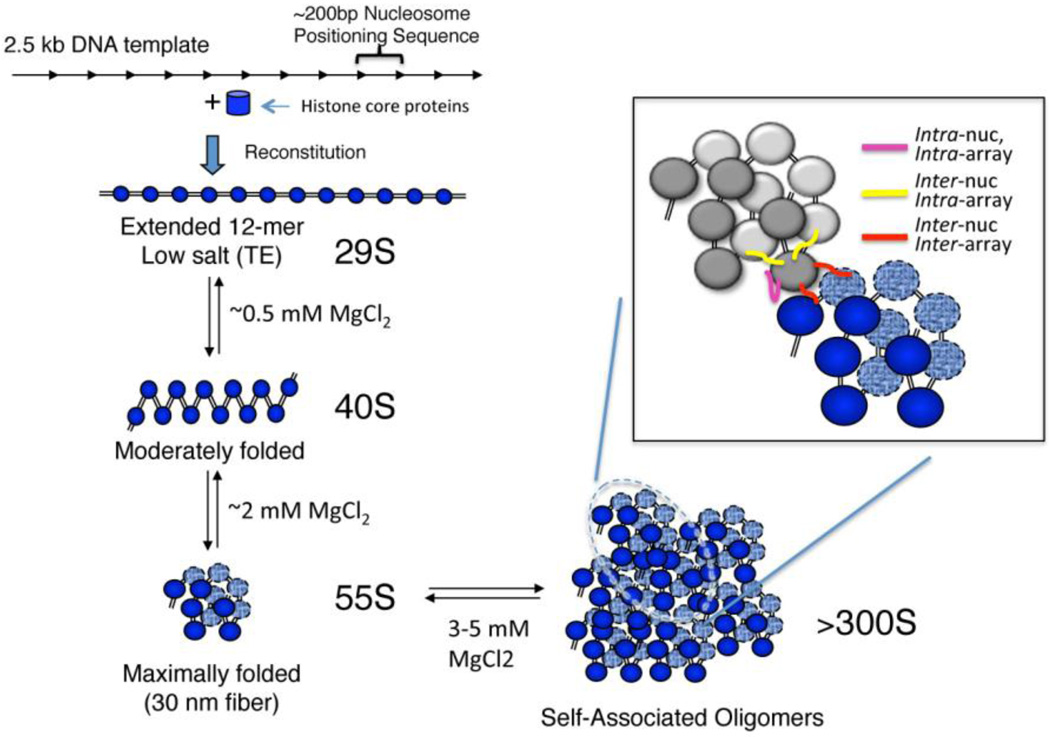
Figure 2. Salt-dependent folding and condensation of model nucleosome arrays.
A model nucleosome array is reconstituted from a template containing 12 tandem repeats of a nucleosome positioning sequence and purified core histone proteins (top right). The salt-dependent folding behavior of the arrays is shown, first forming a partially folded ‘contacting zig-zag’ structure in low Mg2+ and a fully condensed structure in higher salt that has the same hydrodynamic shape as a 30 nm-diameter chromatin fiber. Additional increases in salt induce self-association of arrays to form tertiary chromatin structures (bottom, left). Array folding is facilitated by inter-nucleosome, intra-array interactions while self-association is driven in part by inter-nucleosome, inter-array contacts (see inset).
Reconstituted nucleosome arrays, containing only the core histone proteins, can reversibly fold into secondary and tertiary chromatin structures identical to structures formed by native chromatin lacking H1 and as monitored by analytical ultracentrifugation, and resembling structures observed in vivo (Garcia-Ramirez, Dong and Ausio 1992; Hansen, Ausio, Stanik and van Holde 1989; Schwarz, Felthauser, Fletcher and Hansen 1996). Arrays lacking H1 exist as extended beads-on-a-string structures in TE but form a moderately folded, contacting zigzag structure in ~0.5 mM MgCl2, reflecting the behavior of native chromatin (Fig. 2) (Garcia-Ramirez, Dong and Ausio 1992; Hansen, Ausio, Stanik and van Holde 1989; Thoma, Koller and Klug 1979). Further increases in MgCl2 induce reconstituted arrays to fold into a secondary chromatin structure with the same hydrodynamic shape as a fully compact 30 nm fiber (Hansen, Ausio, Stanik and van Holde 1989). In addition, reconstituted arrays reversibly self-associate into higher-order tertiary structures at increased salt concentrations, thus reflecting the behavior of native chromatin (Garcia-Ramirez, Dong and Ausio 1992). These studies demonstrate that nucleosome arrays can fold into and maintain higher-order chromatin structures due solely to the intrinsic properties of core histones, and that the histone tails, along with specific counter-ions, work in concert to direct folding of the chromatin arrays. Similarly, arrays reconstituted with H1 behave identically to native chromatin, folding in the presence of monovalent or multivalent ions (Carruthers, Bednar, Woodcock and Hansen 1998; Huynh, Robinson and Rhodes 2005). Thus, reconstituted model systems reflect the ability of native chromatin to form primary, secondary, and tertiary chromatin structures.
Role of the Core Histone Tail Domains in Higher-Order Chromatin Structures
The N-terminal histone tail domains project to the exterior of the nucleosome and contact extra-nucleosomal constituents in chromatin. Indeed, evidence indicates that core histone tails play a comparatively minor role in nucleosome structure and stability. Core histones in which the majority of the tail domains have been removed still yield bona fide nucleosomes upon reconstitution in vitro with DNA, with high efficiency (Ausio, Dong and van Holde 1989; Whitlock and Stein 1978). Denaturation studies demonstrate that the tails contribute only marginally to the thermal stability of individual mononucleosomes, and have no effect on the salt dependent stability of nucleosomes (Ausio, Dong and van Holde 1989; Hong, Schroth, Matthews, Yau and Bradbury 1993).
Early experiments involving proteolytic removal of the tail domains of the core histones (as well as that of H1) demonstrated that these domains are required for formation of higher-order chromatin structures (Allan, Harborne, Rau and Gould 1982). Later, the Ausio and Hansen groups used both native and reconstituted nucleosome arrays to show that the core histone tail domains are essential for salt-dependent folding and self-association of these arrays into secondary and tertiary chromatin structures. (Garcia-Ramirez, Dong and Ausio 1992; Schwarz, Felthauser, Fletcher and Hansen 1996). Each of the core histone tails provides distinct contributions to the overall stability of chromatin folding. Investigations utilizing hybrid nucleosome arrays in which either the H2A/H2B N-terminal tails or the H3/H4 N-terminal tails were lacking showed that neither array was able to form the 30 nm fiber even at salt concentrations above that needed to fold standard arrays into the same structures (Schwarz, Felthauser, Fletcher and Hansen 1996). However, both sets of hybrid arrays were observed to undergo array oligomerization, albeit at elevated divalent cation concentrations, and both eventually reach the same level of compaction (Tse and Hansen 1997). Interestingly, arrays containing only the H3 or H4 tail domains can undergo oligomerization, while those containing only H2A or H2B tails are unable to oligomerize (Gordon, Luger and Hansen 2005). Taken together, these data indicate that the histone tails work cooperatively for full oligomerization of nucleosomal arrays, with a greater contribution made by the H3/H4 tails.
Acetylation: Direct Modification of Chromatin Structure
The tail domains are posttranslationally modified in conjunction with a multitude of nuclear processes, in a manner consistent with their role as important regulators of chromatin folding and condensation, and thus are a repository of epigenetic information. These modifications include but are not limited to arginine and lysine methylation, serine phosphorylation and lysine acetylation. Posttranslational modifications (PTMs) of the tails in specific combinations, sometimes referred to as the “Histone Code,” may act to precisely alter chromatin structure (Strahl and Allis 2000). The effect of PTMs on chromatin structure and gene activity can be indirect, as modifications serve to specifically recruit transcription factors and other activities to the chromatin, or direct, by modifying tail interactions and chromatin structure directly (Annunziato and Hansen 2000; Eberharter and Becker 2002; Shahbazian and Grunstein 2007).
Acetylation is perhaps the best characterized posttranslational modification, and as mentioned above, can elicit both direct and indirect effects on chromatin structure (only direct effects will be discussed in this review). Early experiments indicated that acetylation directly alters interactions of the histones with DNA and/or protein in arrays, to destabilize the chromatin structure and facilitate transcription (Allfrey, Faulkner and Mirsky 1964). Core histone tail acetylation is associated with open and active regions of euchromatin (Shahbazian and Grunstein 2007). General acetylation of the tail domains increases the ability of RNA polymerase III to transcribe an oligonucleosome template about 15-fold, and results in decondensation of the highly acetylated arrays (Tse, Sera, Wolffe and Hansen 1998). Interestingly, this effect appears to be primarily at the level of chromatin folding as acetylation does not significantly increase transcription through individual nucleosomes (Roberge, O'Neill and Bradbury 1991). Consistent with this observation, acetylation only marginally decreases the overall stability of mononucleosomes (Hong, Schroth, Matthews, Yau and Bradbury 1993; Simpson 1978), but oligonucleosome arrays containing hyperacetylated histone octamers exhibit a significantly reduced propensity to fold into higher-order structures (Garcia-Ramirez, Rocchini and Ausio 1995; Tse, Sera, Wolffe and Hansen 1998) and to self-associate into tertiary structures compared to non-acetylated arrays (Ridsdale, Hendzel, Delcuve and Davie 1990; Tse, Sera, Wolffe and Hansen 1998; Wang and Hayes 2008).
The effect of acetylation of individual histone tail domains, as well as acetylation of combinations of the tails, has been investigated with regard to secondary chromatin structure formation. By constructing arrays containing native core histone proteins with increasing levels of acetylation, Hansen and colleagues found that nucleosomes with an average of 12 acetyllysines (46% of potential sites acetylated) were completely unfolded in buffers containing 5 mM MgCl2 and 50 mM KCl and exhibited 15-fold greater transcription activity than unacetylated arrays (Tse, Sera, Wolffe and Hansen 1998). In contrast, arrays with 0–6 acetyllysines per nucleosome were completely folded and oligomerized. This work indicates a threshold effect in the disruption of higher-order chromatin structure. Interestingly, disruption of folding was observed with acetylation mimics located only on the H4 tail domain, and acetylation of H4K16 was shown to reduce MgCl2-dependent array folding and self-association as much as acetylation at lysines 5, 8, 12 and 16 within this protein (Allahverdi, Yang, Korolev, Fan, Davey, Liu and Nordenskiold 2011; Robinson, An, Routh, Martino, Chapman, Roeder and Rhodes 2008; Shogren-Knaak, Ishii, Sun, Pazin, Davie and Peterson 2006). Interestingly, this effect may be due to specific cation binding to the pocket within the H2A acidic patch thought to be occupied by H4K16 in condensed chromatin structures (Allahverdi, Yang, Korolev, Fan, Davey, Liu and Nordenskiold 2011).
Likewise, acetylation disrupts the ability of chromatin to form tertiary chromatin structures. Arrays containing recombinant histone proteins with lysine → glutamine substitutions to mimic acetylation in the tails showed that a reduction in the propensity for array self-association was most prominent for arrays with modified H4 tails (Wang and Hayes 2008). Indeed, acetylation only at H4K16 can disrupt self-association (Shogren-Knaak, Ishii, Sun, Pazin, Davie and Peterson 2006). Importantly, while acetylation mimics may not completely mimic the effect of specific acetylated lysines within the H4 tail domain with respect to chromatin folding, glutamine has been show to be an adequate mimic of acetylated lysine with regard to effects on array self-association into tertiary chromatin structures (Allahverdi, Yang, Korolev, Fan, Davey, Liu and Nordenskiold 2011; Wang and Hayes 2008). Surprisingly, and in contrast to the tail deletion studies mentioned above, acetylation mimics with the H2B tail domain had nearly a commensurate effect on self-association compared to H4 tail modification, while H3 acetylation had a much more modest effect (Wang and Hayes 2008). Modification of the H2A tail resulted in either a reduction or an increase in the propensity for salt-dependent array self-association, dependent on the modification state of the other tails (Wang and Hayes 2008). Additionally, only arrays containing modified H3 tails demonstrated an interesting decrease in stability of DNA wrapping at the edge of the nucleosome, as observed by an increase in DNA susceptibility to micrococcal-nuclease (MNase) digestion in this region. Thus, acetylation within each tail domain does not induce equivalent effects on nucleosome DNA wrapping and tertiary chromatin structure formation.
Lysine acetylation results in the loss of a positive charge, suggesting that charge neutralization plays a role in the direct effect of acetylation on chromatin folding. However, the available data indicate that a simple charge neutralization mechanism does not adequately explain the effect of this modification. Acetylation-induced reduction of only ~10% of the total positive charge in the tail domains results in a substantial inhibition of higher-order chromatin folding (Tse, Sera, Wolffe and Hansen 1998). Additionally, as mentioned above, acetylation mimics have distinct effects on nucleosome array self-association, dependent on their location in the nucleosome (Wang and Hayes 2008). These results are consistent with a model in which specific tail domains (and perhaps regions within each tail) mediate distinct interactions in condensed chromatin.
Intra- and Inter-nucleosome Interactions of the Tail Domains
The interactions of the tails pertinent to chromatin structure may be broadly parsed into intra-nucleosome interactions, where the tails interact with the DNA/protein of their own nucleosome, and inter-nucleosomal contacts, whereby the tails presumably mediate interactions between nucleosomes within condensed chromatin (Fig. 2, inset, Table 1). Indeed, it is likely that most core histone tail domains participate in both intra- and internucleosome contacts. It is important to note that distinct inter-nucleosome interactions may be cis with respect to local nucleosome arrays, relevant to folding of secondary structures, or trans, mediating long-range interactions to stabilize tertiary chromatin structures (Fig. 2, inset, Table 1). Determining how the tails interact within and between nucleosome arrays is crucial to the understanding of chromatin folding and genetic regulation.
Table 1. Summary of documented interactions of the H3 and H4 N-terminal tail domains in nucleosome arrays.
Interactions are divided into tail contacts with DNA (left) and protein (right).
| Array Conformation |
H3 N-tail - DNA | H4 N-tail - DNA | H3 N-tail - Protein |
H4 N-tail - Protein |
|---|---|---|---|---|
| Extended arrays | Intra-nucleosome Only (Kan et al., 2007). | Distribution of intra/inter nucleosome contacts to DNA to be determined | Not reported | H2A Acidic Patch (Kan et al., 2009) |
| Secondary chromatin structures (Intra-array contacts) | Inter-nucleosome only (Kan et al., 2007). | Distribution of intra/inter nucleosome contacts to DNA to be determined | Not reported | Inter-nucleosome H2A acidic patch (Dorigo, et al., ’03 JMB; Dorigo et al., ’04 Science). H4 K16 acetylation disrupts salt-dependent array folding (Shogren-Knaak, 2006, Allahverdi, 2010) |
| Tertiary structures (Inter-array contacts) | Inter-array contacts detected in salt-condensed nucleosome arrays (Kan et al., 2007). ~25% of inter-nucleosome contacts to DNA in condensed chromatin are inter-array | Inter-array contacts detected in salt-condensed nucleosome arrays (Kan et al., 2009). Note that total H4 contacts to DNA are significantly reduced upon salt-dependent condensation of nucleosome arrays (Kan, 2009). | Not reported | Inter-array contacts to H2A acidic patch detected in salt-condensed nucleosome arrays (Sinha et al., 2010). Interactions between H4 tails also detected (Sinha, 2010). |
Some intra-nucleosome contacts of the tail domains have been characterized. For example, site-specific crosslinking shows the C-terminal tail of histone H2A interacts with the DNA near the nucleosome dyad in nucleosome core particles and near the edge of the nucleosome core region when linker DNA is present (Lee and Hayes 1998; Usachenko, Bavykin, Gavin and Bradbury 1994). Crosslinking studies show the N-terminal tails of H4 and H2A interact with nucleosome DNA about 1.5 and 4.5 helical turns from the dyad, respectively (Ebralidse, Grachev and Mirzabekov 1988; Lee and Hayes 1997). Furthermore, histone H3 and H4 tails have been observed to interact with DNA 35–40 bps from the nucleosome dyad axis in mononucleosomes, and the histone H3 tail has been observed to form specific intra-nucleosomal interactions with DNA in extended nucleosomal arrays (Ausio, Dong and van Holde 1989; Zheng, Lu, Hansen and Hayes 2005). While the majority of tail domains are not visible in most crystal structures of nucleosome cores, a recent crystal structure of a nucleosome core containing the 601 nucleosome positioning sequence shows intra-nucleosome contacts of the tail domains that are, in part, in agreement with the crosslinking data (Davey, Sargent, Luger, Maeder and Richmond 2002).
Interestingly, as noted above, the availability of linker DNA alters tail interactions within nucleosomes. For example, total UV-dependent crosslinking of tails to DNA increases upon addition of linker DNA to nucleosome core particles (Angelov, Vitolo, Mutskov, Dimitrov and Hayes 2001). The H2A C-terminal tail domain contacts DNA near the nucleosome dyad in core particles, but interacts with DNA near the nucleosome periphery when linker DNA is present (Lee and Hayes 1998; Usachenko, Bavykin, Gavin and Bradbury 1994). Moreover, tail-dependent inhibition of DNA ligase activity within a nucleosome core is relieved by tail removal or addition of linker DNA, which results in redistribution of tail-DNA interactions (Chafin, Vitolo, Henricksen, Bambara and Hayes 2000). This suggests that tail interactions observed in nucleosome core crystal structures, which lack linker DNA, may not completely reflect actual tail interactions within native chromatin.
The core histone tail domains also mediate interactions between nucleosomes via inter-nucleosome contacts. An inter-nucleosome contact between the H4 tail domain and a surface of the H2A/H2B dimer interface of a nearby nucleosome was noted in the first X-ray crystal structure of a nucleosome core (Luger, Mader, Richmond, Sargent and Richmond 1997). Residues 16–25 within one histone H4 N-terminal tail contact an acidic patch on the surface of H2A/H2B on a neighboring nucleosome (Luger, Mader, Richmond, Sargent and Richmond 1997). Evidence for this interaction outside of the crystal lattice was provided by experiments showing that deletion of the region within the H4 tail observed to interact with the H2A/H2B acidic patch reduces MgCl2-dependent folding and self-association of reconstituted nucleosome arrays (Dorigo, Schalch, Bystricky and Richmond 2003). Moreover, crosslinking studies show that the H4 tail contacts with the H2A/H2B acidic patch occur commensurate with salt-dependent folding of nucleosome arrays into secondary chromatin structures and self-association of arrays into higher-order tertiary structures (Dorigo, Schalch, Kulangara, Duda, Schroeder and Richmond 2004; Kan, Caterino and Hayes 2009; Sinha and Shogren-Knaak 2010).
Interestingly, acetylation of H4K16, which is associated with actively transcribed chromatin, inhibits both salt-dependent intra-array folding/secondary structure formation as well as inter-array self-association (Allahverdi, Yang, Korolev, Fan, Davey, Liu and Nordenskiold 2011; Shogren-Knaak, Ishii, Sun, Pazin, Davie and Peterson 2006). Furthermore, evidence suggests ~30% acetylation of H4K16 in nucleosome arrays in vitro reduces compaction to a greater extent than deletion of the H4 tail (Robinson, An, Routh, Martino, Chapman, Roeder and Rhodes 2008). In S. cerevisiae, an H4K16Q acetylation mimic induces unique gene expression changes compared to acetylation mimics substituted for lysines 5, 8, or 12, which instead result in cumulative, non-specific transcriptional changes (Dion, Altschuler, Wu and Rando 2005). Additionally, while in vitro acetylation of H4K16 disrupts salt-dependent array folding, combinatorial acetylation of lysines 5, 8 and 12 does not disrupt 30 nm fiber folding to a similar extent (Allahverdi, Yang, Korolev, Fan, Davey, Liu and Nordenskiold 2011). It is therefore likely that inter-nucleosome contacts between the H4 tail and the H2A acidic patch play a major role in directing secondary and tertiary structure formation, while H4K16 acetylation provides a distinct regulatory mechanism for the stability of higher-order structures.
Incorporation of non-allelic variants of the major core histone proteins can alter the folding and stability of higher-order chromatin structures. For example, the H2A variant H2A.Z has a patch that is more acidic than that of major H2A histone due to replacement of asparagine and lysine residues in the patch with aspartic acid and serine residues, respectively (Fan, Rangasamy, Luger and Tremethick 2004). This intrinsic property of H2A.Z contributes to the formation of more compact secondary structures than observed with arrays containing H2A, (Fan, Gordon, Luger, Hansen and Tremethick 2002), a property lost when the relevant residues are mutated back to that of H2A. Interestingly, the more acidic patch of H2A.Z also inhibits array self-association into tertiary structures, suggesting competing functions for the H4 tail domain. Conversely, the acidic patch of the H2A variant H2A.Bbd, which is associated with more actively transcribed regions of chromatin, is less acidic than canonical H2A and results in a less stably folded secondary structure that is more transcriptionally competent than that formed with arrays containing H2A (Zhou, Fan, Rangasamy and Tremethick 2007). Moreover, H2A.Bbd-containing arrays appear to more efficiently undergo salt dependent self-association into higher-order structures. These results suggest that the H4 tail domain can contribute to either intra-array folding or inter-array self-association interactions. More broadly, such work provides a potential mechanism whereby the H2A/H2B acidic patch regulates array folding and gene expression.
Importantly, the acidic patch has also been shown to interact with several non-histone proteins, many of which are thought to modify chromatin structure. These include the chromatin architectural factor HMGN2, which binds to nucleosome cores and generates a more open and transcriptionally permissible chromatin secondary structure. HMGN2 has recently been shown to interact directly with the acidic patch within nucleosome cores (Kato, van Ingen, Zhou, Feng, Bustin, Kay and Bai 2011). Likewise, HP1, a heterochromatin-associated protein instrumental in the formation of highly compact heterochromatin to maintain a relatively silent transcriptional state (James and Elgin 1986; Kalashnikova, Porter-Goff, Muthurajan, Luger and Hansen 2013), is thought to interact with the acidic patch. When H2A within nucleosome arrays is replaced in vitro with H2A.Z, a histone variant with a slightly more acidic patch than H2A, HP1 binds with higher affinity to the nucleosome array (Zhou, Fan, Rangasamy and Tremethick 2007). These proteins in part compete with the H4 tail domain for binding to the patch and alter chromatin folding and self-association. The herpes virus latency-associated nuclear antigen (LANA) helps tether the viral genome to the host genome by binding with high affinity to the acidic patch (Barbera et al., 2006). Notably, a peptide from the LANA protein containing only the interacting region competes with the H4 tail for binding to the patch and results in an increased propensity for salt-dependent chromatin folding (Chodaparambil, Barbera, Lu, Kaye, Hansen and Luger 2007). Additional evidence provided by these investigators indicates that LANA (or H4 tail) association with the patch neutralizes repulsive forces between nucleosomes thereby stabilizing secondary structures. Interestingly, peptide-dependent displacement of the H4 tail from the acidic pocket also stimulates tertiary structure formation, perhaps by making more H4 tail domains available for inter-array contacts. Finally, several other chromatin-associated proteins contact the nucleosome partially through the H2A/H2B acidic patch, suggesting this may be a more general mode of interaction (Armache, Garlick, Canzio, Narlikar and Kingston 2011; Makde, England, Yennawar and Tan 2010).
Consistent with expectation, both the H3 and H4 tail domains have been shown to mediate inter-nucleosome interactions by contacting the DNA of neighboring nucleosomes. The H3 tail domain contacts only the DNA of its own nucleosome in extended nucleosome arrays, but rearranges to exclusively contact the DNA of neighboring nucleosomes in fully folded and self-associated arrays (Zheng, Lu, Hansen and Hayes 2005) (Fig. 2, inset). Thus the H3 tail seems ‘designed’ to mediate nucleosome-nucleosome interactions in condensed chromatin. Further work allowed discrimination between short-range inter-nucleosome interactions that likely facilitate folding of nucleosome arrays, and long-range inter-array interactions that facilitate array self-association and formation of tertiary chromatin structures (Kan and Hayes 2007). Specifically, crosslinking showed that about 20% of inter-nucleosome interactions of the H3 tail in condensed chromatin were inter-array, while 80% were intra-array. As expected, inter-array interactions are primarily mediated by residues near the exposed region of the H3 tail domain, while positions near the histone fold domain do not contribute to such interactions. Interestingly, while H1 stimulated inter-array interactions at lower salt concentrations, the extent of inter-nucleosome interactions that was comprised of inter-array contacts did not change in maximally condensed chromatin structures (Kan, Lu, Hansen and Hayes 2007). This result is consistent with a model whereby H1 stabilizes higher-order chromatin structures but does not fundamentally alter the folded and condensed structures. Moreover, it was found that while acetylation mimics cause reduction in inter-array interaction by the H3 tail, such mimics almost completely eliminate the stimulation of these interactions induced by H1, suggesting that the effect of acetylation is dominant over that of H1 with regard to inter-array interactions of the H3 tail domain.
Similar to the H3 tail domain, the H4 tail also participates in inter-array interactions with DNA. Inter-array contacts are observed for positions near the beginning of the H4 tail but are substantially reduced for positions near the histone fold domain. Of note, the total extent of inter-nucleosome interactions for the H4 tail appears to be much lower, perhaps due to the propensity of this tail domain to contact the H2A acidic pocket in addition to DNA (Kan, Caterino and Hayes 2009). Nevertheless, crosslinking studies indicate that some fraction of the H4 tail domains mediate long-range inter-array interactions with DNA, thus stabilizing tertiary chromatin structures (Kan, 2009).
SUMMARY
The core histone tail domains, particularly the H3 and H4 tails, mediate both intra- and inter-nucleosome interactions in chromatin (Table 1). While intra-nucleosome contacts stabilize the wrapping of DNA around nucleosome cores, inter-nucleosome interactions mediate formation of secondary and tertiary chromatin structures. Inter-nucleosome interactions can thus be parsed into short-range ‘intra-array’ and longer-range ‘inter-array’ interactions. Interestingly, while the bulk of interactions of the H3 tail involve contacts to DNA, the H4 tail contacts both DNA and the histone surface of nucleosomes. The H3 tail undergoes a transition from primarily intra-nucleosome interactions in extended arrays, to primarily inter-nucleosome contacts in condensed chromatin. While the H4 tail participates in similar intra-nucleosome contacts in extended chromatin, the data indicate that this tail participates in a diverse range of contacts including intranucleosome contacts to DNA, inter-nucleosome, intra-array contacts to the H2A/H2B acidic pocket, and inter-nucleosome, inter-array contacts to DNA to facilitate array-array self-association. Many more details of these interactions remain to be elucidated (Table 1).
References
- Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, Nordenskiold L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J, Harborne N, Rau DC, Gould H. Participation of the core histone tails in the stabilization of the chromatin solenoid. J Cell Biol. 1982;93:285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ. Preferential interaction of the core histone tail domains with linker DNA. Proc Natl Acad Sci USA. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT, Hansen JC. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Bai L, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2010;18:544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DL, Butler JG, Pearson EC, Thomas JO. Stability of the higher-order structure of chicken erythrocyte chromatin in solution. Eur J Biochem. 1981;119:469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm L, Crane-Robinson C. Proteases as structural probes for chromatin: the domain structure of histones. Biosci Rep. 1984;4:365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero RD, Sollner-Webb B, Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976;8:333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- Caterino TL, Fang H, Hayes JJ. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered h1 carboxyl-terminal domain. Mol Cell Biol. 2011;31:2341–2348. doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafin DR, Vitolo JM, Henricksen LA, Bambara RA, Hayes JJ. Human DNA ligase I efficiently seals nicks in nucleosomes. EMBO J. 2000;19:5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat Struct Mol Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DJ, Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990;211:883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- Daban JR. Electron microscopy and atomic force microscopy studies of chromatin and metaphase chromosome structure. Micron. 2011;42:733–750. doi: 10.1016/j.micron.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin Fiber Folding: Requirement for the Histone H4 N-terminal Tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebralidse KK, Grachev SA, Mirzabekov AD. A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nature. 1988;331:365–367. doi: 10.1038/331365a0. [DOI] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M, Dong F, Ausio J. Role of the histone "tails" in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- Graziano V, Gerchman SE, Ramakrishnan V. Reconstitution of chromatin higher-order structure from histone H5 and depleted chromatin. J Mol Biol. 1988;203:997–1007. doi: 10.1016/0022-2836(88)90124-6. [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Exp Cell Res. 2012;318:1448–1455. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Hansen JC. Conformational Dynamics of the Chromatin Fiber in Solution: Determinants, Mechanisms, and Functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Hansen JC, Ausio J, Stanik VH, van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined "30 nm" chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface. 2013;10 doi: 10.1098/rsif.2012.1022. 20121022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan PY, Hayes JJ. Detection of interactions between nucleosome arrays mediated by specific core histone tail domains. Methods. 2007;41:278–285. doi: 10.1016/j.ymeth.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol Cell Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci U S A. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Lee KM, Hayes JJ. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc Natl Acad Sci USA. 1997;94:8959–8964. doi: 10.1073/pnas.94.17.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Hayes JJ. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry. 1998;37:8622–8628. doi: 10.1021/bi980499y. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzabekov AD, Bavykin SG, Karpov VL, Preobrazhenskaya OV, Ebralidze KK, Tuneev VM, Melnikova AF, Goguadze EG, Chenchick AA, Beabealashvili RS. Structure of nucleosomes, chromatin, and RNA polymerase-promoter complex as revealed by DNA-protein cross-linking. Cold Spring Harb Symp Quant Biol. 1983;47:503–510. doi: 10.1101/sqb.1983.047.01.060. [DOI] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Lowary PT, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 2000;298:211–233. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–224. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Ridsdale JA, Hendzel MJ, Delcuve GP, Davie JR. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990;265:5150–5156. [PubMed] [Google Scholar]
- Roberge M, O'Neill TE, Bradbury EM. Inhibition of 5S RNA transcription in vitro by nucleosome cores with low or high levels of histone acetylation. FEBS Lett. 1991;288:215–218. doi: 10.1016/0014-5793(91)81037-9. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Simpson RT. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Sinha D, Shogren-Knaak MA. Role of direct interactions between the histone H4 Tail and the H2A core in long range nucleosome contacts. J Biol Chem. 2010;285:16572–16581. doi: 10.1074/jbc.M109.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Rill RL. Mobile histone tails in nucleosomes. Assignments of mobile segments and investigations of their role in chromatin folding. J Biol Chem. 1989;264:10574–10581. [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci U S A. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Tse C, Hansen JC. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usachenko SI, Bavykin SG, Gavin IM, Bradbury EM. Rearrangement of the histone H2A C-terminal domain in the nucleosome. Proc Natl Acad Sci USA. 1994;91:6845–6849. doi: 10.1073/pnas.91.15.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde KE. Chromatin. New York: Springer Verlag; 1989. [Google Scholar]
- Vettese-Dadey M, Walter P, Chen H, Juan LJ, Workman JL. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo JM, Yang Z, Basavappa R, Hayes JJ. Structural features of transcription factor IIIA bound to a nucleosome in solution. Mol Cell Biol. 2004;24:697–707. doi: 10.1128/MCB.24.2.697-707.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker IO. Differential Dissociation of Histone Tails from Core Chromatin. Biochemistry. 1984;23:5622–5628. doi: 10.1021/bi00318a037. [DOI] [PubMed] [Google Scholar]
- Wang X, Hayes JJ. Site-specific binding affinities within the H2B tail domain indicate specific effects of lysine acetylation. J Biol Chem. 2007;282:32867–32876. doi: 10.1074/jbc.M706035200. [DOI] [PubMed] [Google Scholar]
- Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JP, Jr, Stein A. Folding of DNA by histones which lack their NH2-terminal regions. J Biol Chem. 1978;253:3857–3861. [PubMed] [Google Scholar]
- Widom J. Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. J Mol Biol. 1986;190:411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- Widom J, Finch JT, Thomas JO. Higher-order struction of long repeat chromatin. EMBO J. 1985;4:3189–3194. doi: 10.1002/j.1460-2075.1985.tb04064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SP, Langmore JP. Small angle x-ray scattering of chromatin. Radius and mass per unit length depend on linker length. Biophys J. 1991;59:606–618. doi: 10.1016/S0006-3495(91)82276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]