Abstract
The extracellular matrix proteoglycan (ECM) perlecan, also known as heparan sulfate proteoglycan 2 or HSPG2, is one of the largest (>200 nm) and oldest (>550M years) extracellular matrix molecules. In vertebrates, perlecan’s five-domain structure contains numerous independently folding modules with sequence similarities to other ECM proteins, all connected like cars into one long, diverse complex train following a unique N-terminal domain I decorated with three long glycosaminoglycan chains, and an additional glycosaminoglycan attachment site in the C-terminal domain V. In lower invertebrates, perlecan is not typically a proteoglycan, possessing the majority of the core protein modules, but lacking domain I where the attachment sites for glycosaminoglycan chains are located. This suggests that uniting the heparan sulfate binding growth factor functions of domain I and the core protein functions of the rest of the molecule in domains II-V occurred later in evolution for a new functional purpose. In this review, we surveyed several decades of pertinent literature to ask a fundamental question: Why did nature design this protein uniquely as an extraordinarily long multifunctional proteoglycan with a single promoter regulating expression, rather than separating these functions into individual proteins that could be independently regulated? We arrived at the conclusion that the concentration of perlecan at functional borders separating tissues and tissue layers is an ancient key function of the core protein. The addition of the heparan sulfate chains in domain I likely occurred as an additional means of binding the core protein to other ECM proteins in territorial matrices and basement membranes, and as a means to reserve growth factors in an on-site depot to assist with rapid repair of those borders when compromised, such as would occur during wounding. We propose a function for perlecan that extends its role from that of an extracellular scaffold, as we previously suggested, to that of a critical agent for establishing and patrolling tissue borders in complex tissues in metazoans. We also propose that understanding these unique functions of the individual portions of the perlecan molecule can provide new insights and tools for engineering of complex multi-layered tissues including providing the necessary cues for establishing neotissue borders.
Keywords: perlecan, HSPG2, basement membrane, basal lamina, heparan sulfate, proteoglycan, tissue structure, tissue borders
1. Introduction
Perlecan, or heparan sulfate proteoglycan 2 (HSPG2), is an exceptionally large, secreted extracellular matrix (ECM) proteoglycan with a modular structure that supports its many functions in complex tissues. Human perlecan consists of 4391 amino acids including a twenty one amino acid long signal sequence that is absent in the mature protein. The protein core of perlecan consists of a series of individual folding motifs or “modules” that include one SEA domain, three each of laminin G-like domains and laminin B domains, four each of LDL-receptor class A domains and EGF-like domains, eleven laminin EGF-like domains including two that are split (1st and 5th), and one that is incomplete (4th), and twenty two Ig-like C2 type immunoglobulin-like domains, all but one of which are in domain IV. The structural features of these modules and their organization into five domains have been well reviewed previously and the interested reader is referred to any of these excellent articles for more information (Farach-Carson and Carson, 2007; Gomes et al., 2002; Hassell et al., 2002; Ida-Yonemochi et al., 2011; Iozzo, 2005; Iozzo et al., 1994; Iozzo et al., 2009; Ishijima et al., 2012; Kaneko et al., 2013; Knox and Whitelock, 2006; Melrose et al., 2002).
One fascinating aspect of perlecan in vertebrates is its almost invariable existence as a long modular multi-functional protein. While a few variants of the human protein have been reported (i.e. miniperl GenBank AAL79552.1), vertebrate perlecan almost always is produced as a single chain core protein with the biological variability largely attributable to the nature and extent of its decoration with glycosaminoglycan chains, specifically some combination of heparan sulfate (HS) and chondroitin sulfate, added to the core protein at specific sites in modular domains I (see annotation in UniProt P98160) and sometimes V (Tapanadechopone et al., 1999). A single promoter drives and regulates perlecan expression. Then, like train cars linked behind one engine, the expression of the forty eight independently folding modules of human perlecan follows an encoded signal peptide locomotive that directs the entire sequence into the secretory path for carbohydrate decoration. Beginning with the SEA module as the coal car and with the final laminin G-like 3 (LG3) module as the caboose, the diverse modules of perlecan are hitched together intracellularly by their assembly as a single polypeptide chain (Figure 1). After modification during intracellular trafficking through the secretory route, the whole assembly then is deposited into the extracellular space where the individual modules can interact with a variety of other ECM proteins, heparan sulfate binding proteins, and other protein modifiers (Farach-Carson and Carson, 2007; Whitelock et al., 2008). While some cells, such as chondrocytes and endothelial cells have the capacity, chaperones, and sugar nucleotide pools to make large amounts of fully decorated perlecan, the size and complexity of the protein prevent it from being produced easily as a full length recombinant protein. Most laboratories that study perlecan thus rely on purification protocols using large volumes of medium from cultured cells or tissue extracts, and take advantage of its large size and charge to separate it from other ECM proteins (Castillo et al., 1996; Kaji et al., 2000; Whitelock, 2001). Other approaches include the recombinant production of individual perlecan domains or clusters of modules (Chakravarti et al., 1995; Clarke et al., 2012; Decarlo et al., 2012; French et al., 2002; Hopf et al., 1999), or synthesis of perlecan sequence-containing peptides (Farach-Carson et al., 2008a). The human HSPG2 gene is located on the reverse strand of chromosome 1 and covers 115,000 base pairs. There are 97 exons in the transcript, which is 14,327 base pairs in length.
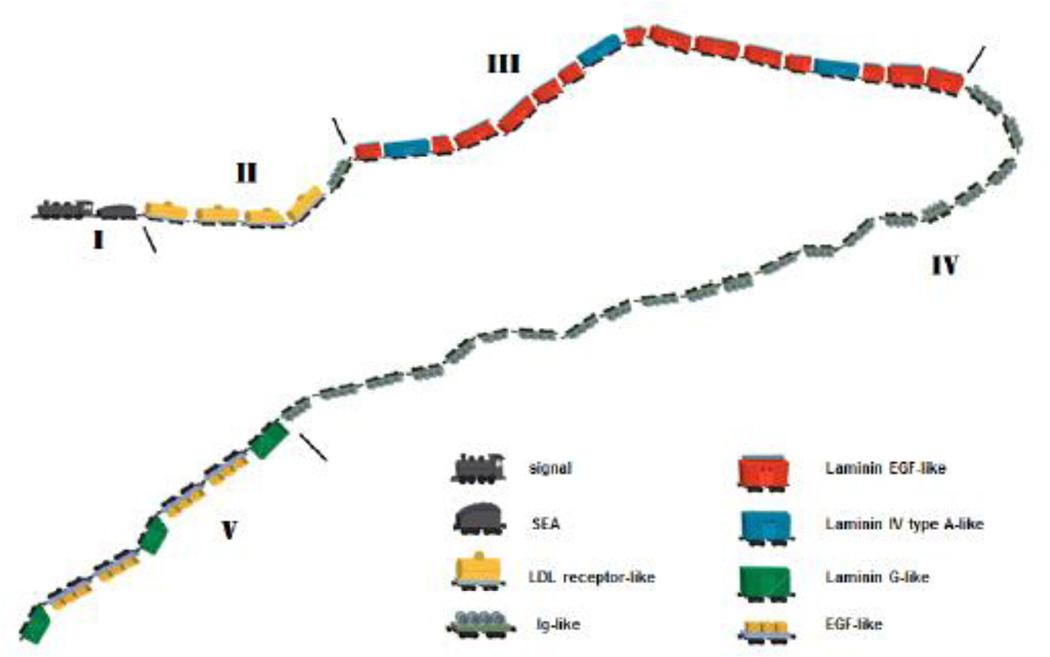
Figure 1. Modular structure of perlecan chain depicted as train cars assembled in tandem behind a common “engine” that is unique to perlecan.
The individual modules are self-folding, joined by linker sequences between. The locomotive depicts the signal sequence driving the train into the cellular secretory path for secretion into the extracellular space. The SEA module is envisioned as the coal car, and is near the region where the majority of the glycosylation occurs in domain I (sugars not shown). Features of note include the “split cars” such as the three laminin EGF-like modules split by a laminin IV type A-like module (red split by blue). Note the one Ig-like module at the end of domain II, with the rest in long domain IV. The curvature of the train is suggested by the visualization of the perlecan monomer by atomic force microscopy (see text).
The HSPG2 gene promoter that drives and modulates perlecan expression in vertebrates has been defined as the 2.7 kilobase (kb) long region lying immediately upstream of the start site of transcription (Iozzo et al., 1997). The proximal promoter region has the features of a CpG island promoter, lacking a TATA box. There are five apparent transcriptional start sites, and a short 5′ UTR consisting of 80 base pairs. Putative cis-responsive elements (CREs) were identified previously in the HSPG2 promoter (Iozzo et al., 1997). To better understand the evolutionary conservation of this promoter, we performed an in silico analysis using the PhastCons (Siepel et al., 2005) function of the UCSC genome browser, along with a series of complementary analyses using publically available programs. This analysis revealed evolutionary conservation in the noncoding region upstream of the HSPG2 gene amongst human, chimpanzee, rhesus monkey and mouse genomes, with much less conservation between the upstream regions of the HSPG2 gene in human and zebrafish genomes. The 20kb upstream region of the HSPG2 gene includes 78 putative cis-responsive elements (CREs) as identified by the presence of known conserved sequences that interact with known transcription factors. Nineteen of these conserved putative CREs reside in the proximal 2.7 kb. Upstream of this conserved region lie approximately three kb of non-conserved sequence, followed by the next highly conserved upstream region. This indicates that the majority of functional cis elements in the HSPG2 promoter likely are found within 2.7 kb upstream of the transcriptional start site. Although additional putative CREs can be identified in the promoter region, when investigating the human genome alone, we suggest that the nineteen conserved CREs proximal to the transcriptional start site of HSPG2 are the most likely to function in primary gene regulation to produce the primary perlecan transcript (Figure 2A).
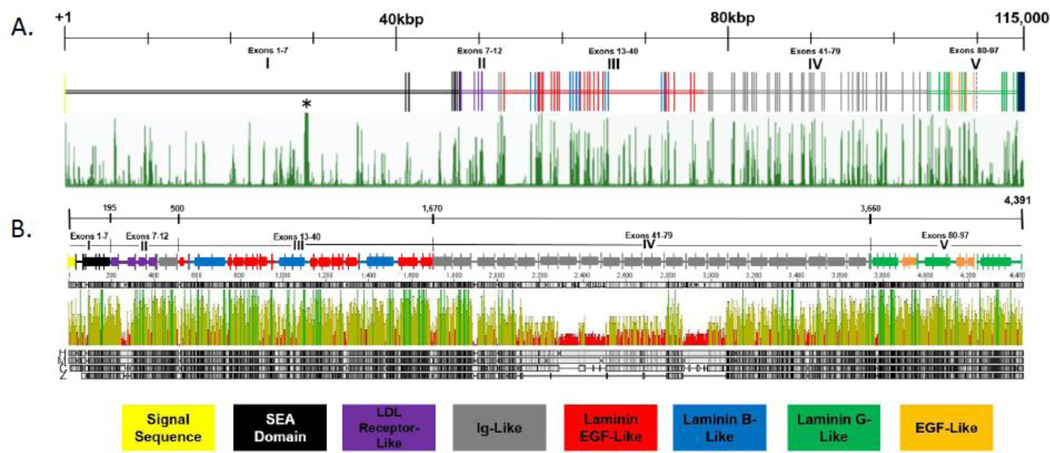
Figure 2. Conservation of the perlecan gene and protein.
(A) The perlecan gene is represented in schematic form. Vertical hashes represent coding exons, color coded to represent what type of folding module they encode. Numbering represents how many base pairs of the gene are covered; small hashes on this line represent 10,000 base pairs. Green vertical lines below the gene schematic represent conservation based upon the PhastCons statistical model, using the perlecan gene of 46 vertebrates as comparison (http://genome.ucsc.edu/). Asterisk notes the location of a pseudogene, RPL21P29, similar to ribosomal proteins. (B) The perlecan protein is represented in schematic form. Numbering on the top line represents amino acids at certain positions. Hashes on the second black line denotes where the arbitrary perlecan domains begin and end. Vertical hashes on the schematic represent exon-exon junctions, and rectangles represent independently folding modules, color coded as described at the bottom of the figure. Below the schematic is displayed conservation of the protein amongst four vertebrates, human, mouse, chicken and zebrafish (denoted by H, M, C, Z next to the grey bars). Green lines represent 4-way protein identity, yellow lines represent 2 or 3-way identity, and red lines indicate no match between species. The 4 grey lines at the bottom represent the protein sequences of perlecan from each species, and how well they match to the human protein. Note that the line in domain IV of the chicken and zebrafish proteins, unmarked by an attending rectangle, means there is in fact no protein at those locations (domain IV in zebrafish is much shorter than in humans).
Despite the complexity of the perlecan gene and the number of exons, the perlecan protein is surprisingly conserved (Figure 2B). This retention of protein structure throughout millions of years of evolution suggests an accompanying retention of function. In a previous article (Farach-Carson and Carson, 2007), we proposed a function for perlecan as an extracellular scaffold capable of modulating local intracellular signaling. In this review, we will extend that idea and further present a role for perlecan as a key molecule designed by nature to patrol her cell and tissue boundaries, where it can not only regulate local signals, but also help physically separate signals and confine them to individual cell and tissue compartments: hence, a role in “border patrol”.
2. Unique Modular Structure Creates Unique Functions
As described above, the perlecan core protein historically has been described in terms of five functional domains, each of which contains a group of distinct functional modules that can interact with a variety of other proteins and molecules present in the ECM (Table I). These modules, especially those nearer the C-terminus, arose early in evolution, suggesting a critical function that was needed early to establish the boundaries of functional tissues. We previously suggested that intact perlecan with all functional modules linked together could serve as an extracellular scaffold large enough to hold together several lipid rafts in the plasma membrane of cells exposed to the ECM (Farach-Carson and Carson, 2007). An analysis of the assembly of perlecan’s multi-modular structure in lower species compared to that in humans provides a unique way to examine the evolution of perlecan’s function in tissues of multicellular organisms.
Table I.
Perlecan Binding Factors Involved in Tissue Structure and Function
| Binding Partner | Binding domain involved |
Affinity (Kdapp) | Bioactivity | References |
|---|---|---|---|---|
| collagen type V alpha 4 (rat) |
HS | unreported | Schwann cell binding |
(Rothblum et al.,2004) |
| elastin | HS and protein core |
unreported | elastic fiber assembly |
(Hayes et al.,2011) |
| laminin | HS | 5 nM | BM structure and integrity |
(Ettner et al., 1998) |
| nephronectin (mouse) |
HS | unreported | development, BM assembly |
(Sato et al., 2013) |
| PRELP (mouse) | HS | 3 nM | connective tissue/BM integration |
(Bengtsson et al., 2002) |
| fibrillin-1 | II | 5–10 nM | connective tissue microfibril organization |
(Tiedemann et al., 2005) |
| WARP | III - second Laminin-EGF repeat |
23 nM | chondrocyte pericellular matrix |
(Allen et al., 2006) |
| fibronectin | unknown | 2 nM | pericellular matrix assembly |
(Heremans et al., 1990) |
| collagen IV | IV | 233 nM | basement membrane integrity |
(Hopf et al., 1999; Whitelock et al., 1999) |
| nidogen-1 | IV-1 | 10 nM | structural integrity of BM |
(Hopf et al., 1999) |
| nidogen-2 (human) | IV-1 (mouse) | 10 nM | structural integrity of BM |
(Hopf et al., 1999) |
| alpha-dystroglycan | V | 3 nM | epithelial cell polarity |
(Talts et al., 1999) |
| alpha-2 – beta-1 integrin |
V (mouse) | 80 nM | anti-angiogenic signaling |
(Bix et al., 2004) |
| alpha-5 - beta-1 integrin (mouse) |
V | 160 nM | cell adhesion, anti-angiogenic signaling |
(Lee et al., 2011; Nystrom et al., 2009) |
| ECM1 | V | unreported | basement membrane organization |
(Mongiat et al., 2003) |
| fibulin-2 (mouse) | V | 196 nM | BM regulation | (Brown et al., 1997) |
The perlecan protein is composed of a chain of modular motifs homologous to those of other ECM proteins (Murdoch et al., 1992; Noonan et al., 1991). This structural organization is conserved amongst vertebrates and invertebrates alike (Hummel et al., 2004; Noonan et al., 1991; Rogalski et al., 1993; Voigt et al., 2002). The domains of perlecan are arbitrarily divided based upon shared homology with other proteins, with human perlecan comprising five domains and perlecan in nematodes comprising only domains II, III, and IV (Rogalski et al., 1993). The nature of mammalian perlecan’s modular structure has been reviewed at length (Farach-Carson and Carson, 2007) Here we will describe the known and putative functions of each domain in perlecan, and the extent of knowledge concerning each domain’s contribution to the proposed barrier function of perlecan, with a focus on evolutionary insights.
2.1 Domains and their functions
At some point during the evolution of organisms with multiple cell layers, it appears that selective pressure forced perlecan’s organization as a single several-hundred kiloDalton chain of modular repeating motifs. Speculation that the protein serves as an ECM scaffold is borne out by the variety of binding partners for the protein core and heparan sulfate chains. Recent work has added molecular details to show how perlecan links the two separate protein networks of collagen IV and laminin which together constitute the basement membrane (Behrens et al., 2012). This work demonstrated binding of the laminin network by the perlecan core protein and binding of the collagen IV network by perlecan’s heparan sulfate chains, which corroborates previous studies (Brown et al., 1994; Tillet et al., 1994). Despite this, heparan sulfate-dependent binding of collagen IV has little bearing on perlecan’s function in early metazoans, as invertebrate perlecan molecules lack domain I and thus most of the heparan sulfate attachment sites (Hummel et al., 2004), although these species have the machinery to make heparan sulfate (Toyoda et al., 2000). Certain of perlecan’s interactions are crucial for maintaining tissue stability during development, and we will describe later in this article cases where loss of or deficiencies in perlecan protein can abrogate the barrier function of the molecule.
2.2 Domain I
Perlecan domain I consists of a module that shares some homology with the Sperm, Enterokinase and Agrin (SEA) fold of other ECM and cell surface proteins. The function that perlecan domain I plays in vertebrate tissue integrity and development cannot be overstated. A large part of this function is owed to perlecan’s glycosaminoglycan chains, typically a mixture of heparan sulfate and chondroitin sulfate chains usually with a majority of heparan sulfate, attached at three sites that immediately precede the SEA fold, the coal car in Figure 1 (Murdoch et al., 1992). Domain I is the only domain of perlecan unique to the molecule – modules of domains II through V have high sequence homology to modules present in other ECM proteins (Murdoch et al., 1992). These three glycosaminoglycan attachment sites share the consensus site S-G-D in mammalian perlecan. In Gallus gallus perlecan the third site is conserved, while the first two are modified to S-A-D and S-G-E, respectively. In Danio rerio, the SEA fold of domain I is conserved, but there exists no homology of the glycosaminoglycan attachment sites observed in higher vertebrates. Domain I is not present in Drosophila or C. elegans perlecan and appears to have been linked to the perlecan core protein later in evolution (Hummel et al., 2004).
2.3 Domain II
Human perlecan domain II consists of four repeats with homology to low-density lipoprotein (LDL) receptor and one folding module with homology to the immunoglobulin (Ig) V-set domain. This is the only isolated Ig module in the perlecan core protein. Each of these folding modules shares greater than 80% pair-wise identity with other mammalian perlecan proteins. Perlecan domain II in Gallus gallus and Danio rerio is structured similarly to mammalian domain II, and their folding modules share 50 to 75 percent homology with human perlecan. In Drosophila perlecan, or trol, domain II diverges from the vertebrate molecule by containing twenty one LDL-receptor domains, and one additional I-set Ig repeat. The final two hundred and fifteen amino acids of Drosophila perlecan domain II share 43% pair-wise identity with human perlecan and the folding modules in this region are spaced very similarly to those in human perlecan. C. elegans perlecan, or Unc-52, domain II, is similar to that of Drosophila, contains two Ig folds, but in contrast to Drosophila and similar to vertebrate perlecan, contains only three LDL-receptor-like folds. Unc-52 type III mutant occurs via transposon element insertion into the first Ig repeat of domain II, resulting in creation of a premature stop codon (Rogalski et al., 1995).
2.4 Domain III
Human perlecan domain III encompasses an interesting stretch of the protein, containing three laminin B domains that break up a longer stretch of eleven laminin EGF-like repeats. An interesting structural feature of the laminin B domains is that their insertions split three of the laminin EGF-like repeats, hinting that they were inserted sometime during evolution into what was likely a contiguous stretch of EGF-repeats. This is easily seen by looking at the blue and red cars in Figure 1. Also note that one of the laminin EGF-like repeats is shortened (the 4th). All vertebrates including zebrafish share this splitting of the laminin EGF-like folding modules, and it appears to be the same in Drosophila perlecan. In C. elegans perlecan, these “split” laminin EGF-like modules also exist, but as described below, C. elegans domain III is shortened by one third. The similarities between human domain III and that of other vertebrates mirror that of domain II; domain III in mammals shares roughly 90% protein identity whereas Gallus gallus and Danio rerio share roughly 60% protein sequence identity. Overall domain III protein identity with invertebrates is lower, 32% in Drosophila and 36.8% in C. elegans. While protein sequence identity within folding modules can be as high as 50%, the Drosophila domain III appears to encode one less laminin-EGF repeat than human perlecan at the C-terminal, and C. elegans domain III is absent the 200 amino acid C-terminal stretch containing a laminin B module and several laminin EGF-like repeats. This variability no doubt provides clues to the complex evolution of this domain over the hundreds of millions of years that perlecan has existed in the metazoan genome.
Interestingly, in Unc-52, the homologue of perlecan in C. elegans, a type II mutation (st549) occurs in the second laminin B repeat of domain III – resulting in a premature stop codon. This mutation creates a terminal paralyzed arrest at embryonic two cell stage (PAT) phenotype characterized by immobility and arrested elongation at the two-fold stage of embryonic development (Rogalski et al., 1995), a phenotype attributed to incomplete development of body-wall muscle cells.
2.5 Domain IV
Human perlecan domain IV is the most homogeneous region of perlecan, and consists of twenty one tandem Ig domain repeats, numbered as repeats 2–22 in human perlecan. Many mammalian perlecan genes encode 21 Ig modules in perlecan domain domain IV, but mouse perlecan includes only fourteen repeats (Hopf et al., 1999), a result of splicing. A mouse splice variant form that added human Ig module-equivalents 8–10 has been reported (Noonan and Hassell, 1993). The number and composition of the Ig repeats in domain IV also varies throughout evolution. Gallus gallus perlecan contains eighteen Ig repeats, Danio rerio perlecan contains twelve, C. elegans perlecan contains fourteen, and Drosophila contains only ten. Protein identity for domain IV amongst mammalian species is near 90%,, but in Gallus gallus protein identity is reduced to close to 60%, and among Danio rerio and invertebrate perlecan there is less than 20% protein amino acid sequence identity when compared to human perlecan. All protein alignments and analytics were performed using the sequence analysis software package Geneious (Auckland, New Zealand) and GenBank reference sequences.
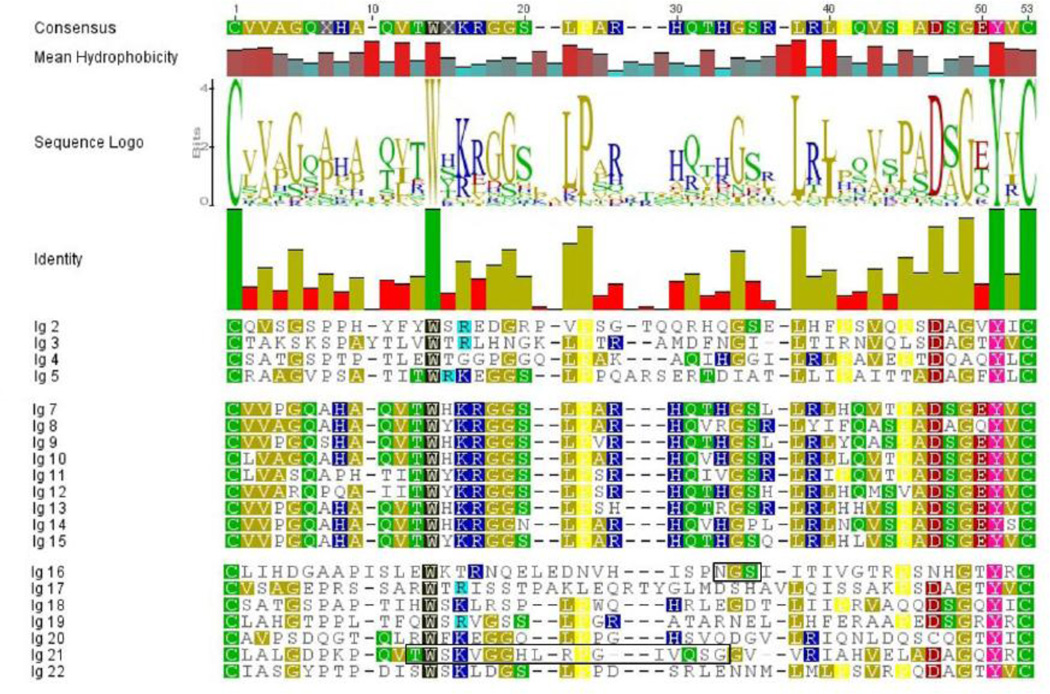
A colorized analysis of the individual sequences of the modules contained within the disulfide bonded regions of each the Ig repeats of human domain IV is shown in Figure 3. The two bonded cysteines are shown in green. All repeats contain a common tyrosine two amino acids from the C-terminal cysteine, shown in pink, and a common invariant tryptophan about a third of the way into the sequence shown in black. Proline (yellow) also is a common feature, with minor variation seen. From visual inspection, it is immediately apparent that two patterns appear: repeats 2–5 and 16–22 characterized by a more variable pattern for the Igs nearer the domain termini, and a tighter and more conserved pattern within the central repeats 6–15. All of these central repeats contain a doublet of basic amino acids (dark blue) followed by a glycine couplet (gold). It is intriguing to speculate that two common ancestral Ig folding patterns account for this structure. The blocked sequence in repeat 21 is the sequence of the adhesive peptide reported previously (Farach-Carson et al., 2008b).
Figure 3. Colorized analysis of Ig-modules in domain IV showing two clear patterns.
The amino acid sequences from the human perlecan core protein that lie in between the cysteines involved in disulfide bond formation of the Ig-modules in domain IV (green) were extracted, aligned, and colorized by polarity of amino acid (green – polar, gold – nonpolar, blue – basic, red – acidic). Highly conserved residues are given unique coloring (tryptophan – black, proline – yellow, tyrosine – pink). The terminal module sets (2–5) and (16–22) are similar to one another, and are split by (6–15) showing a distinct pattern (see text). The consensus sequence represents the modal amino acid at that position. Mean hydrophobicity is a representation of the overall hydrophobic nature of the amino acids at that position in all 21 repeats (higher, redder bars represent more hydrophobic residues at that position). The sequence logo readout indicates what amino acids are present at that position, and in what proportions. Finally the height of the bars in the sequence identity graph indicates the degree of identity at the position in these 21 sequences.
Unc-52 mutations e1421, e1012, e669, e444, e998, st196 occur in domain IV (Rogalski et al., 1995), in exons that can be spliced out of alternative transcripts, rescuing the worm phenotype. This suggests that mutations in the Ig repeat domains can act as dominant negatives, and that having fewer repeats and a shorter domain IV is better than having misfolded proteins with regard to tissue structure and function. It is intriguing to speculate that the length of the Ig repeat-rich region thus plays another function, such as specification of tissue stiffness or elasticity at the tissue border.
2.6 Domain V
Human perlecan domain V contains three folding modules with homology to the laminin G chain, separated by two modules that share homology to EGF-like protein domains. Protein identity with vertebrate orthologs mirrors that of domain III, but in Drosophila and C. elegans, protein identity of folding modules is between fifteen and thirty-five percent. The C-terminal EGF-like fold is missing from C. elegans perlecan.
In mammals, the functions of this portion of the perlecan protein are associated with modulation of blood vessel growth and structural integrity (Mongiat et al., 2003). Ongoing work in several labs focuses on dissecting the complex structural functional relationships of the motifs in domain V that provide angiogenic control. Like a border patrol agent, endorepellin, one of the active fragments created by proteolytic cleavage of perlecan domain V (Gonzalez et al., 2005), was reported to inhibit four aspects of angiogenesis: 1) endothelial cell migration; 2) collagen-induced endothelial tube morphogenesis; 3) blood vessel growth in the chorioallantoic membrane and; 4) angiogenesis in Matrigel™ plug assays (Mongiat et al., 2003). Further work by this group showed that endorepellin requires both the α2β1 integrin and VEGFR2 to demonstrate its angiostatic activity (Goyal et al., 2011). In brain, perlecan domain V (DV) promotes brain angiogenesis by triggering release of VEGF from brain endothelial cells as shown in an in vivo stroke model (Clarke et al., 2012). Interestingly, in this same study, the LG3 portion of DV, which is reported to possess the majority of DV's “angio-modulatory” activity outside of brain, binds poorly to α5β1 and induces less endothelial cell proliferation than does complete domain V (Clarke et al., 2012). This combined work provides experimental strength to the notion that perlecan can act as a context driven “switch” that can, by virtue of local proteolytic processing, patrol borders where endothelial growth and tissue penetration is under strict regulation.
3. Enzymatic Cleavage Releases Latent Activities
It is interesting to speculate that nature hitched together the perlecan modular train progressively during evolution to create a unique multi-functional protein capable of serving as a large, quiet barrier in resting tissues whose potential latent activities could be unleashed immediately during tissue activation. An obvious advantage of this design would be the immediate readiness for action of perlecan’s many latent functions upon enzymatic activation, which would be available in seconds rather than the hours it would take if the individual functions had to be synthesized and then released in response to an activating stimulus such as a tissue wound or ischemic stroke. Additionally, these functions could be released coordinately in the ECM rather than through a complex series of signaling events and associated changes in gene expression involving many nuclear transcription factors. Finally, if indeed perlecan scaffolds signaling complexes in resting tissues and their unperturbed matrices, then proteolytic cleavage might “unraft” these signaling complexes, allowing them to diffuse through the plasma membrane to find productive binding partners for signal initiation. An idea of this nature would intimately link perlecan’s domain structure and the evolution of its structure as a long monomeric protein present in the ECM, and further place it as a key player in innate host immunity. In essence, perlecan would provide a tissue “ECM patrol” to accompany the “cellular patrol” of innate immune systems.
To follow this line of thinking, consider that wholesale catabolic turnover of human perlecan by a combination of proteases and glycosaminoglycanases would immediately release a diffusable trainload of cargo of heparan sulfate bound factors and all of the potential protein binding interactions contained in the 48 modules of the perlecan core protein. Many heparan sulfate binding factors associate with perlecan’s glycosaminoglycan (GAG) chains, and many of these factors are involved in tissue development and wound healing (Table 1). It seems possible that this feature of perlecan evolved as a normal mechanism to create and border areas of new tissue formation. For this reason, perlecan’s many structural features can be exploited in tissue engineering of new tissues, a concept discussed further later in this article.
Table II shows a representative list of molecules whose binding to perlecan, either to the core protein or to the HS chains, would add them to the depot made available for immediately delivery to a wound or site of tissue remodeling or turnover. In addition, the enzymatic degradation of the ECM in such sites also would release potentially bioactive fragments from the ECM core proteins themselves, as has been shown for collagen XVIII (angiostatin), perlecan (endorepellin, DMV, LG3), and laminin (Gately et al., 1997; Gonzalez et al., 2005; Mann et al., 1988). This active turnover can create a positive feed forward loop in which the fragments can recruit patrolling white blood cells that release locally active cytokines, which in turn up-regulate ECM, which is first a boundary and then can be further turned over until the wound is healed. For normal tissues, balanced tissue homeostasis is eventually restored. In some cases, an overshoot of repair can lead to fibrosis or scar (Czubryt, 2012). In the case of solid cancers, the “wound that never heals”, continuous remodeling and inflammation around the expanding tumor produces an unending tissue repair cycle that the cancer cells hijack to support their own growth. An example of this is the activation of the transcription factor, SMAD3, that activates the perlecan promoter in colon cancer cells (Iozzo et al., 1997). Treatment with interferon-gamma can inactivate the production of perlecan at the promoter level as demonstrated in reporter assays (Sharma and Iozzo, 1998). Other ongoing studies in our laboratory show up-regulation of perlecan by inflammatory cytokines in the reactive stroma of prostate cancer cells (Warren and Farach-Carson, in preparation).
Table II.
Bound Active Molecules that can be Released by Perlecan Turnover
| Binding Partner | Binding Domain |
Affinity | Bioactivity | References |
|---|---|---|---|---|
| acetylcholinesterase | HS (avian) | unreported | NMJ function | (Peng et al., 1999) |
| activin A | HS | unreported | tissue growth and repair | (Li et al., 2010) |
| BMP-1 | HS | 573 pM | osteogenesis | (Decarlo et al., 2012) |
| BMP-2 | HS | 600 pM | osteogenesis/ chondrogenesis |
(Decarlo et al., 2012) |
| FGF-1 | HS | unreported | tissue growth and repair | (Melrose et al., 2006) |
| FGF-2 | HS | 0.6–2 nM; 12 nM | tissue growth and repair | (Chu et al., 2005; Knox et al., 2002) |
| FGF-9 | HS | unreported | tissue growth and repair | (Melrose et al., 2006) |
| HB-EGF | HS | 44 nM; 6.1 nM | tissue growth and repair | (Chu et al., 2005) |
| histone H1 | HS | unreported | tissue growth and repair | (Henriquez et al., 2002) |
| IL-2 (mouse) | HS | unreported | inflammation; immune surveillance |
(Miller et al., 2008) |
| LTBP-2 | HS | unreported | release of TGFβ | (Parsi et al., 2010) |
| Thrombospondin-1 | HS | unreported | tissue remodeling and repair |
(Vischer et al., 1997) |
| VEGF165 | HS | “high” | tissue growth /angiogenesis |
(Zoeller et al., 2009) |
| WNTsa/Wg | HS | 0.01–0.1 nM | tissue development and growth |
(Lin, 2004; Reichsman et al., 1996) |
| Hedgehogs (Hhs)a | HS/Core Protein |
“high” | tissue development and growth |
(Datta et al., 2006; Lin, 2004; Park et al., 2003) |
| TGF beta | HS/Core Protein |
low nM | tissue growth and repair | (Cheifetz and Massague, 1989; Harrison et al., 2011) |
| LDL & VLDL | II | “high” | cholesterol and lipid delivery |
(Hummel et al., 2004) |
| FGF-7 | III | 60 nM | tissue growth and repair | (Mongiat et al., 2000) |
| FGF-18 | III | 27.8 nM | tissue growth/chondrogenesis | (Smith et al., 2007) |
| FGF-bp | III - second laminin-EGF repeat |
18 nM | release of FGF | (Mongiat et al., 2001) |
| PDGF | III - second laminin-EGF repeat |
8–25 nM | tissue growth /angiogenesis |
(Gohring et al., 1998) |
| FGF-7 | V | 60 nM | tissue growth and repair | (Mongiat et al., 2000) |
| myostatin (mouse) | V (mouse) | 11 nM | regulation of muscle mass | (Sengle et al., 2011) |
| Progranulin | V | 1 µM | blood vessel growth | (Gonzalez et al., 2003) |
| VEGFR1 | V | 10.6 nM | VEGF signaling; angiogenesis |
(Goyal et al., 2011) |
| VEGFR2 | V | 2.8 nM | VEGF signaling; angiogenesis |
(Goyal et al., 2011) |
Extrapolated from genetic and diffusion data.
4. Concentration at Tissue Borders in Complex Tissues of Vertebrates
When examining the distribution of perlecan in vertebrate tissues, it quickly becomes obvious that perlecan is concentrated in regions that separate tissues and tissue functions. Its presence at these cell and tissue borders inspired the title of this article, and lead us to carefully examine the tissue functions taking place on each side of a number of these perlecan-rich borders. We asked the question, exactly what activities are perlecan-rich matrices separating? A follow up question clearly is how does its structure allow it to do this? To illustrate these concepts, perlecan functions in six distinct types of normal tissue where it is required for normal tissue formation were considered. In some of these, such as the vasculature and the epithelium, perlecan is found in a basement membrane or basal lamina in association with collagen IV, laminin, nidogen/entactin, and other components of the basement membrane (Timpl et al., 1984). In others such as muscle or bone, perlecan is part of the territorial matrix where it binds to other ECM proteins in tissue specific multi-protein complexes.
4.1 Muscle
Skeletal muscle units are surrounded by a territorial membrane that is rich in perlecan. A mouse model in which levels of perlecan were reduced in skeletal muscle showed muscle hypertrophy, a decrease in myostatin expression, along with alterations in muscle fiber composition (Xu et al., 2010). By studying responses to mechanical overload in control and perlecan deficient mice, these authors concluded that perlecan is needed to maintain fast muscle mass and fiber composition, as well as regulating signaling through myostatin. These observations are completely consistent with a role for perlecan in patrolling and maintaining the borders of skeletal muscle units. Likewise, it has been proposed that perlecan is critical for maintaining the integrity of the heart during cardiac development and further that perlecan supports heart function in the adult heart after injury (Sasse et al., 2008). Complete ablation of perlecan expression is lethal in the mouse, with widespread cardiac defects (Arikawa-Hirasawa et al., 2001a; Costell et al., 1999; Sasse et al., 2008). In the hearts of normal mice, perlecan resides in the basal surface of both myocardium and endocardium, as well as that of the surrounding presumptive neural crest cells (Costell et al., 2002). In one of the most striking examples of the border patrol exerted by perlecan in the developing heart, the work in this same report observed an excess of mesenchyme or “mesenchymal overpopulation” that was attributed to uncontrolled migration of neural crest cells in the absence of perlecan, which would arrive prematurely to the heart and disrupt the formation of the outflow tract. Based on these observations, the authors concluded that perlecan is a key control of the mesenchymal population size and placement in the heart. In developing human heart tissues, perlecan first was found in the heart enlage, then in the basement membranes of the endo- and pericardium, as well as capillaries, through the first twelve weeks of growth (Roediger et al., 2009).
4.2 Bone and Cartilage
In mineralized bone itself, there is very little, if any, perlecan present, as its heparan sulfate chains would be expected to inhibit mineralization (Grant et al., 1989; Thompson et al., 2011). Rather perlecan is found in regions of bone that separate mineralized and non-mineralized compartments at the chondro-osseous junction (Figure 4). In developing long bones, perlecan is found in a tight peri-chondreal band of active growth where bone is both lengthening and widening, and is co-localized with the enzyme heparanase (Brown et al., 2008) (Figure 4). Perlecan is rich in bone vasculature, in bone marrow stroma (Chen et al., 2007) (Figure 5), and in the lacunar-canilicular system (LCS) where it is a key component of the tethering system for osteocytes (Thompson et al., 2011). In fact, the LCS system is compromised in perlecan-deficient mice (Thompson et al., 2011).
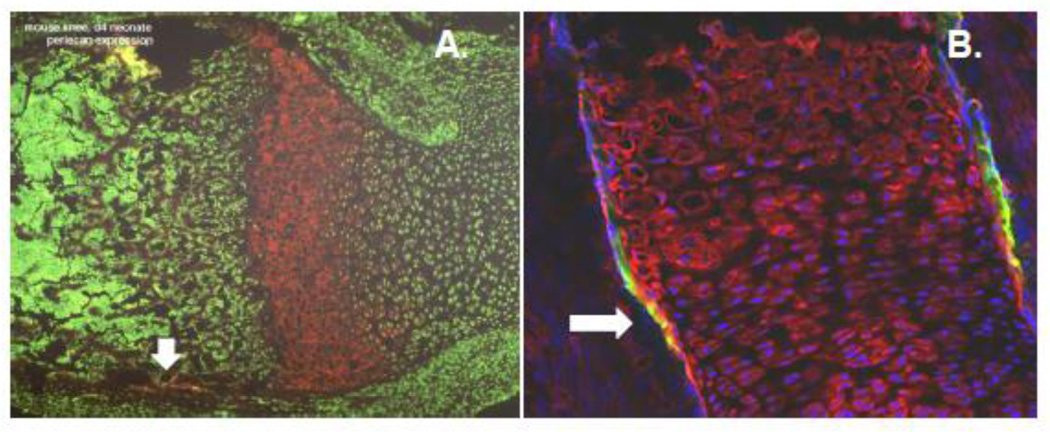
Figure 4. Perlecan expression in the growth plate borders of long bones.
Perlecan (red in both panels) patrols the border at the chondro-osseous junction that separates vascularized bone marrow from avascular cartilage. Panel A shows perlecan concentrated in the hypertrophic zone of the growth plate of a day 4 neonatal mouse. The spicules of mineralized bone are seen to the left (brown), and growing cartilage to the right, seen by nuclear stain (green). Panel B shows strong perlecan staining (red) on embryonic day 16.5 in avascular cartilage in both pre-hypertrophic and hypertrophic (top) regions. Note the strong yellow signal (arrows) that represents a ring of staining in the peri-chondrium that co-localizes with the enzyme, heparanase (green). This is an area where the bone is actively widening. Blue is nuclear staining.
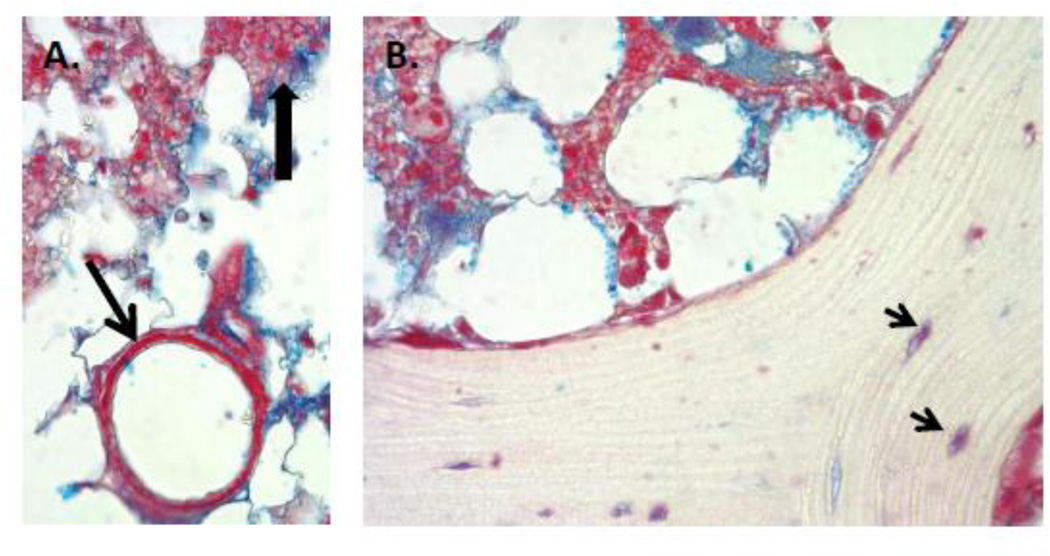
Figure 5. Perlecan staining in human bone, bone marrow and basement membrane of bone marrow endothelial cells.
A. Perlecan staining (red) is clearly seen in bone marrow, particularly in some cells (dark arrowhead), and intense in the basement membrane surrounding bone marrow capillaries (arrow). Blue is GAG staining with Alcian Blue. Any cells entering or leaving the circulation would need to traverse the perlecan rich border underlying the capillary endothelial cells. B. Perlecan staining is seen at the endosteal border, surrounding osteocytes in dense bone (arrows), and in the reticular network supporting hematopoiesis in bone marrow, but not in mineralized bone. (Images courtesy George Dodge).
Interestingly, in fractured bone, perlecan was found to be one of the earliest new genes to be expressed in the new fracture callous (Wang et al., 2006), suggesting it plays a key role in bone healing. In this instance, perlecan is likely to appear both to re-establish tissue boundaries, and also to provide a depot for the timed delivery of growth factors needed for new bone formation. Additionally, perlecan domain I has been used as a vehicle for delivery of BMP2 for healing of early lesions in an induced model of osteoarthritis or in bone repair (Decarlo et al., 2012; Srinivasan et al., 2012).
4.3. Vasculature
Perlecan is a key component of the endothelial BM, where it also helps connect the pericytes to the surrounding connective tissue matrix to stabilize the vasculature (Gustafsson et al., 2013; Stratman and Davis, 2012). In this capacity, perlecan serves as a barrier between the contents of circulating blood and the surrounding tissue (Figure 5). An excellent review of perlecan’s role in angiogenesis and the developing vasculature, including the control of bioactive angiogenic factors has been published (Jiang and Couchman, 2003). For white blood cells to reach sites of a wound or inflammation, the perlecan barrier must be crossed during extravasation. Likewise, metastatic cancer cells cross a perlecan-rich barrier during both intravasation and extravasation. To do this, they express both glycosidases and proteases (Heppner et al., 1996; Walch and Marchetti, 1999; Whitelock et al., 1996) that allow them to degrade the core protein and the associated HS chains. For metastases to brain, the perlecan layer that serves as part of the blood-brain barrier in the vascular basement membrane must be breached (Roberts et al., 2012). Likewise, drugs and nanodelivery systems designed to reach the brain must be made to consider the properties of the perlecan barrier surrounding the tight endothelium in the brain.
4.4 Glandular Epithelium
In glandular epithelia, a secretory epithelium is separated from an underlying connective tissue stroma containing vasculature and nerve. Regulation of transport and secretion of both fluid and solid components of exudate into proper ductwork requires the presence of a highly polarized epithelium that knows “top from bottom”, otherwise cysts are formed. Perlecan plays a key role in this highly ordered structure by its presence in the compact basement membrane underlying the epithelium, where along with collagen IV and laminin, it supports epithelial polarity (Schneider et al., 2006; Yurchenco and Patton, 2009). Along with this, however, it is worth noting that the presence of perlecan acts as a border beneath the epithelium that can restrict nerve and capillary growth to the connective tissue layer. There are several ways this can be accomplished. First, acting as a physical barrier, the long interwoven meshwork containing perlecan can act like a fence through which it is difficult for branching vessels to penetrate. Thus, they tend to spread out underneath the basement membrane sheath rather than push through. Second, the release of the anti-angiogenic C-terminal fragments of perlecan through limited proteolysis can provide a second retardant to vessel ingrowth into secretory epithelia (Goyal et al., 2011). Figure 6 shows a stained section of salivary gland and a cartoon of a single secretory acinus. Note the presence of perlecan (green) as a defining feature of each acinar border separating it from the connective tissue underneath. Similar tissue concentrations of perlecan are seen in breast, prostate, pancreas, uterus, skin and many other organs with glands.
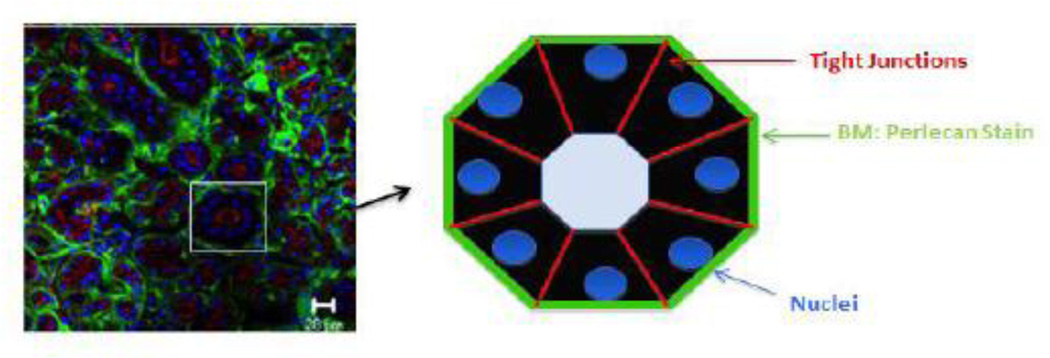
Figure 6. Perlecan expression in the human salivary gland basement membrane.
Primary tissue from a freshly excised patient specimen was stained with an antibody recognizing perlecan (green) or the tight junction marker, ZO-1 (red). The nuclei are shown in blue. The cartoon illustrates the concept that perlecan in the basement membrane separates and surrounds each structural unit, separating structures from stroma and each other, forming discrete borders around each secretory unit. (Figure courtesy Dr. Swati Pradhan-Bhatt).
4.5 Brain, Nervous Tissue, Eye
As a component of the basal lamina underlying the developing neuroendothelium, perlecan helps maintain the structure of the separate brain layers, and also provides a depot of growth factors needed to support key aspects of brain tissue morphogenesis. The spatiotemporal distribution of perlecan transcript and perlecan protein has been studied in the brain and nervous systems of several developing organisms as well as in adults, with a common theme of expression and structure stabilization at key borders and neuromuscular junctions. In mice, the developing central nervous system (CNS) was reported to be largely devoid of perlecan with the exception of the choroid plexus (Handler et al., 1997). In the rat pineal gland, two basal laminae appear to be separated by a perlecan-rich barrier (Bagyura et al., 2010). In Drosophila, trol transcripts and protein appear in the CNS around stage 15 (Friedrich et al., 2000). Studies have suggested that the appearance of perlecan around this stage increases the proliferation of neuroblasts (Datta, 1995). In the peripheral nervous system (PNS), perlecan is concentrated in the basement membranes separating Schwann cells from tissue, and is concentrated in the regions surrounding the nodes of Ranvier (Bangratz et al., 2012). In vitro culture data suggests it can be made by the Schwann cells themselves (Chernousov et al., 1998). A role for perlecan in clustering and stabilizing acetylcholinesterase and alpha dystroglycan at postsynaptic membranes has been reported by several laboratories (Hohenester et al., 1999; Singhal and Martin, 2011; Steen and Froehner, 2003). Studies by Roediger et al (Roediger et al., 2009), using human embryonic tissues, showed perlecan core expression, specifically domain III, in the endoneurium of peripheral ganglia and the basement membranes of neural capillaries from growth weeks 6–12. Neuronal cells themselves were negative for domain III and had little, if any, staining for domain V.
In the eye, perlecan is deposited into the basement membrane of the corneal epithelium (Inomata et al., 2012; Kabosova et al., 2007), where immunohistological analysis of 8 week old mice has shown it clearly separates the corneal epithelium from the underlying stroma. It also is expressed in the basement membrane of the retina (Marneros et al., 2004), where it is commonly used to demarcate the inner limiting membrane or ILM (Keenan et al., 2012). In the lens, the presence of fully glycosaminoglycan substituted perlecan is critical for the development of the capsule, which itself functions as a barrier to insulate the lens from its environment (Rossi et al., 2003). Thus, in multiple structures of the eye, perlecan’s presence delineates the regions separating critical structures and functions, and in some cases is thought to actually determine the critical sizes of these compartments.
4.6 Decidua and Embryo Implantation
In mammalian reproduction, two major structures develop that lead to segregation of the developing embryo from maternal tissues. The deciduum is a maternal tissue that forms very early in the embryo implantation process. It is composed of differentiated stromal cells of the uterus and includes changes in the expression of many genes and gene products, including those of the ECM (Giudice, 2004; Ruiz-Alonso et al., 2012). Perlecan is not expressed by undifferentiated cells of the uterine stroma, but its expression increases rapidly and robustly during decidua formation in both mice and humans (Aplin et al., 1988; Chen et al., 2008; D'Souza et al., 2007; Julian et al., 2001). At least in mice, perlecan appears on the external surface of the blastocyst at the time the blastocyst becomes attachment competent and some evidence indicates that heparan sulfate proteoglycans promote embryo attachment (Carson et al., 1993; Farach et al., 1987, 1988; Smith et al., 1997). Nonetheless, perlecan null embryos do not appear to have implantation defects suggesting that perlecan is not absolutely required in the implantation process (Costell et al., 1999), a unique border where tissues from two separate organisms form a stable boundary. Decidual differentiation occurs to some extent in all mammalian species, but is not as robust in domestic species (sheep, ruminants) in which the placentation occurs in the uterine lumen via superficial attachment to the surface endometrium (Igwebuike, 2009). In rodents and humans placentation is an invasive process, occurring deep in the endometrium (Cha et al., 2012). In these species, perlecan expression is rapidly induced following embryo attachment in stroma subjacent to the embryo attachment site and then radiates from the site into the deep stroma (Aplin et al., 1988; Chen et al., 2008; D'Souza et al., 2007; Julian et al., 2001). Other basal lamina components including laminin and type IV collagen also are induced, although organized basal lamina are not morphologically apparent (Aplin et al., 1988; Wewer et al., 1985). Perlecan also is expressed robustly by the placenta particularly at the points of attachment to maternal tissue (Rohde et al., 1998). In both decidua and placenta, this perlecan-rich matrix forms prior to vascularization and compartmentalizes the developing embryo and placenta from maternal tissue, maintaining the border for the duration of the pregnancy. Nutrient, gas and ion exchange occurs between maternal and fetal/placental tissues and, as is the case in kidney glomeruli, perlecan is likely to play a role as a filter (Rosenzweig and Kanwar, 1982; van den Born et al., 1992). Moreover, decidua and placenta are rich reservoirs of heparin-binding growth factors (Cha et al., 2012; Smith, 2000; Sonderegger et al., 2010). Thus, in addition to providing a barrier, perlecan also likely acts to filter and control delivery of ions and small molecules as well as proteins at the fetal-maternal interface.
5. Loss of Perlecan at Tissue Borders Associated with Pathology in Vertebrates
Given the clear role that perlecan plays in preserving functional borders in vertebrate tissues, what are the consequences of loss or reduction of perlecan levels on tissue integrity? What happens when the border is not being properly patrolled? A variety of mouse, Drosophila, and C. elegans mutants including mice null for perlecan and perlecan hypomorphs provide the answers to some of these questions.
5.1 Schwartz-Jampel Syndrome/ DSSH
Complete absence of functional secreted perlecan is lethal both in mice (Costell et al., 1999) and in humans (Arikawa-Hirasawa et al., 2001a; Arikawa-Hirasawa et al., 2001b), where it is associated with the syndrome Dyssegmental dysplasia, Silverman-Handmaker type (DSSH) (OMIM entry 224410), an autosomal recessive condition in which truncated perlecan is retained intracellularly (Arikawa-Hirasawa et al., 2001a). A similar phenotype could occur where mutated forms of perlecan, acting as a co-aggregating dominant negative, prevent secretion of normal perlecan creating a functional absence of the proteoglycan (Arikawa-Hirasawa et al., 2001a). In contrast, more minor reductions in perlecan in tissues lead to a syndrome called Schwartz-Jampel (SJS) (OMIM entry 255800), which can range from very mild to quite severe. SJS, like DSSH, is thought to be an autosomal recessive trait, but cases of dominant inheritance also have been reported, likely owing to dominant negative actions of mutant proteins (Pascuzzi et al., 1990).
The most affected tissues are muscle, cartilage and bone, not surprisingly as these are the tissues where perlecan expression is highest. In addition to the disorganization of the growth plate and chondro-osseous junction of bones in the absence of perlecan, there are other distortions in the growth of joints including the temporo-mandibular joint (Stephen and Beighton, 2002), myotonia and electrical disturbances of muscle (Echaniz-Laguna et al., 2009) and modifications to the structures of facial bones (Pinto-Escalante et al., 1997). The common aspect of these features is that they can be attributed to an impairment of the boundary functions of perlecan, allowing tissues to form without clear structural or functional delineations.
The loss of perlecan in the peripheral nervous system, particularly that occurring in the basement membrane of Schwann cells and in neuromuscular junctions, accounts for peripheral nerve hyperexcitability (PNH) associated with SJS. A mouse model of SJS showed Schwann cell dysfunction and neuromuscular junctional changes that were associated with PNH and axonal depolarization (Bangratz et al., 2012).
5.2 Osteoporosis/osteoarthritis
In skeletal tissues, loss of perlecan in bone and cartilage can have devastating consequences. In humans with SJS, the musculoskeletal consequences are among the most severe (Arikawa-Hirasawa et al., 2002). Perlecan loss is associated with loss of normal compartmental structures in bone and cartilage (Costell et al., 1999), and patients with SJS are prone to bone and cartilage loss (Stum et al., 2005; Stum et al., 2006). Changes in perlecan expression have been implicated in the late stages of osteoarthritis in the human knee joint (Tesche and Miosge, 2004). Mice containing functional interruption and mutations in the Hspg2 gene to model SJS showed impaired mineralization, misshapen bones, flat face, and joint dysplasias similar to those seen in osteoarthritis (Rodgers et al., 2007).
In bone, cells called osteocytes project long processes throughout the mineralized bone matrix, forming an interactive living network that coordinates and communicates signals throughout the tissue. This network lies within a network (LCN) that is vital to fluid movement, nutrient and waste exchange, as well as passage of signaling molecules (Knothe Tate, 2003). Perlecan plays a key role in maintaining the boundary surrounding osteocytic processes including the integrity of the LCN (Thompson et al., 2011). Ultrastructural measurements of images obtained using electron microscopy of perlecan/Hspg2-deficient mice showed a diminished pericellular area in the LCN compared to wild type littermates with normal perlecan levels (Thompson et al., 2011). This loss of osteocyte function and communication can lead to bone loss, osteopenia, and eventually osteoporosis (Bonewald, 2004). Hspg2 is among those genes listed as those related to risk of osteoporosis (Valdés-Flores et al., 2013).
5.3 Cardiac and vasculature malfunction; stroke
In normal vascular tissues, perlecan present in basement membranes of blood vessels stabilizes their structures while anchoring them with other ECM molecules to the surrounding connective or neural tissue. Recent work showed that during ischemic stroke in the CNS, rapid processing of perlecan to release domain V is both neuroprotective and pro-angiogenic (Lee et al., 2011). This occurs without changes in permeability of the blood-brain barrier, suggesting the tissue barrier function of the perlecan core protein lies in modules N-terminal to domain V, likely in domains II-IV that also are present in invertebrates with distinct body compartments but lacking domain I. Consistent with this notion, further degradation of perlecan in endothelial basement membranes leads to compromise of the blood-brain barrier (Kahle et al., 2012; Roberts et al., 2012). During arterial injury, perlecan turnover again plays a role in tissue recovery. Uncompromised perlecan in normal arterial tissue maintains separation from smooth muscle layers, and is a potent inhibitor of smooth muscle cell proliferation while storing growth factors needed for rapid vascular repair (Chen et al., 2001; Segev et al., 2004). After damage, perlecan fragments and bound pro-angiogenic fragments released from damaged ECM can support endothelial cell proliferation to restore the vascular structure and prevent hypoxic damage (Segev et al., 2004).
Perlecan also is critical for maintaining the stability of vital structures in the heart. Perlecan-null mice die en masse on embryonic day 10.5, primarily because of blood leakage into the peri-cardial cavity (Costell et al., 1999), a loss of a vital mechanostable tissue barrier that fails as the heart demand on tissue increases. More recent studies designed to unravel the cause of the leakage showed that basement membranes lacking perlecan deteriorate in the heart with accompanying loss of cell-cell attachment in the ventricle and outflow tract (Sasse et al., 2008).
5.4 Brain and nervous tissue; exencephaly, eye and other tissue failures
In mice lacking perlecan both defects in the border in the brain and the heart appear to contribute to lethality. In a majority (55%) of mice lacking perlecan, disruptions in the basal lamina contribute to a condition of exencephaly; whereas the remainder tends towards micocephaly (Giros et al., 2007). Many mice with cardiac leakage also have exencephaly. Interestingly, cell proliferation decays in the absence of perlecan in those animals that do not become exencephalic (Giros et al., 2007). Because mice and humans with disrupted EXT1, an enzyme involved in heparan sulfate biosynthesis, also show a small cerebral cortex (Inatani et al., 2003; Narvid et al., 2009), it is tempting to speculate that HS contributes to key growth factor binding functions of perlecan domain I. With regard to border patrol functions in the brain, reductions in perlecan allow brain tissue to expand into the overlaying ectoderm, a breach that is reported to allow neuroepithelial expansion, neuronal ectopia, in addition to exencephaly (Costell et al., 1999).
Several features of the eye include thin, delicate epithelial structures supported on a basement membrane. As described above, several labs have demonstrated perlecan’s presence in the basement membrane of the normal corneal epithelium (Dietrich-Ntoukas et al., 2012; Schlötzer-Schrehardt et al., 2007), separating the epithelial and stromal layers, and in Descemet’s membrane, separating the stromal and endothelial layers. Its expression varies with age (Kabosova et al., 2007), and across various locations (Ljubimov et al., 1995), and is known to be expressed by corneal stromal cells, both normally and in response to injury (Brown et al., 1999; Hassell et al., 1992). Additionally, its influence on epithelial structure was confirmed recently, as perlecan-deficient mice were shown to have defects in the structure of this epithelial layer (Inomata et al., 2012). Thus, loss of perlecan is highly detrimental to the structures of the eye.
6. Evolutionary Origins Reveal Association with Establishment of Tissue function
6.1 Uncs
The “uncoordinated” or Unc mutants were first identified by Brenner in 1974 (Brenner, 1974), but Unc-52 was not attributed to a mutation in the C. elegans perlecan gene until 1993 (Rogalski et al., 1993). Loss of Unc-52 results in a PAT phenotype similar to that seen in the loss of nematode collagen IV genes, characterized by loss of embryonic extension concomitant with body wall muscle dysfunction (Gilchrist and Moerman, 1992; Gupta et al., 1997). Loss of functional Unc-52 protein is thought to disrupt formation of body wall muscle cell dense bodies, an analogue to focal adhesions (Waterston et al., 1980). These dense bodies detach from the plasma membrane with age or if pressure is applied to the animal (Waterston, 1988). In certain Unc-52 mutants, electron microscopy reveals that as the animals age, foreign cell types invade the muscle cell layer, which should be contiguous (Waterston et al., 1980). This is not unlike what is seen in the developing heart in the absence of perlecan (Costell et al., 1999; Sasse et al., 2008).
Additional effects of viable Unc-52 mutations include defects in gonad organogenesis. These defects can be suppressed independently of the body-wall muscle defects, indicating that these functions of Unc-52 are separate (Gilchrist and Moerman, 1992). Loss of Unc-52 regulation of growth factor signaling may be responsible for gonad dysgenesis in mutants (Merz et al., 2003). The Unc-52 M isoform is crucial for maintenance of wild-type gonadogenesis, loss of this specific isoform leads to aberrant growth factor signaling (Merz et al., 2003). The barrier function of Unc-52 is apparent in the gonadal sheath rupture phenotype of certain mutant strains. This phenotype includes destruction of the basement membrane surrounding the gonad, sometimes accompanied by “disappearance” of the sheath cells and leaking of oocytes into the body cavity (Gilchrist and Moerman, 1992). It is unclear what drives this effect on sheath cell integrity, but it has been speculated that it is mediated by a loss of adhesion signals to the cells from the Unc-52-depleted basement membrane. Unc-52 mutation or depletion has been tied to developmental defects in the pharynx (Jafari et al., 2010; Morck et al., 2010).
6.2 Trol mutants
Drosophila perlecan is referred to as terribly reduced optic lobes or trol. Mapped to the X chromosome, trol was identified in a genetic screen for mutations affecting imaginal central nervous system (CNS) development (Datta and Kankel, 1992). Certain mutant alleles of trol result in vacuolation of the optic lobe neurons and misguided development of axons in the optic lobe, which occur to varying degrees of severity. These phenotypes are tied to a loss of proliferation of the precursor optic lobe neuroblast cells during the transition from second to third instar larvae, and to a decrease in overall CNS cell proliferation. The functional consequence of this mutant is decreased performance in certain behavioral experiments that were created to test vision (Datta and Kankel, 1992). One allele of trol affects the integrity of the neuropil in the ventral ganglion of third instar larvae: neuropil in these animals is nearly overtaken by neuron cell bodies (Datta and Kankel, 1992). Once again, this is an example of compromised function of a critical barrier that allows cells to move into regions from which they are normally excluded.
Trol, in accord with the signaling role of perlecan in vertebrates, regulates signaling and diffusion of signals by the FGF and hedgehog molecules (Park et al., 2003). This interaction is not clearly understood, as in vertebrates it relies upon heparan sulfate binding to heparin-binding growth factors, and trol protein includes neither GAG attachment sites nor a homologue to the vertebrate domain I. Signaling transforming growth factor beta and WNT, or wingless, in the Drosophila brain also depends upon trol expression (Lindner et al., 2007).
Trol has some roles in tissue development that seem to be related to cell adhesion and tissue polarity. There are three Arg-Gly-Asp (RGD) sequences in trol, and some evidence that they bind to the Drosophila integrin βV in larval hemocytes. This interaction has been suggested to mediate the phagocytosis of apoptotic cells (Nagaosa et al., 2011). Trol and dystroglycan partner to maintain follicle-cell epithelium polarity through basement membrane interactions when flies are under energetic stress. In trol-negative systems this barrier is no longer functional, and dystroglycan is no longer sequestered in the basal membrane, in parallel with apical cell markers migrating to the basal membrane (Schneider et al., 2006). This may contribute to reproductive dysfunction in these fly genotypes. Studies of matrix metalloproteinase (MMP) function in developing flies revealed a function for MMP2 in spatially and temporally dynamic regulation of perlecan and collagen IV deposition in the BM. This barrier, when left intact by MMP2 knockout, prohibited the proper formation of the Drosophila air sac primordium, and was characterized by accumulation of collagen IV and perlecan (Guha et al., 2009). Basement membrane-associated trol also defines organ shape in developing wing disc and ventral nerve cord (Pastor-Pareja and Xu, 2011). In trol-knockout larvae, these organs have a narrow and constricted shape in comparison with wild-type organs. This provides yet another example of a key role for perlecan in setting cell boundaries during complex tissue morphogenesis.
6.3 Evolutionary clues about perlecan and basement membrane assembly
The evolutionary origin of the perlecan gene remains speculative, and a multicellular animal genome that does not encode a perlecan-like gene has yet to be discovered. A telling insight into the evolution of the perlecan gene will be provided when an early genome that does not encode a perlecan gene is found. C. elegans Unc-52 is the oldest well studied perlecan gene, and Drosophila melanogaster trol is the second-oldest well studied form of perlecan. Clues to the evolutionary organization of perlecan are found by comparing these ancient genes to the more modern perlecan genes of vertebrates, as described above in section 2. It is interesting to speculate that the inclusion of 18–20 more LDL receptor-like folding modules in trol represents a remnant of the ancestral form of domain II, and these additional LDL receptor-like modules were lost in vertebrate perlecan, but the absence of these additional repeats in the more simple C. elegans makes one wonder if this is so. Interesting too is the presence in Unc-52 of an additional Ig module in domain II, which is present in flies, but not vertebrates. It is unclear whether these differences occur due to duplication of these LDL receptor-like modules in flies or negative selective pressure in the other clades. Also of note is the lack of O-glycosylation sites in domain I of zebrafish, Drosophila, and C. elegans perlecan. There are three O-glycosylation sites in domain I of chicken and mammalian perlecan molecules, at positions where heparan sulfate or chondroitin sulfate typically are added. The evolutionary pressure that led to these complex adaptations, or “rehitchings” of the train cars, is unclear.
Of interest, the RPL21P29 pseudogene resides within the 40,000 base pair intron one of perlecan in all apes, but not in lower mammals or any other species. This pseudogene is found 145 times in the human genome (Zhang et al., 2002). Given that it is conserved in the perlecan gene only in apes, it seems to be a recent addition to this locus. We find variation among the numbers of Ig modules in domain IV (Figure 2), suggesting that insertions or duplications of certain Ig modules in domain IV occurred commonly throughout evolution. Although the central Ig modules are more conserved relative to each other in human perlecan (Figure 3), they are not conserved in evolution (Figure 2), persisting in mouse perlecan, but not entirely present in chicken perlecan, and even more scarcely present in zebrafish perlecan. The reasons for this variation remain unknown.
7. Translating Boundary Functions into Useful Constructs for Tissue Engineering of Multi-layered Tissues
Efforts to regenerate any of the tissue systems described in Section 4 should include some or all of perlecan’s barrier characteristics. In some tissues, such as cartilage and bone, perlecan is present throughout a territorial matrix, demarcating a region of low vascularity. Conversely, in vasculature, perlecan’s presence is reduced to a nearly 2-dimensional sheet within the basal lamina, where it participates in endothelial cell adhesion, preserves pericyte separation, and inhibits pathogen penetration. In lieu of simply trying to implement pure perlecan protein (as is more frequently and easily accomplished with collagen I or fibrin hydrogels), tissue engineers most commonly have used polymeric systems to recapitulate as many of perlecan’s barrier properties as possible for selected applications. In this context, key characteristics to target would include:
physical barrier, optimally with selective permeability to the diffusion of molecular or nanoscale gases, nutrients, and cytokines or growth factors
physical barrier, optimally with selective permeability to cell migration, via cell-directed degradation of the barrier (e.g. MMPs, hyaluronidases, local pH, etc.)
depot for the retention (covalent or non-covalent) of selected growth factors
reference point for cell polarization, via the display of appropriate receptor ligands
Direct efforts to mimic perlecan’s boundary functions to date have been limited, but recent tissue engineering examples demonstrate progress toward this end. Some examples follow.
7.1 Ocular Engineering
Considering perlecan’s role in development of eye structures (Dong et al., 2002; Keenan et al., 2012), including its presence as a common basement membrane component and phenotype in trol mutants in Drosophila discussed above, efforts to regenerate these delicate eye structures would benefit from incorporation of one or more of perlecan’s barrier functions. Many recent efforts to grow corneal layers artificially have attempted to replicate some aspects of the basement membrane interface. Researchers may instinctively employ biologically-derived substrates to support cell proliferation and organization. For example, amniotic membrane is frequently employed, as it contains many basement membrane components, can be easily harvested and preserved, and may be sufficiently anti-inflammatory and anti-immunogenic for this use (Ahn et al., 2007; Dietrich-Ntoukas et al., 2012). However, the chief disadvantage of such biologically-derived membranes is their inherent variability, which may vary with the site of harvest, the age of the donor, post-processing methods, and the duration of pregnancy at the time of harvest. Such compositional properties impact the consistency in the desired performance parameters, such as nutrient diffusion, optical clarity, and mechanical strength. Processed single-protein films greatly reduces this variability, at the expense of reduced complexity compared to native tissue (Reichl et al., 2011; Shadforth et al., 2012).
Alternately, researchers use thin polymer layers, which offer far higher reproducibility and often easier processing and characterization. For the ocular application, a ∼100 µm thick film might be used, with added porosity to enable nutrient movement through the membrane. Evans et al. implemented multiple polycarbonate-based materials as a surrogate BM, with particular recent focus on a biostable perfluoropolyether to support epithelial overgrowth (Evans et al., 2011). Their work examined the impact of pore size on this parameter, and demonstrated that cell overgrowth was improved with smaller pores, likely due to the greater surface roughness associated with larger pore sizes. Wang et al. (Wang et al., 2012) employed chitosan-poly(ε-caprolactone) blends to create films for corneal endothelium transplantation, and Treharne et al. used methacrylate-based copolymers as artificial Bruch’s membranes for retinal pigment epithelia (Treharne et al., 2012).
7.2 Salivary Gland Engineering
Functional reconstitution of a 3D glandular structure such as the human salivary gland is difficult, owing to a complex tissue structure of distinct cell types that must reassemble to provide vectorial secretion of a complex biological fluid. As in other multilayered glandular tissues, the basement membrane guides and supports both the structure and function of the salivary gland, and as noted in Section 4.4, perlecan’s role in this organization is integral. Perlecan’s involvement as a depot for influential heparin-binding growth factors in the submandibular gland has been well-documented by Hoffman and co-workers (Makarenkova et al., 2009; Patel et al., 2007; Patel et al., 2008). Heparan sulfate significantly modulates the diffusion and availability of many FGFs, when then differentially affect branching morphogenesis.
Despite extensive recent research in salivary gland development, efforts at regeneration using the classic tissue engineering models are few, and suffer from insufficient sources of cells and scaffold materials (Scheller et al., 2009). Some of the earliest efforts employed biodegradable poly(α-esters) (PGA/PLGA/PLLA), fashioned into microporous scaffolds through traditional porogen leaching or microfiber methods. (Aframian et al., 2000; Aframian et al., 2002; Baum et al., 1999; Joraku et al., 2005) Other materials, which can present necessary protein cues and more relevant mechanical properties, have been employed more recently. Collagen/Matrigel™ hybrids (Joraku et al., 2007) are closer matches to native tissue, both in terms of gel modulus and composition, but murine Matrigel™ cannot be used for human tissue repair. Recent work (Chen et al., 2005; Yang and Young, 2008a, b, 2009) suggests that well-defined polysaccharide gels may be a potential solution. Naturally-derived hyaluronic acid (HA) hydrogels also have been employed and have a long history of biocompatibility coupled with HA’s known role in promoting tissue regeneration over fibrosis (Pradhan-Bhatt et al., 2013; Pradhan et al., 2010). Incorporation of a small peptide sequence from perlecan domain IV (Farach-Carson et al., 2008b) into HA hydrogels induces 3D acinus formation and expression of salivary-specific markers (Pradhan et al., 2009). Future work should include perlecan’s incorporation into 3D hyaluronan-based hydrogels to further guide salivary gland regeneration, and mediate the interaction between acinar and myoepithelial cell types.
7.3 Cartilage Engineering
In contrast to its presence in the thin basement membranes of the eye and the salivary gland, perlecan is established as a broad barrier in the osteochondral junction (as described in 4.2 and observed in Fig 4). We have previously described the integral role of perlecan’s tethered glycosaminoglycan chains in directing and supporting cartilage development (French et al., 2002; French et al., 1999; Gomes et al., 2002). While perlecan in cartilage is localized in the close pericellular space around chondrocytes, it is also generated in great supply throughout the growth plate by hypertrophic chondrocytes. The abundance of heparan sulfate in this region serves to prevent mineralization, and retain potent growth factors that influence cell fate.
Researchers have coined the field of “interfacial tissue engineering” (Lu and Jiang, 2006; Seidi et al., 2011; Yang and Temenoff, 2009) as a description of regenerative scaffolds that target the osteochondral junction. Examples of this strategy cross many fields, as shown in a tri-laminate cardiac heart valve application with varying stiffnesses of each poly(ethylene glycol) hydrogel layer (Tseng et al., 2013). Some systems may consider the mechanical interface between the relatively “hard” bone and “soft” cartilage, but most inevitably mimic either the barrier or cytokine depot functions of perlecan (Santo et al., 2013). In cartilage-specific applications, we have described a perlecan-based system for the retention and gradual release of BMP-2 from the heparan sulfate chains of perlecan domain I, tethered to hyaluronic acid-based hydrogel particles (HGPs) (Jha et al., 2009) or to polylactic acid scaffolds (Yang et al., 2006). This system significantly enhanced the chondrogenic differentiation of C3H10T1/2 cells in high-density micromass culture. Conversely, a recent example of 3D chondrocyte culture on a silk scaffold lends credence to perlecan’s role as a barrier to slow the progression of heparin-binding growth factors (Chen et al., 2012). When these 3D scaffolds were cultured in media over a 2D layer of osteoblasts, the outer chondrocytes in closest contact to the osteoblasts demonstrated reduced expression of chondrocytic markers (aggrecan, collagen II), and upregulation of collagen X and MMP13, markers of hypertrophic chondrocytes at the osteochondral junction. In this case, without a proteoglycan barrier (such as perlecan) to retard the diffusion of osteogenic cytokines, chondrocytes were driven to an alternate differentiation program.
As these and other examples demonstrate, researchers in interfacial tissue engineering have focused predominantly on creating scaffolds with biphasic or gradient features, such as elastic modulus, macroscale pore size, or chondrogenic/osteogenic growth factor release. True biomimicry of the osteochondral junction would additionally include a proteoglycan-based barrier to mediate cell phenotype at the osteochondral interface. In many of the most prominent examples of this work, perlecan may indeed be present at this junction, but yet to be discovered. Deliberate incorporation of perlecan into these scaffold designs may alter cell response and improve the overall regeneration of these skeletal structures.
8. Summary and Future Directions
It is exciting to consider the roles of a complex proteoglycan such as perlecan, a large and ancient molecule with many diverse functions. We believe that only a few of the many potential functions of this molecule have been discovered, and suggest that there are many more to be uncovered. Insights into the functions of the five domains, the individual modules or cars of its long train, the relationship between core protein and glycosaminoglycan, and the roles of the intact and degraded molecule lie on the horizon. Studies of the core protein, as presented in the literature, and of domain IV in particular, have led us to propose a key role for perlecan as a key player in the border patrol of unique regions of complex tissues in metazoans. Compromise of the border often is associated with tissue pathologies and developmental anomalies. Understanding the border functions and their molecular basis can provide new ways to engineer tissues for repair of tissue lost to damage or disease. There are many more rail ties for perlecan biologists to lay down to achieve a full understanding of this proteoglycan in health and disease.
Highlights.
Perlecan’s function in maintaining tissue borders is examined.
Perlecan’s functions during degradation and as an intact proteoglycan are compared.
Perlecan’s use and potential in tissue engineering is assessed.
Evolution of the perlecan gene is considered.
The HSPG2 gene’s structure and perlecan’s protein domain functions are described
Acknowledgements
The authors thank the many members of their lab groups over the years who have contributed to this work by providing data, ideas, and helpful discussions. A special thanks is extended to the many colleagues, family and friends who have endured hours of discussion with the authors about the virtues of perlecan in health and disease. Much of this work in the authors’ labs was supported by the NIH, including current support from the NCI program project PO1 CA98912 and from the NIDCR DE022969.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aframian DJ, Cukierman E, Nikolovski J, Mooney DJ, Yamada KM, Baum BJ. The growth and morphological behavior of salivary epithelial cells on matrix protein-coated biodegradable substrata. Tissue engineering. 2000;6:209–216. doi: 10.1089/10763270050044380. [DOI] [PubMed] [Google Scholar]
- Aframian DJ, Redman RS, Yamano S, Nikolovski J, Cukierman E, Yamada KM, Kriete MF, Swaim WD, Mooney DJ, Baum BJ. Tissue compatibility of two biodegradable tubular scaffolds implanted adjacent to skin or buccal mucosa in mice. Tissue engineering. 2002;8:649–659. doi: 10.1089/107632702760240562. [DOI] [PubMed] [Google Scholar]
- Ahn J-I, Lee D-H, Ryu Y-H, Jang I-K, Yoon M-Y, Shin YH, Seo Y-K, Yoon H-H, Kim J-C, Song K-Y, Yang E-K, Kim K-H, Park J-K. Reconstruction of rabbit corneal epithelium on lyophilized amniotic membrane using the tilting dynamic culture method. Artificial organs. 2007;31:711–721. doi: 10.1111/j.1525-1594.2007.00441.x. [DOI] [PubMed] [Google Scholar]
- Allen JM, Bateman JF, Hansen U, Wilson R, Bruckner P, Owens RT, Sasaki T, Timpl R, Fitzgerald J. WARP is a novel multimeric component of the chondrocyte pericellular matrix that interacts with perlecan. The Journal of biological chemistry. 2006;281:7341–7349. doi: 10.1074/jbc.M513746200. [DOI] [PubMed] [Google Scholar]
- Aplin JD, Charlton AK, Ayad S. An immunohistochemical study of human endometrial extracellular matrix during the menstrual cycle and first trimester of pregnancy. Cell and tissue research. 1988;253:231–240. doi: 10.1007/BF00221758. [DOI] [PubMed] [Google Scholar]
- Arikawa-Hirasawa E, Le AH, Nishino I, Nonaka I, Ho NC, Francomano CA, Govindraj P, Hassell JR, Devaney JM, Spranger J, Stevenson RE, Iannaccone S, Dalakas MC, Yamada Y. Structural and functional mutations of the perlecan gene cause Schwartz-Jampel syndrome, with myotonic myopathy and chondrodysplasia. Am J Hum Genet. 2002;70:1368–1375. doi: 10.1086/340390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikawa-Hirasawa E, Wilcox WR, Le AH, Silverman N, Govindraj P, Hassell JR, Yamada Y. Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat Genet. 2001a;27:431–434. doi: 10.1038/86941. [DOI] [PubMed] [Google Scholar]
- Arikawa-Hirasawa E, Wilcox WR, Yamada Y. Dyssegmental dysplasia, Silverman-Handmaker type: unexpected role of perlecan in cartilage development. Am J Med Genet. 2001b;106:254–257. doi: 10.1002/ajmg.10229. [DOI] [PubMed] [Google Scholar]
- Bagyura Z, Pocsai K, Kalman M. Distribution of components of basal lamina and dystrophin-dystroglycan complex in the rat pineal gland: differences from the brain tissue and between the subdivisions of the gland. Histol Histopathol. 2010;25:1–14. doi: 10.14670/HH-25.1. [DOI] [PubMed] [Google Scholar]
- Bangratz M, Sarrazin N, Devaux J, Zambroni D, Echaniz-Laguna A, Rene F, Boerio D, Davoine CS, Fontaine B, Feltri ML, Benoit E, Nicole S. A mouse model of Schwartz-Jampel syndrome reveals myelinating Schwann cell dysfunction with persistent axonal depolarization in vitro and distal peripheral nerve hyperexcitability when perlecan is lacking. Am J Pathol. 2012;180:2040–2055. doi: 10.1016/j.ajpath.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum BJ, Wang S, Cukierman E, Delporte C, Kagami H, Marmary Y, Fox PC, Mooney DJ, Yamada KM. Re-engineering the functions of a terminally differentiated epithelial cell in vivo. Annals of the New York Academy of Sciences. 1999;875:294–300. doi: 10.1111/j.1749-6632.1999.tb08512.x. [DOI] [PubMed] [Google Scholar]
- Behrens DT, Villone D, Koch M, Brunner G, Sorokin L, Robenek H, Bruckner-Tuderman L, Bruckner P, Hansen U. The Epidermal Basement Membrane Is a Composite of Separate Laminin- or Collagen IV-containing Networks Connected by Aggregated Perlecan, but Not by Nidogens. The Journal of biological chemistry. 2012;287:18700–18709. doi: 10.1074/jbc.M111.336073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson E, Morgelin M, Sasaki T, Timpl R, Heinegard D, Aspberg A. The leucine-rich repeat protein PRELP binds perlecan and collagens and may function as a basement membrane anchor. The Journal of biological chemistry. 2002;277:15061–15068. doi: 10.1074/jbc.M108285200. [DOI] [PubMed] [Google Scholar]
- Bix G, Fu J, Gonzalez EM, Macro L, Barker A, Campbell S, Zutter MM, Santoro SA, Kim JK, Hook M, Reed CC, Iozzo RV. Endorepellin causes endothelial cell disassembly of actin cytoskeleton and focal adhesions through alpha2beta1 integrin. The Journal of cell biology. 2004;166:97–109. doi: 10.1083/jcb.200401150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonewald LF. Osteocyte biology: its implications for osteoporosis. J Musculoskelet Neuronal Interact. 2004;4:101–104. [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Alicknavitch M, D'Souza SS, Daikoku T, Kirn-Safran CB, Marchetti D, Carson DD, Farach-Carson MC. Heparanase expression and activity influences chondrogenic and osteogenic processes during endochondral bone formation. Bone. 2008;43:689–699. doi: 10.1016/j.bone.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Nugent MA, Lau FW, Trinkaus-Randall V. Characterization of proteoglycans synthesized by cultured corneal fibroblasts in response to transforming growth factor beta and fetal calf serum. The Journal of biological chemistry. 1999;274:7111–7119. doi: 10.1074/jbc.274.11.7111. [DOI] [PubMed] [Google Scholar]
- Brown JC, Sasaki T, Gohring W, Yamada Y, Timpl R. The C-terminal domain V of perlecan promotes beta1 integrin-mediated cell adhesion, binds heparin, nidogen and fibulin-2 and can be modified by glycosaminoglycans. European journal of biochemistry / FEBS. 1997;250:39–46. doi: 10.1111/j.1432-1033.1997.t01-1-00039.x. [DOI] [PubMed] [Google Scholar]
- Brown JC, Wiedemann H, Timpl R. Protein binding and cell adhesion properties of two laminin isoforms (AmB1eB2e, AmB1sB2e) from human placenta. Journal of cell science. 1994;107(Pt 1):329–338. doi: 10.1242/jcs.107.1.329. [DOI] [PubMed] [Google Scholar]
- Carson DD, Tang JP, Julian J. Heparan sulfate proteoglycan (perlecan) expression by mouse embryos during acquisition of attachment competence. Developmental biology. 1993;155:97–106. doi: 10.1006/dbio.1993.1010. [DOI] [PubMed] [Google Scholar]
- Castillo GM, Cummings JA, Ngo C, Yang W, Snow AD. Novel purification and detailed characterization of perlecan isolated from the Engelbreth-Holm-Swarm tumor for use in an animal model of fibrillar A beta amyloid persistence in brain. J Biochem. 1996;120:433–444. doi: 10.1093/oxfordjournals.jbchem.a021430. [DOI] [PubMed] [Google Scholar]
- Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nature medicine. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti S, Horchar T, Jefferson B, Laurie GW, Hassell JR. Recombinant domain III of perlecan promotes cell attachment through its RGDS sequence. The Journal of biological chemistry. 1995;270:404–409. doi: 10.1074/jbc.270.1.404. [DOI] [PubMed] [Google Scholar]
- Cheifetz S, Massague J. Transforming growth factor-beta (TGF-beta) receptor proteoglycan. Cell surface expression and ligand binding in the absence of glycosaminoglycan chains. The Journal of biological chemistry. 1989;264:12025–12028. [PubMed] [Google Scholar]
- Chen CP, Liu SH, Lee MY, Chen YY. Heparan sulfate proteoglycans in the basement membranes of the human placenta and decidua. Placenta. 2008;29:309–316. doi: 10.1016/j.placenta.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Chen G, Paka L, Kako Y, Singhal P, Duan W, Pillarisetti S. A protective role for kidney apolipoprotein E. Regulation of mesangial cell proliferation and matrix expansion. The Journal of biological chemistry. 2001;276:49142–49147. doi: 10.1074/jbc.M104879200. [DOI] [PubMed] [Google Scholar]
- Chen K, Teh TKH, Ravi S, Toh SL, Goh JCH. Osteochondral interface generation by rabbit bone marrow stromal cells and osteoblasts coculture. Tissue engineering Part A. 2012;18:1902–1911. doi: 10.1089/ten.TEA.2011.0580. [DOI] [PubMed] [Google Scholar]
- Chen M, Chen R, Hsu YH, Chen Y, Young TH. Proliferation and phenotypic preservation of rat parotid acinar cells. Tissue engineering. 2005;11:526–534. doi: 10.1089/ten.2005.11.526. [DOI] [PubMed] [Google Scholar]
- Chen XD, Dusevich V, Feng JQ, Manolagas SC, Jilka RL. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2007;22:1943–1956. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- Chernousov MA, Stahl RC, Carey DJ. Schwann cells use a novel collagen-dependent mechanism for fibronectin fibril assembly. Journal of cell science. 1998;111(Pt 18):2763–2777. doi: 10.1242/jcs.111.18.2763. [DOI] [PubMed] [Google Scholar]
- Chu CL, Goerges AL, Nugent MA. Identification of common and specific growth factor binding sites in heparan sulfate proteoglycans. Biochemistry. 2005;44:12203–12213. doi: 10.1021/bi050241p. [DOI] [PubMed] [Google Scholar]
- Clarke DN, Al Ahmad A, Lee B, Parham C, Auckland L, Fertala A, Kahle M, Shaw CS, Roberts J, Bix GJ. Perlecan Domain V induces VEGf secretion in brain endothelial cells through integrin alpha5beta1 and ERK-dependent signaling pathways. PloS one. 2012;7:e45257. doi: 10.1371/journal.pone.0045257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costell M, Carmona R, Gustafsson E, Gonzalez-Iriarte M, Fassler R, Munoz-Chapuli R. Hyperplastic conotruncal endocardial cushions and transposition of great arteries in perlecan-null mice. Circ Res. 2002;91:158–164. doi: 10.1161/01.res.0000026056.81424.da. [DOI] [PubMed] [Google Scholar]
- Costell M, Gustafsson E, Aszodi A, Morgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fassler R. Perlecan maintains the integrity of cartilage and some basement membranes. The Journal of cell biology. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czubryt MP. Common threads in cardiac fibrosis, infarct scar formation, and wound healing. Fibrogenesis Tissue Repair. 2012;5:19. doi: 10.1186/1755-1536-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza SS, Daikoku T, Farach-Carson MC, Carson DD. Heparanase expression and function during early pregnancy in mice. Biology of reproduction. 2007;77:433–441. doi: 10.1095/biolreprod.107.061317. [DOI] [PubMed] [Google Scholar]
- Datta S. Control of proliferation activation in quiescent neuroblasts of the Drosophila central nervous system. Development. 1995;121:1173–1182. doi: 10.1242/dev.121.4.1173. [DOI] [PubMed] [Google Scholar]
- Datta S, Kankel DR. l(1)trol and l(1)devl, loci affecting the development of the adult central nervous system in Drosophila melanogaster. Genetics. 1992;130:523–537. doi: 10.1093/genetics/130.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Pierce M, Datta MW. Perlecan signaling: helping hedgehog stimulate prostate cancer growth. The international journal of biochemistry & cell biology. 2006;38:1855–1861. doi: 10.1016/j.biocel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Decarlo AA, Belousova M, Ellis AL, Petersen D, Grenett H, Hardigan P, O'Grady R, Lord M, Whitelock JM. Perlecan domain 1 recombinant proteoglycan augments BMP-2 activity and osteogenesis. BMC biotechnology. 2012;12:60. doi: 10.1186/1472-6750-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich-Ntoukas T, Hofmann-Rummelt C, Kruse FE, Schlötzer-Schrehardt U. Comparative analysis of the basement membrane composition of the human limbus epithelium and amniotic membrane epithelium. Cornea. 2012;31:564–569. doi: 10.1097/ICO.0b013e3182254b78. [DOI] [PubMed] [Google Scholar]
- Dong S, Landfair J, Balasubramani M, Bier ME, Cole G, Halfter W. Expression of basal lamina protein mRNAs in the early embryonic chick eye. J Comp Neurol. 2002;447:261–273. doi: 10.1002/cne.10245. [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Rene F, Marcel C, Bangratz M, Fontaine B, Loeffler JP, Nicole S. Electrophysiological studies in a mouse model of Schwartz-Jampel syndrome demonstrate muscle fiber hyperactivity of peripheral nerve origin. Muscle Nerve. 2009;40:55–61. doi: 10.1002/mus.21253. [DOI] [PubMed] [Google Scholar]
- Ettner N, Gohring W, Sasaki T, Mann K, Timpl R. The N-terminal globular domain of the laminin alpha1 chain binds to alpha1beta1 and alpha2beta1 integrins and to the heparan sulfate-containing domains of perlecan. FEBS letters. 1998;430:217–221. doi: 10.1016/s0014-5793(98)00601-2. [DOI] [PubMed] [Google Scholar]
- Evans MDM, Chaouk H, Wilkie JS, Dalton BA, Taylor S, Xie RZ, Hughes TC, Johnson G, McFarland GA, Griesser HH, Steele JG, Meijs GF, Sweeney DF, McLean KM. The influence of surface topography of a porous perfluoropolyether polymer on corneal epithelial tissue growth and adhesion. Biomaterials. 2011;32:8870–8879. doi: 10.1016/j.biomaterials.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Farach-Carson MC, Brown AJ, Lynam M, Safran JB, Carson DD. A novel peptide sequence in perlecan domain IV supports cell adhesion, spreading and FAK activation. Matrix biology : journal of the International Society for Matrix Biology. 2008a;27:150–160. doi: 10.1016/j.matbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farach-Carson MC, Brown AJ, Lynam M, Safran JB, Carson DD. A novel peptide sequence in perlecan domain IV supports cell adhesion, spreading and FAK activation. Matrix biology : journal of the International Society for Matrix Biology. 2008b;27:150–160. doi: 10.1016/j.matbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farach-Carson MC, Carson DD. Perlecan--a multifunctional extracellular proteoglycan scaffold. Glycobiology. 2007;17:897–905. doi: 10.1093/glycob/cwm043. [DOI] [PubMed] [Google Scholar]
- Farach MC, Tang JP, Decker GL, Carson DD. Heparin/heparan sulfate is involved in attachment and spreading of mouse embryos in vitro. Developmental biology. 1987;123:401–410. doi: 10.1016/0012-1606(87)90398-8. [DOI] [PubMed] [Google Scholar]
- Farach MC, Tang JP, Decker GL, Carson DD. Differential effects of p-nitrophenyl-D-xylosides on mouse blastocysts and uterine epithelial cells. Biology of reproduction. 1988;39:443–455. doi: 10.1095/biolreprod39.2.443. [DOI] [PubMed] [Google Scholar]
- French MM, Gomes RR, Timpl R, Höök M, Czymmek K, Farach-Carson MC, Carson DD. Chondrogenic activity of the heparan sulfate proteoglycan perlecan maps to the N-terminal domain I. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2002;17:48–55. doi: 10.1359/jbmr.2002.17.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French MM, Smith SE, Akanbi K, Sanford T, Hecht J, Farach-Carson MC, Carson DD. Expression of the heparan sulfate proteoglycan, perlecan, during mouse embryogenesis and perlecan chondrogenic activity in vitro. The Journal of cell biology. 1999;145:1103–1115. doi: 10.1083/jcb.145.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich MV, Schneider M, Timpl R, Baumgartner S. Perlecan domain V of Drosophila melanogaster. Sequence, recombinant analysis and tissue expression. European journal of biochemistry / FEBS. 2000;267:3149–3159. doi: 10.1046/j.1432-1327.2000.01337.x. [DOI] [PubMed] [Google Scholar]
- Gately S, Twardowski P, Stack MS, Cundiff DL, Grella D, Castellino FJ, Enghild J, Kwaan HC, Lee F, Kramer RA, Volpert O, Bouck N, Soff GA. The mechanism of cancer-mediated conversion of plasminogen to the angiogenesis inhibitor angiostatin. Proc Natl Acad Sci U S A. 1997;94:10868–10872. doi: 10.1073/pnas.94.20.10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist EJ, Moerman DG. Mutations in the sup-38 gene of Caenorhabditis elegans suppress muscle-attachment defects in unc-52 mutants. Genetics. 1992;132:431–442. doi: 10.1093/genetics/132.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giros A, Morante J, Gil-Sanz C, Fairen A, Costell M. Perlecan controls neurogenesis in the developing telencephalon. BMC Dev Biol. 2007;7:29. doi: 10.1186/1471-213X-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice LC. Microarray expression profiling reveals candidate genes for human uterine receptivity. American journal of pharmacogenomics : genomics-related research in drug development and clinical practice. 2004;4:299–312. doi: 10.2165/00129785-200404050-00003. [DOI] [PubMed] [Google Scholar]
- Gohring W, Sasaki T, Heldin CH, Timpl R. Mapping of the binding of platelet-derived growth factor to distinct domains of the basement membrane proteins BM-40 and perlecan and distinction from the BM-40 collagen-binding epitope. European journal of biochemistry / FEBS. 1998;255:60–66. doi: 10.1046/j.1432-1327.1998.2550060.x. [DOI] [PubMed] [Google Scholar]
- Gomes R, Kirn-Safran C, Farach-Carson MC, Carson DD. Perlecan: an important component of the cartilage pericellular matrix. Journal of musculoskeletal & neuronal interactions. 2002;2:511–516. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez EM, Mongiat M, Slater SJ, Baffa R, Iozzo RV. A novel interaction between perlecan protein core and progranulin: potential effects on tumor growth. The Journal of biological chemistry. 2003;278:38113–38116. doi: 10.1074/jbc.C300310200. [DOI] [PubMed] [Google Scholar]
- Gonzalez EM, Reed CC, Bix G, Fu J, Zhang Y, Gopalakrishnan B, Greenspan DS, Iozzo RV. BMP-1/Tolloid-like metalloproteases process endorepellin, the angiostatic C-terminal fragment of perlecan. The Journal of biological chemistry. 2005;280:7080–7087. doi: 10.1074/jbc.M409841200. [DOI] [PubMed] [Google Scholar]
- Goyal A, Pal N, Concannon M, Paul M, Doran M, Poluzzi C, Sekiguchi K, Whitelock JM, Neill T, Iozzo RV. Endorepellin, the angiostatic module of perlecan, interacts with both the alpha2beta1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2): a dual receptor antagonism. The Journal of biological chemistry. 2011;286:25947–25962. doi: 10.1074/jbc.M111.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D, Long WF, Williamson FB. Inhibition by glycosaminoglycans of CaCO3 (calcite) crystallization. Biochem J. 1989;259:41–45. doi: 10.1042/bj2590041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Lin L, Kornberg TB. Regulation of Drosophila matrix metalloprotease Mmp2 is essential for wing imaginal disc:trachea association and air sac tubulogenesis. Developmental biology. 2009;335:317–326. doi: 10.1016/j.ydbio.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MC, Graham PL, Kramer JM. Characterization of alpha1(IV) collagen mutations in Caenorhabditis elegans and the effects of alpha1 and alpha2(IV) mutations on type IV collagen distribution. The Journal of cell biology. 1997;137:1185–1196. doi: 10.1083/jcb.137.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson E, Almonte-Becerril M, Bloch W, Costell M. Perlecan maintains microvessel integrity in vivo and modulates their formation in vitro. PloS one. 2013;8:53715. doi: 10.1371/journal.pone.0053715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler M, Yurchenco PD, Iozzo RV. Developmental expression of perlecan during murine embryogenesis. Dev Dyn. 1997;210:130–145. doi: 10.1002/(SICI)1097-0177(199710)210:2<130::AID-AJA6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Harrison CA, Al-Musawi SL, Walton KL. Prodomains regulate the synthesis, extracellular localisation and activity of TGF-beta superfamily ligands. Growth factors. 2011;29:174–186. doi: 10.3109/08977194.2011.608666. [DOI] [PubMed] [Google Scholar]
- Hassell J, Yamada Y, Arikawa-Hirasawa E. Role of perlecan in skeletal development and diseases. Glycoconj J. 2002;19:263–267. doi: 10.1023/A:1025340215261. [DOI] [PubMed] [Google Scholar]
- Hassell JR, Schrecengost PK, Rada JA, Sundarraj N, Sossi G, Thoft RA. Biosynthesis of stromal matrix proteoglycans and basement membrane components by human corneal fibroblasts. Investigative ophthalmology & visual science. 1992;33:547–557. [PubMed] [Google Scholar]
- Hayes AJ, Lord MS, Smith SM, Smith MM, Whitelock JM, Weiss AS, Melrose J. Colocalization in vivo and association in vitro of perlecan and elastin. Histochemistry and cell biology. 2011;136:437–454. doi: 10.1007/s00418-011-0854-7. [DOI] [PubMed] [Google Scholar]
- Henriquez JP, Casar JC, Fuentealba L, Carey DJ, Brandan E. Extracellular matrix histone H1 binds to perlecan, is present in regenerating skeletal muscle and stimulates myoblast proliferation. Journal of cell science. 2002;115:2041–2051. doi: 10.1242/jcs.115.10.2041. [DOI] [PubMed] [Google Scholar]
- Heppner KJ, Matrisian LM, Jensen RA, Rodgers WH. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. Am J Pathol. 1996;149:273–282. [PMC free article] [PubMed] [Google Scholar]
- Heremans A, De Cock B, Cassiman JJ, Van den Berghe H, David G. The core protein of the matrix-associated heparan sulfate proteoglycan binds to fibronectin. The Journal of biological chemistry. 1990;265:8716–8724. [PubMed] [Google Scholar]
- Hohenester E, Tisi D, Talts JF, Timpl R. The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin. Mol Cell. 1999;4:783–792. doi: 10.1016/s1097-2765(00)80388-3. [DOI] [PubMed] [Google Scholar]
- Hopf M, Gohring W, Kohfeldt E, Yamada Y, Timpl R. Recombinant domain IV of perlecan binds to nidogens, laminin-nidogen complex, fibronectin, fibulin-2 and heparin. European journal of biochemistry / FEBS. 1999;259:917–925. doi: 10.1046/j.1432-1327.1999.00127.x. [DOI] [PubMed] [Google Scholar]
- Hummel S, Osanger A, Bajari TM, Balasubramani M, Halfter W, Nimpf J, Schneider WJ. Extracellular matrices of the avian ovarian follicle. Molecular characterization of chicken perlecan. The Journal of biological chemistry. 2004;279:23486–23494. doi: 10.1074/jbc.M312694200. [DOI] [PubMed] [Google Scholar]
- Ida-Yonemochi H, Satokata I, Ohshima H, Sato T, Yokoyama M, Yamada Y, Saku T. Morphogenetic roles of perlecan in the tooth enamel organ: an analysis of overexpression using transgenic mice. Matrix biology : journal of the International Society for Matrix Biology. 2011;30:379–388. doi: 10.1016/j.matbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Igwebuike UM. A review of uterine structural modifications that influence conceptus implantation and development in sheep and goats. Animal reproduction science. 2009;112:1–7. doi: 10.1016/j.anireprosci.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- Inomata T, Ebihara N, Funaki T, Matsuda A, Watanabe Y, Ning L, Xu Z, Murakami A, Arikawa-Hirasawa E. Perlecan-deficient mutation impairs corneal epithelial structure. Investigative ophthalmology & visual science. 2012;53:1277–1284. doi: 10.1167/iovs.11-8742. [DOI] [PubMed] [Google Scholar]
- Iozzo RV. Basement membrane proteoglycans: from cellar to ceiling. Nature reviews. Molecular cell biology. 2005;6:646–656. doi: 10.1038/nrm1702. [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Cohen IR, Grassel S, Murdoch AD. The biology of perlecan: the multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem J. 1994;302(Pt 3):625–639. doi: 10.1042/bj3020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV, Pillarisetti J, Sharma B, Murdoch AD, Danielson KG, Uitto J, Mauviel A. Structural and functional characterization of the human perlecan gene promoter. Transcriptional activation by transforming growth factor-beta via a nuclear factor 1-binding element. The Journal of biological chemistry. 1997;272:5219–5228. doi: 10.1074/jbc.272.8.5219. [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Zoeller JJ, Nystrom A. Basement membrane proteoglycans: modulators Par Excellence of cancer growth and angiogenesis. Molecules and cells. 2009;27:503–513. doi: 10.1007/s10059-009-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima M, Suzuki N, Hozumi K, Matsunobu T, Kosaki K, Kaneko H, Hassell JR, Arikawa-Hirasawa E, Yamada Y. Perlecan modulates VEGF signaling and is essential for vascularization in endochondral bone formation. Matrix biology : journal of the International Society for Matrix Biology. 2012;31:234–245. doi: 10.1016/j.matbio.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari G, Burghoorn J, Kawano T, Mathew M, Morck C, Axang C, Ailion M, Thomas JH, Culotti JG, Swoboda P, Pilon M. Genetics of extracellular matrix remodeling during organ growth using the Caenorhabditis elegans pharynx model. Genetics. 2010;186:969–982. doi: 10.1534/genetics.110.120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Yang W, Kirn-Safran CB, Farach-Carson MC, Jia X. Perlecan domain I-conjugated, hyaluronic acid-based hydrogel particles for enhanced chondrogenic differentiation via BMP-2 release. Biomaterials. 2009;30:6964–6975. doi: 10.1016/j.biomaterials.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Couchman JR. Perlecan and tumor angiogenesis. J Histochem Cytochem. 2003;51:1393–1410. doi: 10.1177/002215540305101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joraku A, Sullivan CA, Yoo J, Atala A. In-vitro reconstitution of three-dimensional human salivary gland tissue structures. Differentiation. 2007;75:318–324. doi: 10.1111/j.1432-0436.2006.00138.x. [DOI] [PubMed] [Google Scholar]
- Joraku A, Sullivan CA, Yoo JJ, Atala A. Tissue engineering of functional salivary gland tissue. Laryngoscope. 2005;115:244–248. doi: 10.1097/01.mlg.0000154726.77915.cc. [DOI] [PubMed] [Google Scholar]
- Julian J, Das SK, Dey SK, Baraniak D, Ta VT, Carson DD. Expression of heparin/heparan sulfate interacting protein/ribosomal protein l29 during the estrous cycle and early pregnancy in the mouse. Biology of reproduction. 2001;64:1165–1175. doi: 10.1095/biolreprod64.4.1165. [DOI] [PubMed] [Google Scholar]
- Kabosova A, Azar DT, Bannikov GA, Campbell KP, Durbeej M, Ghohestani RF, Jones JCR, Kenney MC, Koch M, Ninomiya Y, Patton BL, Paulsson M, Sado Y, Sage EH, Sasaki T, Sorokin LM, Steiner-Champliaud M-F, Sun T-T, Sundarraj N, Timpl R, Virtanen I, Ljubimov AV. Compositional differences between infant and adult human corneal basement membranes. Investigative ophthalmology & visual science. 2007;48:4989–4999. doi: 10.1167/iovs.07-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle MP, Lee B, Pourmohamad T, Cunningham A, Su H, Kim H, Chen Y, McCulloch CE, Barbaro NM, Lawton MT, Young WL, Bix GJ. Perlecan domain V is upregulated in human brain arteriovenous malformation and could mediate the vascular endothelial growth factor effect in lesional tissue. Neuroreport. 2012;23:627–630. doi: 10.1097/WNR.0b013e3283554c5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji T, Yamada A, Miyajima S, Yamamoto C, Fujiwara Y, Wight TN, Kinsella MG. Cell density-dependent regulation of proteoglycan synthesis by transforming growth factor-beta(1) in cultured bovine aortic endothelial cells. The Journal of biological chemistry. 2000;275:1463–1470. doi: 10.1074/jbc.275.2.1463. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Ishijima M, Futami I, Tomikawa-Ichikawa N, Kosaki K, Sadatsuki R, Yamada Y, Kurosawa H, Kaneko K, Arikawa-Hirasawa E. Synovial perlecan is required for osteophyte formation in knee osteoarthritis. Matrix biology : journal of the International Society for Matrix Biology. 2013;32:178–187. doi: 10.1016/j.matbio.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan TD, Clark SJ, Unwin RD, Ridge LA, Day AJ, Bishop PN. Mapping the differential distribution of proteoglycan core proteins in the adult human retina, choroid, and sclera. Invest Ophthalmol Vis Sci. 2012;53:7528–7538. doi: 10.1167/iovs.12-10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knothe Tate ML. "Whither flows the fluid in bone?" An osteocyte's perspective. J Biomech. 2003;36:1409–1424. doi: 10.1016/s0021-9290(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Knox S, Merry C, Stringer S, Melrose J, Whitelock J. Not all perlecans are created equal: interactions with fibroblast growth factor (FGF) 2 and FGF receptors. The Journal of biological chemistry. 2002;277:14657–14665. doi: 10.1074/jbc.M111826200. [DOI] [PubMed] [Google Scholar]
- Knox SM, Whitelock JM. Perlecan: how does one molecule do so many things? Cell Mol Life Sci. 2006;63:2435–2445. doi: 10.1007/s00018-006-6162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Clarke D, Al Ahmad A, Kahle M, Parham C, Auckland L, Shaw C, Fidanboylu M, Orr AW, Ogunshola O, Fertala A, Thomas SA, Bix GJ. Perlecan domain V is neuroprotective and proangiogenic following ischemic stroke in rodents. The Journal of clinical investigation. 2011;121:3005–3023. doi: 10.1172/JCI46358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shimono C, Norioka N, Nakano I, Okubo T, Yagi Y, Hayashi M, Sato Y, Fujisaki H, Hattori S, Sugiura N, Kimata K, Sekiguchi K. Activin A binds to perlecan through its pro-region that has heparin/heparan sulfate binding activity. The Journal of biological chemistry. 2010;285:36645–36655. doi: 10.1074/jbc.M110.177865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Lindner JR, Hillman PR, Barrett AL, Jackson MC, Perry TL, Park Y, Datta S. The Drosophila Perlecan gene trol regulates multiple signaling pathways in different developmental contexts. BMC Dev Biol. 2007;7:121. doi: 10.1186/1471-213X-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubimov AV, Burgeson RE, Butkowski RJ, Michael AF, Sun TT, Kenney MC. Human corneal basement membrane heterogeneity: topographical differences in the expression of type IV collagen and laminin isoforms. Laboratory investigation; a journal of technical methods and pathology. 1995;72:461–473. [PubMed] [Google Scholar]
- Lu HH, Jiang J. Interface tissue engineering and the formulation of multiple-tissue systems. Advances in biochemical engineering/biotechnology. 2006;102:91–111. [PubMed] [Google Scholar]
- Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, Patel VN, Lang RA, Mohammadi M. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Science signaling. 2009;2:1–9. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Deutzmann R, Timpl R. Characterization of proteolytic fragments of the laminin-nidogen complex and their activity in ligand-binding assays. European journal of biochemistry / FEBS. 1988;178:71–80. doi: 10.1111/j.1432-1033.1988.tb14430.x. [DOI] [PubMed] [Google Scholar]
- Marneros AG, Keene DR, Hansen U, Fukai N, Moulton K, Goletz PL, Moiseyev G, Pawlyk BS, Halfter W, Dong S, Shibata M, Li T, Crouch RK, Bruckner P, Olsen BR. Collagen XVIII/endostatin is essential for vision and retinal pigment epithelial function. The EMBO journal. 2004;23:89–99. doi: 10.1038/sj.emboj.7600014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melrose J, Roughley P, Knox S, Smith S, Lord M, Whitelock J. The structure, location, and function of perlecan, a prominent pericellular proteoglycan of fetal, postnatal, and mature hyaline cartilages. The Journal of biological chemistry. 2006;281:36905–36914. doi: 10.1074/jbc.M608462200. [DOI] [PubMed] [Google Scholar]
- Melrose J, Smith S, Knox S, Whitelock J. Perlecan, the multidomain HS-proteoglycan of basement membranes, is a prominent pericellular component of ovine hypertrophic vertebral growth plate and cartilaginous endplate chondrocytes. Histochemistry and cell biology. 2002;118:269–280. doi: 10.1007/s00418-002-0449-4. [DOI] [PubMed] [Google Scholar]
- Merz DC, Alves G, Kawano T, Zheng H, Culotti JG. UNC-52/perlecan affects gonadal leader cell migrations in C. elegans hermaphrodites through alterations in growth factor signaling. Developmental biology. 2003;256:173–186. doi: 10.1016/s0012-1606(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Miller JD, Stevens ET, Smith DR, Wight TN, Wrenshall LE. Perlecan: a major IL-2-binding proteoglycan in murine spleen. Immunology and cell biology. 2008;86:192–199. doi: 10.1038/sj.icb.7100128. [DOI] [PubMed] [Google Scholar]
- Mongiat M, Fu J, Oldershaw R, Greenhalgh R, Gown AM, Iozzo RV. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. The Journal of biological chemistry. 2003;278:17491–17499. doi: 10.1074/jbc.M210529200. [DOI] [PubMed] [Google Scholar]
- Mongiat M, Otto J, Oldershaw R, Ferrer F, Sato JD, Iozzo RV. Fibroblast growth factor-binding protein is a novel partner for perlecan protein core. The Journal of biological chemistry. 2001;276:10263–10271. doi: 10.1074/jbc.M011493200. [DOI] [PubMed] [Google Scholar]
- Mongiat M, Taylor K, Otto J, Aho S, Uitto J, Whitelock JM, Iozzo RV. The protein core of the proteoglycan perlecan binds specifically to fibroblast growth factor-7. The Journal of biological chemistry. 2000;275:7095–7100. doi: 10.1074/jbc.275.10.7095. [DOI] [PubMed] [Google Scholar]
- Morck C, Vivekanand V, Jafari G, Pilon M. C. elegans ten-1 is synthetic lethal with mutations in cytoskeleton regulators, and enhances many axon guidance defective mutants. BMC Dev Biol. 2010;10:55. doi: 10.1186/1471-213X-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch AD, Dodge GR, Cohen I, Tuan RS, Iozzo RV. Primary structure of the human heparan sulfate proteoglycan from basement membrane (HSPG2/perlecan). A chimeric molecule with multiple domains homologous to the low density lipoprotein receptor, laminin, neural cell adhesion molecules, and epidermal growth factor. The Journal of biological chemistry. 1992;267:8544–8557. [PubMed] [Google Scholar]
- Nagaosa K, Okada R, Nonaka S, Takeuchi K, Fujita Y, Miyasaka T, Manaka J, Ando I, Nakanishi Y. Integrin betanu-mediated phagocytosis of apoptotic cells in Drosophila embryos. The Journal of biological chemistry. 2011;286:25770–25777. doi: 10.1074/jbc.M110.204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvid J, Gorno-Tempini ML, Slavotinek A, Dearmond SJ, Cha YH, Miller BL, Rankin K. Of brain and bone: the unusual case of Dr. A. Neurocase. 2009;15:190–205. doi: 10.1080/13554790802632967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan DM, Fulle A, Valente P, Cai S, Horigan E, Sasaki M, Yamada Y, Hassell JR. The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein-receptor, and the neural cell adhesion molecule. The Journal of biological chemistry. 1991;266:22939–22947. [PubMed] [Google Scholar]
- Noonan DM, Hassell JR. Perlecan, the large low-density proteoglycan of basement membranes: structure and variant forms. Kidney international. 1993;43:53–60. doi: 10.1038/ki.1993.10. [DOI] [PubMed] [Google Scholar]
- Nystrom A, Shaik ZP, Gullberg D, Krieg T, Eckes B, Zent R, Pozzi A, Iozzo RV. Role of tyrosine phosphatase SHP-1 in the mechanism of endorepellin angiostatic activity. Blood. 2009;114:4897–4906. doi: 10.1182/blood-2009-02-207134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Rangel C, Reynolds MM, Caldwell MC, Johns M, Nayak M, Welsh CJ, McDermott S, Datta S. Drosophila perlecan modulates FGF and hedgehog signals to activate neural stem cell division. Developmental biology. 2003;253:247–257. doi: 10.1016/s0012-1606(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Parsi MK, Adams JR, Whitelock J, Gibson MA. LTBP-2 has multiple heparin/heparan sulfate binding sites. Matrix biology : journal of the International Society for Matrix Biology. 2010;29:393–401. doi: 10.1016/j.matbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Pascuzzi RM, Gratianne R, Azzarelli B, Kincaid JC. Schwartz-Jampel syndrome with dominant inheritance. Muscle Nerve. 1990;13:1152–1163. doi: 10.1002/mus.880131210. [DOI] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Xu T. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Dev Cell. 2011;21:245–256. doi: 10.1016/j.devcel.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, Eftekhari S, Whitelock JM, Elkin M, Vlodavsky I, Hoffman MP. Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development (Cambridge, England) 2007;134:4177–4186. doi: 10.1242/dev.011171. [DOI] [PubMed] [Google Scholar]
- Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, Yates EA, Turnbull JE, Ron D, Hoffman MP. Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. The Journal of biological chemistry. 2008;283:9308–9317. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HB, Xie H, Rossi SG, Rotundo RL. Acetylcholinesterase clustering at the neuromuscular junction involves perlecan and dystroglycan. The Journal of cell biology. 1999;145:911–921. doi: 10.1083/jcb.145.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Escalante D, Ceballos-Quintal JM, Canto-Herrera J. Identical twins with the classical form of Schwartz-Jampel syndrome. Clin Dysmorphol. 1997;6:45–49. [PubMed] [Google Scholar]
- Pradhan-Bhatt S, Harrington DA, Duncan RL, Jia X, Witt RL, Farach-Carson MC. Implantable Three-Dimensional Salivary Spheroid Assemblies Demonstrate Fluid and Protein Secretory Responses to Neurotransmitters. Tissue engineering Part A. 2013 doi: 10.1089/ten.tea.2012.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Liu C, Zhang C, Jia X, Farach-Carson MC, Witt RL. Lumen formation in three-dimensional cultures of salivary acinar cells. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2010;142:191–195. doi: 10.1016/j.otohns.2009.10.039. [DOI] [PubMed] [Google Scholar]
- Pradhan S, Pradhan S, Zhang C, Zhang C, Jia X, Jia X, Carson DD, Carson DD, Witt R, Witt R, Farach-Carson MC, Farach-Carson MC. Perlecan domain IV peptide stimulates salivary gland cell assembly in vitro. Tissue engineering Part A. 2009;15:3309–3320. doi: 10.1089/ten.tea.2008.0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichl S, Borrelli M, Geerling G. Keratin films for ocular surface reconstruction. Biomaterials. 2011;32:3375–3386. doi: 10.1016/j.biomaterials.2011.01.052. [DOI] [PubMed] [Google Scholar]
- Reichsman F, Smith L, Cumberledge S. Glycosaminoglycans can modulate extracellular localization of the wingless protein and promote signal transduction. The Journal of cell biology. 1996;135:819–827. doi: 10.1083/jcb.135.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J, Kahle MP, Bix GJ. Perlecan and the blood-brain barrier: beneficial proteolysis? Front Pharmacol. 2012;3:155. doi: 10.3389/fphar.2012.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers KD, Sasaki T, Aszodi A, Jacenko O. Reduced perlecan in mice results in chondrodysplasia resembling Schwartz-Jampel syndrome. Hum Mol Genet. 2007;16:515–528. doi: 10.1093/hmg/ddl484. [DOI] [PubMed] [Google Scholar]
- Roediger M, Kruegel J, Miosge N, Gersdorff N. Tissue distribution of perlecan domains III and V during embryonic and fetal human development. Histol Histopathol. 2009;24:859–868. doi: 10.14670/HH-24.859. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Gilchrist EJ, Mullen GP, Moerman DG. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics. 1995;139:159–169. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Williams BD, Mullen GP, Moerman DG. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 1993;7:1471–1484. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- Rohde LH, Janatpore MJ, McMaster MT, Fisher S, Zhou Y, Lim KH, French M, Hoke D, Julian J, Carson DD. Complementary expression of HIP, a cell-surface heparan sulfate binding protein, and perlecan at the human fetal-maternal interface. Biology of reproduction. 1998;58:1075–1083. doi: 10.1095/biolreprod58.4.1075. [DOI] [PubMed] [Google Scholar]
- Rosenzweig LJ, Kanwar YS. Removal of sulfated (heparan sulfate) or nonsulfated (hyaluronic acid) glycosaminoglycans results in increased permeability of the glomerular basement membrane to 125I–bovine serum albumin. Laboratory investigation; a journal of technical methods and pathology. 1982;47:177–184. [PubMed] [Google Scholar]
- Rossi M, Morita H, Sormunen R, Airenne S, Kreivi M, Wang L, Fukai N, Olsen BR, Tryggvason K, Soininen R. Heparan sulfate chains of perlecan are indispensable in the lens capsule but not in the kidney. The EMBO journal. 2003;22:236–245. doi: 10.1093/emboj/cdg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblum K, Stahl RC, Carey DJ. Constitutive release of alpha4 type V collagen N-terminal domain by Schwann cells and binding to cell surface and extracellular matrix heparan sulfate proteoglycans. The Journal of biological chemistry. 2004;279:51282–51288. doi: 10.1074/jbc.M408837200. [DOI] [PubMed] [Google Scholar]
- Ruiz-Alonso M, Blesa D, Simon C. The genomics of the human endometrium. Biochimica et biophysica acta. 2012;1822:1931–1942. doi: 10.1016/j.bbadis.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Santo VE, Gomes ME, Mano JF, Reis RL. Controlled Release Strategies for Bone, Cartilage, and Osteochondral Engineering-Part I: Recapitulation of Native Tissue Healing and Variables for the Design of Delivery Systems. Tissue Engineering Part B-Reviews. 2013 doi: 10.1089/ten.teb.2012.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse P, Malan D, Fleischmann M, Roell W, Gustafsson E, Bostani T, Fan Y, Kolbe T, Breitbach M, Addicks K, Welz A, Brem G, Hescheler J, Aszodi A, Costell M, Bloch W, Fleischmann BK. Perlecan is critical for heart stability. Cardiovasc Res. 2008;80:435–444. doi: 10.1093/cvr/cvn225. [DOI] [PubMed] [Google Scholar]
- Sato Y, Shimono C, Li S, Nakano I, Norioka N, Sugiura N, Kimata K, Yamada M, Sekiguchi K. Nephronectin binds to heparan sulfate proteoglycans via its MAM domain. Matrix biology : journal of the International Society for Matrix Biology. 2013;32:188–195. doi: 10.1016/j.matbio.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Scheller EL, Krebsbach PH, Kohn DH. Tissue engineering: state of the art in oral rehabilitation. Journal of oral rehabilitation. 2009;36:368–389. doi: 10.1111/j.1365-2842.2009.01939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötzer-Schrehardt U, Dietrich T, Saito K, Sorokin L, Sasaki T, Paulsson M, Kruse FE. Characterization of extracellular matrix components in the limbal epithelial stem cell compartment. Experimental eye research. 2007;85:845–860. doi: 10.1016/j.exer.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Schneider M, Khalil AA, Poulton J, Castillejo-Lopez C, Egger-Adam D, Wodarz A, Deng WM, Baumgartner S. Perlecan and Dystroglycan act at the basal side of the Drosophila follicular epithelium to maintain epithelial organization. Development. 2006;133:3805–3815. doi: 10.1242/dev.02549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev A, Nili N, Strauss BH. The role of perlecan in arterial injury and angiogenesis. Cardiovasc Res. 2004;63:603–610. doi: 10.1016/j.cardiores.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Seidi A, Ramalingam M, Elloumi-Hannachi I, Ostrovidov S, Khademhosseini A. Gradient biomaterials for soft-to-hard interface tissue engineering. Acta Biomaterialia. 2011;7:1441–1451. doi: 10.1016/j.actbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. The Journal of biological chemistry. 2011;286:5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadforth AMA, George KA, Kwan AS, Chirila TV, Harkin DG. The cultivation of human retinal pigment epithelial cells on Bombyx mori silk fibroin. Biomaterials. 2012;33:4110–4117. doi: 10.1016/j.biomaterials.2012.02.040. [DOI] [PubMed] [Google Scholar]
- Sharma B, Iozzo RV. Transcriptional silencing of perlecan gene expression by interferon-gamma. The Journal of biological chemistry. 1998;273:4642–4646. doi: 10.1074/jbc.273.8.4642. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Martin PT. Role of extracellular matrix proteins and their receptors in the development of the vertebrate neuromuscular junction. Dev Neurobiol. 2011;71:982–1005. doi: 10.1002/dneu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, French MM, Julian J, Paria BC, Dey SK, Carson DD. Expression of heparan sulfate proteoglycan (perlecan) in the mouse blastocyst is regulated during normal and delayed implantation. Developmental biology. 1997;184:38–47. doi: 10.1006/dbio.1997.8521. [DOI] [PubMed] [Google Scholar]
- Smith SK. Angiogenesis and implantation. Human reproduction. 2000;15(Suppl 6):59–66. [PubMed] [Google Scholar]
- Smith SM, West LA, Hassell JR. The core protein of growth plate perlecan binds FGF-18 and alters its mitogenic effect on chondrocytes. Archives of biochemistry and biophysics. 2007;468:244–251. doi: 10.1016/j.abb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderegger S, Pollheimer J, Knofler M. Wnt signalling in implantation, decidualisation and placental differentiation--review. Placenta. 2010;31:839–847. doi: 10.1016/j.placenta.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan PP, McCoy SY, Jha AK, Yang W, Jia X, Farach-Carson MC, Kirn-Safran CB. Injectable perlecan domain 1-hyaluronan microgels potentiate the cartilage repair effect of BMP2 in a murine model of early osteoarthritis. Biomed Mater. 2012;7:024109. doi: 10.1088/1748-6041/7/2/024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen MS, Froehner SC. PerleCan fix your muscle AChEs. Trends Neurosci. 2003;26:241–242. doi: 10.1016/S0166-2236(03)00077-8. [DOI] [PubMed] [Google Scholar]
- Stephen LX, Beighton PH. Oro-dental manifestations of the Schwartz-Jampel syndrome. J Clin Pediatr Dent. 2002;27:67–70. doi: 10.17796/jcpd.27.1.923863w781k61p83. [DOI] [PubMed] [Google Scholar]
- Stratman AN, Davis GE. Endothelial cell-pericyte interactions stimulate basement membrane matrix assembly: influence on vascular tube remodeling, maturation, and stabilization. Microsc Microanal. 2012;18:68–80. doi: 10.1017/S1431927611012402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stum M, Davoine CS, Fontaine B, Nicole S. Schwartz-Jampel syndrome and perlecan deficiency. Acta Myol. 2005;24:89–92. [PubMed] [Google Scholar]
- Stum M, Davoine CS, Vicart S, Guillot-Noel L, Topaloglu H, Carod-Artal FJ, Kayserili H, Hentati F, Merlini L, Urtizberea JA, Hammouda el H, Quan PC, Fontaine B, Nicole S. Spectrum of HSPG2 (Perlecan) mutations in patients with Schwartz-Jampel syndrome. Hum Mutat. 2006;27:1082–1091. doi: 10.1002/humu.20388. [DOI] [PubMed] [Google Scholar]
- Talts JF, Andac Z, Gohring W, Brancaccio A, Timpl R. Binding of the G domains of laminin alpha1 and alpha2 chains and perlecan to heparin, sulfatides, alpha-dystroglycan and several extracellular matrix proteins. The EMBO journal. 1999;18:863–870. doi: 10.1093/emboj/18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapanadechopone P, Hassell JR, Rigatti B, Couchman JR. Localization of glycosaminoglycan substitution sites on domain V of mouse perlecan. Biochem Biophys Res Commun. 1999;265:680–690. doi: 10.1006/bbrc.1999.1714. [DOI] [PubMed] [Google Scholar]
- Tesche F, Miosge N. Perlecan in late stages of osteoarthritis of the human knee joint. Osteoarthritis Cartilage. 2004;12:852–862. doi: 10.1016/j.joca.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Thompson WR, Modla S, Grindel BJ, Czymmek KJ, Kirn-Safran CB, Wang L, Duncan RL, Farach-Carson MC. Perlecan/Hspg2 deficiency alters the pericellular space of the lacunocanalicular system surrounding osteocytic processes in cortical bone. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011;26:618–629. doi: 10.1002/jbmr.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann K, Sasaki T, Gustafsson E, Gohring W, Batge B, Notbohm H, Timpl R, Wedel T, Schlotzer-Schrehardt U, Reinhardt DP. Microfibrils at basement membrane zones interact with perlecan via fibrillin-1. The Journal of biological chemistry. 2005;280:11404–11412. doi: 10.1074/jbc.M409882200. [DOI] [PubMed] [Google Scholar]
- Tillet E, Wiedemann H, Golbik R, Pan TC, Zhang RZ, Mann K, Chu ML, Timpl R. Recombinant expression and structural and binding properties of alpha 1(VI) and alpha 2(VI) chains of human collagen type VI. European journal of biochemistry / FEBS. 1994;221:177–185. doi: 10.1111/j.1432-1033.1994.tb18727.x. [DOI] [PubMed] [Google Scholar]
- Timpl R, Fujiwara S, Dziadek M, Aumailley M, Weber S, Engel J. Laminin, proteoglycan, nidogen and collagen IV: structural models and molecular interactions. Ciba Found Symp. 1984;108:25–43. doi: 10.1002/9780470720899.ch3. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Kinoshita-Toyoda A, Selleck SB. Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. The Journal of biological chemistry. 2000;275:2269–2275. doi: 10.1074/jbc.275.4.2269. [DOI] [PubMed] [Google Scholar]
- Treharne AJ, Thomson HAJ, Grossel MC, Lotery AJ. Developing methacrylate-based copolymers as an artificial Bruch's membrane substitute. Journal of biomedical materials research Part A. 2012;100:2358–2364. doi: 10.1002/jbm.a.34178. [DOI] [PubMed] [Google Scholar]
- Tseng H, Cuchiara ML, Durst CA, Cuchiara MP, Lin CJ, West JL, Grande-Allen KJ. Fabrication and mechanical evaluation of anatomically-inspired quasilaminate hydrogel structures with layer-specific formulations. Annals of biomedical engineering. 2013;41:398–407. doi: 10.1007/s10439-012-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Flores M, Casas-Avila L, Ponce de León-Suárez V. Genetic Diseases Related with Osteoporosis. In: Valdés-Flores DM, editor. Topics in Osteoporosis. InTech, Chapters Under CC BY 3.0: License; 2013. [Google Scholar]
- van den Born J, van den Heuvel LP, Bakker MA, Veerkamp JH, Assmann KJ, Berden JH. A monoclonal antibody against GBM heparan sulfate induces an acute selective proteinuria in rats. Kidney international. 1992;41:115–123. doi: 10.1038/ki.1992.15. [DOI] [PubMed] [Google Scholar]
- Vischer P, Feitsma K, Schon P, Volker W. Perlecan is responsible for thrombospondin 1 binding on the cell surface of cultured porcine endothelial cells. European journal of cell biology. 1997;73:332–343. [PubMed] [Google Scholar]
- Voigt A, Pflanz R, Schafer U, Jackle H. Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev Dyn. 2002;224:403–412. doi: 10.1002/dvdy.10120. [DOI] [PubMed] [Google Scholar]
- Walch ET, Marchetti D. Role of neurotrophins and neurotrophins receptors in the in vitro invasion and heparanase production of human prostate cancer cells. Clin Exp Metastasis. 1999;17:307–314. doi: 10.1023/a:1006652605568. [DOI] [PubMed] [Google Scholar]
- Wang K, Vishwanath P, Eichler GS, Al-Sebaei MO, Edgar CM, Einhorn TA, Smith TF, Gerstenfeld LC. Analysis of fracture healing by large-scale transcriptional profile identified temporal relationships between metalloproteinase and ADAMTS mRNA expression. Matrix biology : journal of the International Society for Matrix Biology. 2006;25:271–281. doi: 10.1016/j.matbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Wang IJ, Lu JN, Young TH. Novel chitosan-polycaprolactone blends as potential scaffold and carrier for corneal endothelial transplantation. Mol Vis. 2012;18:255–264. [PMC free article] [PubMed] [Google Scholar]
- Waterston RH. Muscle. In: Wood WB, editor. The Nematode Caenorhabditis Elegans. Cold Spring Harbor Laboratory: Cold Spring Harbor; 1988. pp. 281–335. [Google Scholar]
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure of Caenorhabditis elegans. Developmental biology. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Wewer UM, Faber M, Liotta LA, Albrechtsen R. Immunochemical and ultrastructural assessment of the nature of the pericellular basement membrane of human decidual cells. Laboratory investigation; a journal of technical methods and pathology. 1985;53:624–633. [PubMed] [Google Scholar]
- Whitelock J. Purification of perlecan from endothelial cells. Methods Mol Biol. 2001;171:27–34. doi: 10.1385/1-59259-209-0:027. [DOI] [PubMed] [Google Scholar]
- Whitelock JM, Graham LD, Melrose J, Murdoch AD, Iozzo RV, Underwood PA. Human perlecan immunopurified from different endothelial cell sources has different adhesive properties for vascular cells. Matrix biology : journal of the International Society for Matrix Biology. 1999;18:163–178. doi: 10.1016/s0945-053x(99)00014-1. [DOI] [PubMed] [Google Scholar]
- Whitelock JM, Melrose J, Iozzo RV. Diverse cell signaling events modulated by perlecan. Biochemistry. 2008;47:11174–11183. doi: 10.1021/bi8013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA. The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. The Journal of biological chemistry. 1996;271:10079–10086. doi: 10.1074/jbc.271.17.10079. [DOI] [PubMed] [Google Scholar]
- Xu Z, Ichikawa N, Kosaki K, Yamada Y, Sasaki T, Sakai LY, Kurosawa H, Hattori N, Arikawa-Hirasawa E. Perlecan deficiency causes muscle hypertrophy, a decrease in myostatin expression, and changes in muscle fiber composition. Matrix biology : journal of the International Society for Matrix Biology. 2010;29:461–470. doi: 10.1016/j.matbio.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PJ, Temenoff JS. Engineering orthopedic tissue interfaces. Tissue Engineering Part B-Reviews. 2009;15:127–141. doi: 10.1089/ten.teb.2008.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TL, Young TH. Chitosan cooperates with mesenchyme-derived factors in regulating salivary gland epithelial morphogenesis. Journal of Cellular and Molecular Medicine. 2008a doi: 10.1111/j.1582-4934.2008.00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TL, Young TH. The enhancement of submandibular gland branch formation on chitosan membranes. Biomaterials. 2008b;29:2501–2508. doi: 10.1016/j.biomaterials.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Yang TL, Young TH. The specificity of chitosan in promoting branching morphogenesis of progenitor salivary tissue. Biochemical and Biophysical Research Communications. 2009;381:466–470. doi: 10.1016/j.bbrc.2008.10.116. [DOI] [PubMed] [Google Scholar]
- Yang W, Gomes RR, Brown AJ, Burdett AR, Alicknavitch M, Farach-Carson MC, Carson DD. Chondrogenic differentiation on perlecan domain I, collagen II, and bone morphogenetic protein-2-based matrices. Tissue engineering. 2006;12:2009–2024. doi: 10.1089/ten.2006.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des. 2009;15:1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Harrison P, Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome research. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller JJ, Whitelock JM, Iozzo RV. Perlecan regulates developmental angiogenesis by modulating the VEGF-VEGFR2 axis. Matrix biology : journal of the International Society for Matrix Biology. 2009;28:284–291. doi: 10.1016/j.matbio.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]