Abstract
The structure of the Bacillus anthracis spore-binding phage 8a was determined by cryo-electron tomography. The phage capsid forms a T=16 icosahedron attached to a contractile tail via a head–tail connector protein. The tail consists of a six-start helical sheath surrounding a central tail tube, and a structurally novel baseplate at the distal end of the tail that recognizes and attaches to host cells. The parameters of the icosahedral capsid lattice and the helical tail sheath suggest protein folds for the capsid and tail-sheath proteins, respectively, and indicate evolutionary relationships to other dsDNA viruses. Analysis of 2518 intact phage particles show four distinct conformations that likely correspond to four sequential states of the DNA ejection process during infection. Comparison of the four observed conformations suggests a mechanism for DNA ejection, including the molecular basis underlying coordination of tail sheath contraction and genome release from the capsid.
Keywords: Bacteriophage, Myoviridae, DNA ejection, Contractile tail, Tail contraction, Bacillus anthracis, Cryo-electron tomography, Phage infection, Tail sheath, Base plate
Introduction
Bacteriophages are extraordinarily efficient nanomachines (de Frutos et al., 2005; Fokine et al., 2007; Inamdar et al., 2006; Leiman et al., 2003; Morais et al., 2005a; Muller et al., 1997; Sun et al., 2008), and often virtually every phage particle is infectious. All dsDNA tailed bacteriophages share a similar structural blueprint: an icosahedral, or prolate-icosahedral head (Tao et al., 1998) (capsid) encapsidates the viral genome, and is attached to a six-fold symmetric tail at a unique vertex via a dodecameric adaptor protein called the head–tail connector, or portal protein (Kostyuchenko et al., 2005). During phage morphogenesis, an empty head, or procapsid, is first assembled, followed by packaging of the viral genome through a central channel in the portal into the empty procapsid. A phage-encoded ATPase provides the energy to overcome the entropic and enthalpic costs that accompany compaction of the DNA within the confined space of the prohead. At the conclusion of genome packaging, the DNA translocation machinery is released, and the tail assembles at the portal vertex to complete viral maturation. Mature phages are typically released via cell lysis. Upon infection of a new bacterium, the packaged DNA is injected through the tail and into the cytoplasm of the host cell. Although it is widely believed that the genome injection process is passive, driven only by the potential energy stored in the packaged DNA (Leiman et al., 2004), recent experiments indicate that an active, energy-driven packaging mechanism is required for complete genome ejection in phage T7 (Chang et al., 2010; Kemp et al., 2004). The lifecycles of bacteriophages thus mirror many fundamental biological processes including macromolecular recognition; coordinated conformational switching; protein–protein, protein–membrane and protein–DNA interactions; nucleic acid remodeling; and conversion of chemical energy into mechanical motion.
The tails of bacteriophages are multifunctional organelles that recognize and attach to the host bacterium, and also provide the conduit through which both the host recognition signal is transmitted to the head, and the viral genome is delivered into the host cell’s cytoplasm. The morphology of the tail also provides the basis for classification of bacteriophages into three families: 1) Myoviridae are characterized by long contractile tails (e.g. bacteriophage T4 (Leiman et al., 2003)); 2) Podoviridae are equipped with short non-contractile tails (e.g. bacteriophages T7 (Kemp et al., 2005), phi29 (Guasch et al., 1998; Morais et al., 2001), and N4 (Choi et al., 2008); and 3) Siphoviridae have long, flexible non-contractile tails (bacteriophages λ (Grayson et al., 2007) and SPP1 (Plisson et al., 2007)). Although different DNA ejection mechanisms have been proposed for these three families, they all feature rearrangement of tail protein subunits as the means of relaying the host cell attachment signal to the capsid (Aksyuk et al., 2009; Plisson et al., 2007; Takeda et al., 2004).
The clinical potential of phage therapy was first recognized nearly a century ago by D’Herelle; due to their exquisite host specificity, extraordinary infection efficiency, ease of production/manipulation, and remarkable durability, bacteriophages are nearly ideal anti-microbial agents. In a related application, phages labeled with sensitive imaging agents have been proposed as clinical diagnostics for bacterial infection (Edgar et al., 2006; Tang et al., 2009) and recently, the use of phages as environmental detection/decontamination tools is being explored (Schofield and Westwater, 2009; Schofield et al., 2009; Walter, 2003; Walter and Baker, 2003).
Despite the great promise of phage-based nano-tools, their potential as anti-microbial therapeutics and detection tools has been limited to vegetative bacteria, since phage are typically ineffective against bacterial spores. However, the recent discovery of Spore-binding phage 8a, a Bacillus anthracis-specific Myoviridae phage that not only destroys vegetative B. anthracis cells, but also recognizes and adheres to anthrax spores, suggests that phage-based therapeutics or detection/decontamination tools effective against sporulating bacteria may be developed. Here, we describe the three-dimensional structure of SBP8a as determined by cryo-electron tomography (cryo-ET).
Cryo-ET is an emerging technique for visualization of the 3-D structures of cells and large macromolecular complexes at nanometer resolution (Lucic et al., 2005; Medalia et al., 2002). A strength of cryo-ET is that it is possible to investigate cellular components in their native state without fixation, dehydration, embedding or sectioning artifacts. Cryo-ET also has great potential for structural analysis of dynamic processes in biology. Unlike single particle cryo-electron microscopy where only a single 2-D projection of a particle is recorded, cryo-ET provides direct 3-D information, and thus conformational heterogeneity can be more easily evaluated. The resolution of cryo-ET has significantly improved with the advent of high-throughput data acquisition (Liu et al., 2009a) and 3-D sub-volume averaging techniques (Liu et al., 2006, 2008; Winkler, 2007).
In this study, high throughput cryo-ET was used to visualize intact SBP8a phage under near native conditions. Four distinct conformations were present: 1) DNA-filled particles with extended tails; 2) DNA-filled particles with contracted tails; 3) partially DNA-filled particles with contracted tails; and 4) empty particles with contracted tails. Nanometer resolution structures of each of these conformations were independently determined using cryo-ET and advanced 3-D image analysis. These structures provide a series of 3-D snapshots corresponding to four distinct states of DNA ejection and suggest a mechanism by which conformational changes initiated at the baseplate are propagated along the tail to the head, resulting in translocation of the viral genome from the capsid into the cytoplasm of the B. anthracis host cell.
Results and discussion
Cryo-ET of SBP8a reveals four distinct morphologies of SBP8a
At least four types of phage particles were present in cryo-ET tomograms of SBP8a (Figs. 1A, B, C, D, and E; Table 1) that differ primarily in the amount of DNA encapsidated in head and in the length/conformational state of tail. These conformations likely correspond to four distinct sequential states in the DNA ejection process, and can be ordered as follows: 1) DNA-filled capsid, extended tail (Fig. 1B); 2) DNA-filled capsid, contracted tail (Fig. 1C); 3) capsid partially filled with either protein or DNA, contracted tail (Fig. 1D); and 4) DNA-emptied capsid, contracted tail (Fig. 1E). Observation of the same phage preparation over time indicates that an increasing number of phage particles undergo spontaneous ejection (data not shown). Hence, although the majority of particles observed in our tomographic data set are in state 4, this was not a fresh phage preparation and thus the observed distribution of the different states is not necessarily representative.
Fig. 1.
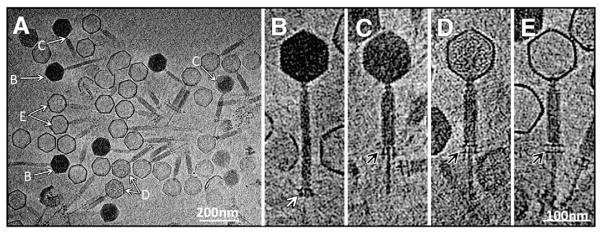
Cryo-ET structure of SPB8a. Four distinct states of the phage (A), are denoted by white arrows and labeled B, C, D and E. These four particles likely represent different intermediates during DNA ejection, and are shown in zoom-in views in B–E respectively. The baseplates at the distal end of tails are indicated by a white arrow in B and black in C, D, and E.
Table 1.
Number of particles averaged for the reconstructions of the four observed states of SBP8a.
| Head (capsid) | Tail sheath | No. of phage | |
|---|---|---|---|
| State 1 | Filled with complete of DNA | Extended | 332 |
| State 2 | Filled with complete of DNA | Contracted | 224 |
| State 3 | Filled with part of DNA | Contracted | 118 |
| State 4 | Empty | Contracted | 1854 |
In addition to contraction of the tail sheath, the baseplate also undergoes a significant conformational change upon tail contraction (compare Fig. 1B with Figs. 1C, D, and E). Improved structures of these four distinct conformational states were determined by 3-D sub-volume averaging of each structurally homogeneous population (Figs. 2A, B, C, and D). The resulting structures provide a series of “snapshots” documenting the conformational changes that accompany genome ejection in bacteriophage SBP8a.
Fig. 2.
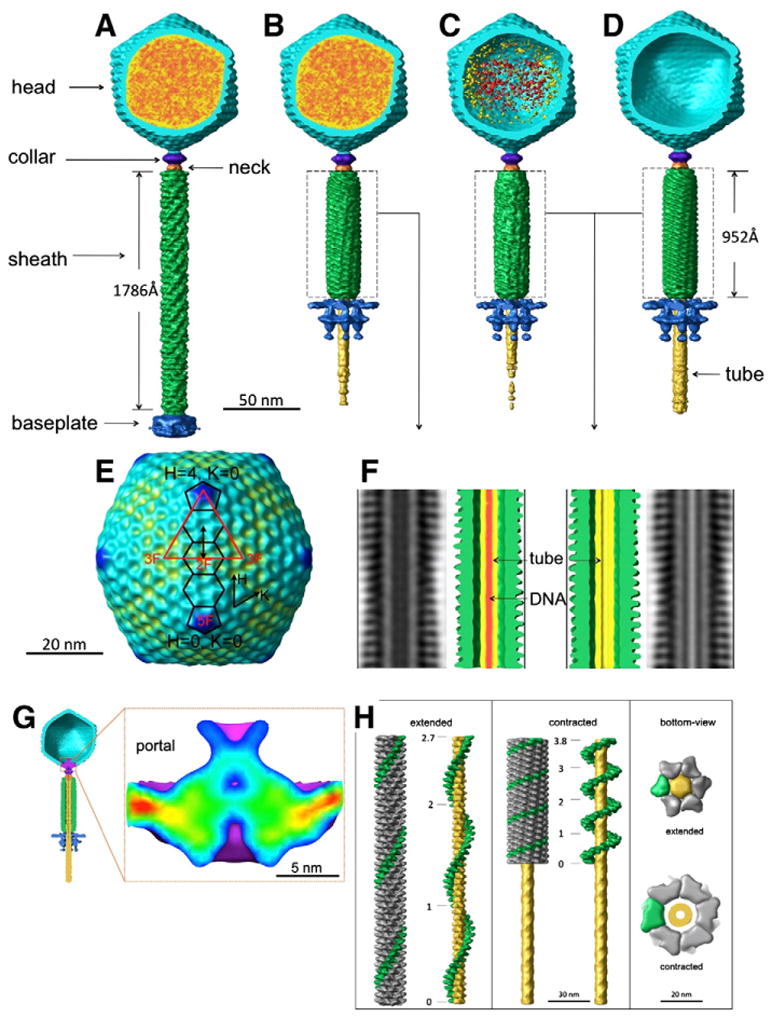
Three dimensional reconstructions of four distinct states of SBP8a (A–D). The head is colored in cyan, DNA in red and orange, the tail in green, the baseplate in blue, the tail tube in yellow, the collar in purple, and the neck in orange. A reconstruction of the SBP8a capsid with icosahedral symmetry imposed is shown in E. The icosahedral asymmetric unit is shown as a red triangle with 5-, 3- and 2-fold symmetry axes labeled. Two pentamers and three hexamers along the H-direction are outlined in black. The approximately 140 Å distance between neighboring hexamers is shown by a double-headed black arrow. F shows helical reconstructions of the contracted tails from B (left) and C, and D (right) respectively. DNA is shown as red density in the tail tube on F (left). G. Structure of the SBP8a portal. A cut-away view of the phage particle with an empty head and contracted tail is shown, with density corresponding to the portal protein colored in purple (left). A close-up of the portal of SBP8a, colored blue (low) to red (high) by density height is shown in the panel on the right. H. Two conformations of the tail sheath. The reconstructions of the extended tail (left) and contracted tail (middle) were carried out using helical reconstruction techniques modified for tomographic data. One of the six helical strands is colored in green. The tail tube in the middle of the tail sheath is colored in yellow. Total twist angles are shown along a single-strand of the sheath. Bottom-views of extended and contracted tails are shown in the right panel.
Structure of the phage head and portal
Other than the presence or absence of DNA, the structures of the capsids in each conformational state are very similar at the resolution of 3.6 nm. After application of icosahedral symmetry to the capsids (excluding density corresponding to the tail-vertex from the averaging calculations), it is clear that the SBP8a shell has T=16 quasi-symmetry with H=4, and K=0. In addition to the 12 pentons present in all icosahedra, this arrangement of subunits gives rise to three different types of hexons, one located at icosahedral 2-fold axes, and the other two residing at general positions; one of the general position hexons shares an edge with pentons, the other is adjacent to icosahedral 3-fold axes (Fig. 2E). The distance between the centers of a penton and the nearest hexon is approximately 130 Å, whereas the distance between neighboring hexons is 135 Å. These distances, along with an observed shell thickness of approximately 31 Å, are consistent with an icosahedral lattice constructed from protein subunits having an HK97 capsid protein-like fold (Fokine et al., 2005; Wikoff et al., 2000). Compared to hexons, additional electron density forming a star-like structure is present at the center of pentons (Fig. 2E). It is not yet known whether this density arises from attachment of a penton-specific decorating protein, or whether pentameric capsomers are made from a minor capsid protein that has an extra domain in addition to the HK97-like domain. A protruding ridge-like density outlines the edges of each capsomer as well (Fig. 2E). Like the extra densities present on pentamers, it is not clear whether these densities are additional domains of the major capsid protein, or separate decorating proteins. Comparable densities are not present in the canonical HK97 fold, which lies flat on the icosahedral surface. Situated at capsomeric borders, this density is well positioned to function as a molecular buckle, stabilizing capsomer–capsomer interactions. There is no apparent difference in the head structure when comparing states 1 and 2. Comparing states 2 and 3, it is apparent that the head contains density corresponding to either DNA or protein in state 3 (Figs. 2B, and C). DNA is completely absent from state 4 (Fig. 2D).
In each of the four states examined, a cone-shaped density is present at a unique five-fold vertex of the pseudo icosahedral head. Based on its easily recognized shape and its position in the phage, this density almost certainly corresponds to the SBP8a portal protein. Portal proteins are typically dodecameric macromolecular assemblies that are essential for phage assembly, genome packaging, and DNA ejection (Muller et al., 1997; Simpson et al., 2000). Although the portal is clearly visible in each of the four reconstructions, its molecular envelope is most easily delineated in the reconstruction of the emptied contracted-tail phages (Fig. 2G). As in other phages, the SBP8a portal has a wide-end located just inside the capsid head, a central region, narrow end, and a central channel just wide enough to accommodate double-stranded DNA. Unlike other phages, the SBP8a portal has an additional goblet-like structure protruding from the SBP8a equivalent of the portal crown domain (Lebedev et al., 2007; Simpson et al., 2000) and extending inward toward the center of the capsid.
Structure of the contractile tail
Comparison of the tail regions of the four structures indicates that the SBP8a tail consists of a six-start helical tail sheath that surrounds a central coaxial tail tube, a protein complex consisting of the collar and neck that connects the head and tail regions, and a “claw”-like baseplate structure at the distal end of the tail. The virus tail is present in at least two different conformations. The most apparent conformational change is the contraction of the tail sheath that occurs upon transitioning from state 1 to state 2, where the tail sheath shortens from 1786 Å to 953 Å in length, and broadens from 195 Å to 280 Å in diameter. Both extended and contracted tails clearly possess helical symmetry that was not initially imposed in the reconstructions (Figs. 2A, B, C and D). Therefore, helical parameters were determined and imposed to improve the 3-D structures of the extended and contracted tail sheaths (Fig. 2H). The contracted tail sheath is similar in states 2, 3 and 4 as shown in Figs. 2E, F. However, the middle of the tail tube in states 3 and 4 is empty, but appears occupied in state 2, indicating the presence of DNA in state 2, but not in states 3 or 4. This suggests that two distinct signals may be required to enable DNA ejection, one to allow DNA to pass from the head into the tail, and a second to release DNA from the tail tube into the cytoplasm of the host cell. The absence of DNA density in the tail tube of state 3 might reflect particles that packaged two segments of DNA but only ejected one, or it’s possible that the residual density within the capsids of these particles is protein rather than DNA. The baseplate also undergoes a dramatic rearrangement from a “closed” conformation into a more “open” structure (see below). Examination of all the particles processed unambiguously showed that all particles with extended tail sheathes had “closed” base-plates, whereas all particles with contracted tail sheathes had “open” baseplates, indicating that the conformations of the tail sheath and base-plate are strictly correlated.
The extended tail sheath in state 1 is assembled around the 78-Å-diameter rigid tail tube as a right-handed, six-stranded helical structure measuring 195 Å in diameter and 1786 Å in length. Each helical strand consists of 45 subunits related to each other by a 39.5 Å translation (helical pitch) and a 21.7° rotation (helical twist). Thus, there is a total of twist angle 954.8° (2.7 turns) going from the first to last subunit in each strand in the extended sheath (Fig. 2H; Table 2). In contrast, the 45 subunits in each helical strand of the contracted tail are related by a 19.1 Å helical pitch and a 31.3° helical twist, resulting in a total twist of 1377.2° (3.8 turns) in the contracted tail (Fig. 2H; Table 2). Assuming that each helical strand of the sheath is fixed at one end to the head/neck region and at the other end to the baseplate, contraction of the tail sheath must be accompanied by a 422.4° rotation of the head around the long axis of the phage during DNA ejection. The structure of individual tail sheath subunits is similar in both conformations at the current resolution (Fig. 2H), indicating that tail contraction results from rigid body rotations and translations of entire subunits rather than by conformational changes within individual subunits. The helical parameters of SBP8a are similar to those of the bacteriophages T4 and ØKZ (Table 2) (Fokine et al., 2007; Leiman et al., 2004), suggesting that the tail sheath proteins of these three phages, and possibly all Myoviridae phages, share the same fold and likely evolved from a common viral ancestor.
Table 2.
Comparison of helical parameters for extended and contracted sheath of phage T4, ØKZ, and SBP8a.
| Sheath | T4
|
ØKZ
|
SBP8a
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Extended | Contracted | Relationship between the two states | Extended | Contracted | Relationship between the two states | Extended | Contracted | Relationship between the two states | |
| Length (Å) | 925 | 420 | Shortens by 65% | 1593 | a | a | 1786 | 953 | Shortens by 47% |
| Diameter (Å) | 240 | 330 | Broadens by 38% | a | a | a | 195 | 280 | Broadens by 44% |
| Number of strands | 6 | 6 | No change | 6 | a | a | 6 | 6 | No change |
| Number of subunits per helical strand | 23 | 23 | No change | 44 | a | a | 45 | 45 | No change |
| Helical pitch (Å) | 40.6 | 16.4 | Decreases by 60% | 36.2 | a | a | 39.5 | 19.1 | Decreases by 57% |
| Helical twist | 17.2° | 32.9° | Increases by 91% | 22° | a | a | 21.7° | 31.3° | Increases by 44% |
| Total twist angle | 378.4° (1.05a2π) | 723.8° (2.01a2π) | Increase by 345.4° (0.96a2π) | 968° (2.69a2π) | a | a | 954.8° (2.65a2π) | 1377.2° (3.8a2π) | Increases by 442.4° (1.15a2π) |
Data absent.
Structure of the junction between the head and the tail
The phage neck proteins not only serve as an adaptor that connects the head to the tail, but may also actively participate in DNA ejection. Comparison of the neck regions in extended and contracted tails indicates that there are two conformations of the neck (Figs. 3A–F). In the extended state, the neck is assembled into a compact shape that is connected to the six helical strands of the tail sheath (Fig. 3E). In the contracted state, the neck forms a central pore that is large enough to allow the passage of dsDNA (Fig. 3F). Thus, the neck protein may function as a gate that regulates the passage of DNA from the head into the tail, by undergoing a substantial conformational change upon tail contraction.
Fig. 3.
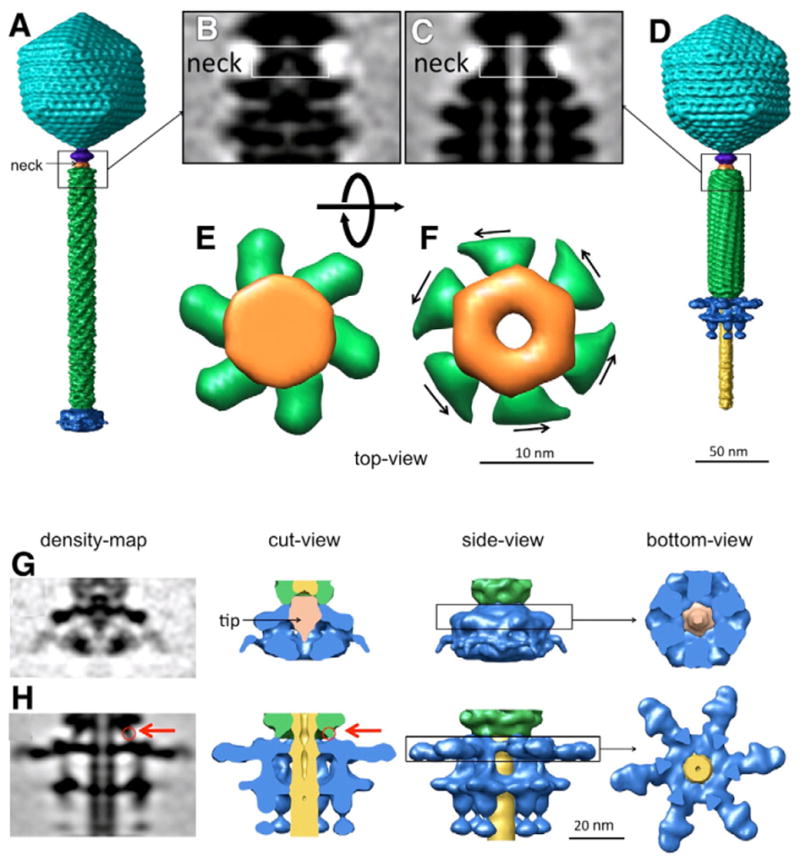
The structure of the neck region before and after the tail contraction. The “closed” conformation of the neck is shown in A, B, and E, and the “open” conformation in C, D, and F. The neck is colored in orange, and the sheath in green. E and F show top-views of neck region noted by the rectangles in B and C; the viewing direction is from the head, looking towards the baseplate. The arrows in F suggest the direction of force imposed on the neck during tail sheath contraction. G. Baseplate density for extended tails. H. Baseplate density for contracted tails. The tip protein, only observed in the closed baseplate associated with extended tails, is colored in orange (G, middle right). The red arrows in H (left, middle left) indicate the connection between the tail sheath and the baseplate. It is clear that the baseplate (blue) in cut-away views (middle right panels) and side-views (middle left panels) undergoes a dramatic conformational change, transforming from a “closed” to an “open” claw-like structure.
Structure of the baseplate
The baseplate forms a complex claw-like structure, with six “talons” that are assembled in a “closed” conformation (Fig. 3G) at the end of extended tail, and in an “open” conformation (Fig. 3H) at the end of the contracted tail. A tail tip structure (colored in orange) is present at the distal end of the tail tube in extended tails (Fig. 3G); no equivalent structure is seen in contracted tails. Short fibers are present at the periphery of the baseplate. These fibers may be analogous to the short tail fibers in T4 that are responsible for recognition of the host cell receptor; no evidence of long fiber is present in either cryo-tomograms or in negative stain images of SBP8a (data not shown). It is clear that the baseplate undergoes a dramatic conformational change, and it is likely that this change is important for cell surface attachment and DNA ejection. By analogy with mechanism of ejection proposed for T4 (Leiman et al., 2004), we hypothesize that the baseplate undergoes a conformational change from a “closed” to an “open” conformation upon binding to the host cell surface. The tail tip may function as a cell-puncturing device as proposed in T4 (Rossmann et al., 2004). Furthermore, the baseplate and tail sheath are physically connected (red arrow in Figs. 3G, H), suggesting that the structural rearrangement of the baseplate could initiate contraction of the tail sheath.
Structural similarities of SBP8a to other phages
The tailed Myoviridae phages T4 and ØKZ have been studied previously using cryo-EM and X-ray crystallography (Aksyuk et al., 2009; Fokine et al., 2004; Kostyuchenko et al., 2005; Leiman et al., 2003, 2004). The structural similarities between these Myoviridae phages and SBP8a are summarized in Table 2. High resolution structural information regarding several SBP8a structural proteins can be inferred by comparison with T4 and other phages: capsomers spaced ~140 Å apart are indicative of an icosahedral lattice built from protein subunits having the HK97 fold (Fokine et al., 2005); the shape of the portal density indicates that it is likely a dodecamer whose monomers have a fold similar to the known structures of the Ø29 and SPP1 portal proteins (Simpson et al., 2000); the similarity of the helical parameters of the SBP8a tail sheath to those determined for T4 suggests that the SBP8a tail sheath protein will be structurally homologous to the known atomic resolution structure of the T4 tail sheath protein (Aksyuk et al., 2009).
Mechanism of DNA ejection
Initial host cell recognition and binding is likely accomplished by the closed conformation of the baseplate since the short tail fibers necessary for receptor recognition are inaccessible in the open conformation. In phage SBP8a, the closed conformation of the baseplate is strictly associated with the extended conformation of the tail. In contrast, the open conformation of the baseplate is only seen in contracted tails, suggesting that either rearrangement of the baseplate protein subunits triggers tail sheath contraction, or vice-versa. The former scenario seems more likely, as this sequence of events provides a means of coupling cellular recognition with genome ejection. Specifically, conformational changes in the baseplate upon host cell binding may trigger tail sheath contraction via the adaptor protein between the baseplate and tail sheath. Furthermore, the open conformation may form a stable interaction between the baseplate and the host cell envelope during DNA ejection, essentially locking the phage into place.
Partially contracted tails were not present in any of the 2518 phage particles imaged, indicating that once contraction is initiated, it always goes to completion. Additionally, neither empty particles with extended tails nor particles with an open baseplate attached to extended tails were observed, strongly suggesting that once contraction occurs, tails are unable to return to the extended state. Furthermore, the apparent amount of buried surface area per subunit increases upon tail sheath contraction. Based on these observations, we speculate that, much like a mouse trap, the SBP8a tail sheath is initially assembled in a high energy state; presumably baseplate recognition of the cellular receptor provides the trigger allowing the tail to irreversibly contract and access its lowest energy conformation. A similar mechanism of contraction was proposed for bacteriophage T4 (Moody, 1967a, b, 1973; Müler et al., 1994), although the magnitude of contraction and rotation is not the same (Table 2). At the current resolution, the conformation of individual subunits is the same in both contracted and extended tails; however, the orientation of a given subunit changes upon switching states, suggesting that coordinated rigid body motion is sufficient to explain tail contraction.
The neck of phage SBP8a is located between the tail and the head, and may function as a gate regulating the passage of DNA from the capsid into the tail. Before tail contraction, the neck is closed, blocking the passage of DNA. The subunits at the capsid-end of the six helical strands are in direct contact with the neck proteins. Thus, as the terminal proteins of the six helical strands rotate around the long axis of the phage during contraction (see above), they will essentially drag the neck proteins into the “open gate” conformation and allow DNA to move from the head into the tail tube.
An additional density is present at the center of the “closed” base-plate in extended tails that is not present in the contracted tails. This “tip” protein may act as a plug that must be dislodged to allow DNA to leave the tail and enter the host cell cytoplasm. The shape and location of the tip protein is similar to the shape and location of the cell puncturing protein gp5 in phage T4 (Rossmann et al., 2004). In T4, the tip of this protein forms a predominantly negatively charged, needle-like structure that is ideally constructed to disrupt the negatively charged phospholipid cell membrane (Kanamaru et al., 2002). Furthermore, the T4 protein gp5 has lysozyme activity that may facilitate infection by degrading the Escherichia coli cell wall. Although the specific enzymatic activity necessary to degrade the B. anthracis cell wall is currently not known, we propose a similar structure, function, and mechanism of action for the SBP8a tip protein. Hence, in addition to functioning as a second gate to regulate DNA ejection, the tip protein may also play an active role in penetrating the host cell.
In summary, we propose the following model DNA ejection by phage SBP8a (Fig. 4): I. The short tail fibers attached to the baseplate recognize the receptor on the host surface, causing it to undergo a conformational change and assume the open conformation. The conformational change in the baseplate triggers tail sheath rotation and contraction, which drives the tail tube through the host cell membrane; II. The contraction signal is propagated to the neck region, causing the neck to open and permitting passage of DNA into the tail tube; III. DNA arrives at the distal end of the tail, and the tail tip is dislodged, allowing DNA to move into the host cell cytoplasm; IV DNA ejection continues until the entire genome has been ejected from the capsid into the host cell cytoplasm. In SBP8a, the length of the tail tube exposed upon contraction of the tail sheath and opening of the baseplate is approximately 60 nm, just long enough to traverse the cell wall and membrane of its B. anthracis host (Matias and Beveridge, 2005). We would expect that the tail lengths of other Myoviridae phages are tuned to the cell wall thickness of their host bacteria.
Fig. 4.
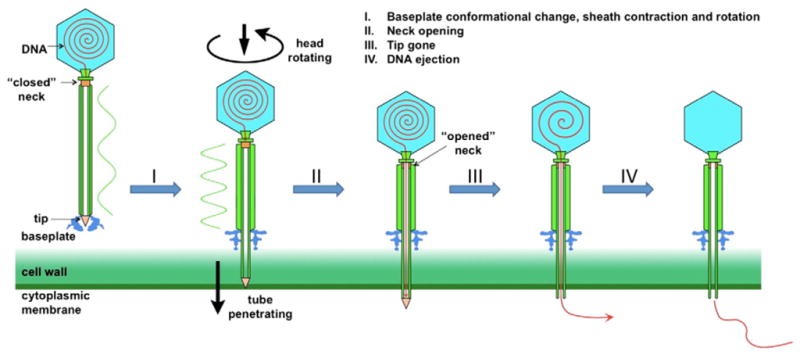
Mechanism of DNA ejection of phage SBP8a. I. The baseplate undergoes a dramatic conformational change after attaching to the host surface; the conformational change of the baseplate triggers tail sheath rotation and contraction, which drives the tail tube through the host cell membrane. II. Tail contraction is propagated to the neck region, forcing the neck to open and permitting passage of DNA into the tail tube. III. DNA arrives at the distal end of the tail, the tail tip is dislodged, and DNA to moves into the host cell cytoplasm. IV. All DNA in head has been ejected.
Materials and methods
Phage preparation and purification and culture
B. anthracis spore production
B. anthracis Sterne spores were grown according to a previously published protocol (Walter, 2003). Briefly, bacteria were incubated in brain–heart infusion broth at 37° until ~109 CFU/ml was reached. Spores were recovered in 10 ml water and incubated for 30 min at 65 °C to kill vegetative cells. Remaining spores were collected by low-speed centrifugation, re-suspended at approximately 108 CFU/ml in 50% glycerol, and stored at 4 °C.
Phage production
Wild assemblages of mixed phages were grown from an urban Iowa topsoil slurry and increased by standard soft-agar plates on B. anthracis Sterne using previously described methods (Walter and Baker, 2003). Phage assemblages or individual phages were suspended and stored (at 4 °C) in sterile PBS at pH 7.3 or in SM buffer (100 mM NaCl, 8 mM MgSO4, Tris–HCl, pH 7.5).
Selection for spore-binding phages
To identify spore-binding phages, 100 μl of mixed phage assembly (at approximately 108 PFU/ml) was mixed with 100 μl of a solution containing spores of B. anthracis Sterne in TSG buffer (10 mM Tris–HCl, 150 mM NaCl, 10 mM MgCl2, 0.3% gelatin, pH 7.4) at 37 °C for 60 min. Phage-spore slurry tubes were then centrifuged and the supernatant containing most phages removed. Spore pellets were washed several times with 500 μl SM buffer. The final 550 μl slurry was transferred to a fresh centrifuge tube, centrifuged as above, and the supernatant removed. SM buffer (100 μl) was then added to the pellet, mixed gently, and the entire process repeated two more times. After the third wash, the 100 μl spore/phage slurries were assayed for bound phage by standard plaque assays on B. anthracis Sterne or B. cereus 569 UM20. Plaque assays for phage bound to spores were indistinguishable in appearance from assays of diluted phage alone on B. anthracis Sterne lawns. Plaques were picked from plaque assays, triple-serially isolated and increased via standard soft-agar plates on B. anthracis Sterne using previously described methods (Walter, 2003). Individual phage isolates, including SBP8a, were suspended and stored at 4 °C in sterile PBS at pH 7.3 or in SM buffer. One of these phages, SBP8a, was selected for structural analysis by cryo-ET.
Cryo-ET data collection, structure determination and refinement
Initial negative-stain TEM images indicated that SBP8a was present in multiple morphological states. At the time, it was not possible to assess the degree of structural heterogeneity in the sample. For example, we could not ascertain the extent of tail contraction in every particle due to ambiguities arising from observing phage particles in projection; more subtle conformational changes would have been more difficult to resolve. Hence, the sample was imaged using cryoET since this technique provides three-dimensional information for each particle imaged, thus facilitating accurate classification of structural variations
Frozen-hydrated specimens were imaged at liquid nitrogen temperature using an FEI Polara electron microscope equipped with a field emission gun and a 4k×4k (16 megapixels) CCD camera (TVIPS; GMBH, Germany). The microscope was operated at 300 kV with a magnification of 23,000×, resulting in an effective pixel size of 3.9 Å. The FEI “batch tomography” program was used to collect low dose single-axis tilt series. Each data set was collected at −4 to −6 μm defocus, with a cumulative dose of ~100 e−/Å2 distributed over 65 images covering an angular range of 128°, with an angular increment of 2°. Marker free alignment was used to align and refine the tilted images (Winkler and Taylor, 2006). 3-D cryo tomograms were reconstructed from the aligned images using a weighted back-projection algorithm implemented in the package Protomo (Winkler and Taylor, 2006). To minimize the size of the data sets, and thus increase the speed of image processing, the data were binned by a factor of two, resulting in an effective pixel size of 7.8 Å.
Subvolume averaging of individual phage particles
A typical cryo-tomogram has low contrast and a poor signal-to-noise ratio, and also suffered by the effects of the “missing wedge”, which arises due to the fact that the data is collected from a limited angular tilt range in the microscope (e.g. from −65° to +65°). Subvolume averaging is therefore a crucial step to improve the signal-to-noise ratio and structural detail as described (Liu et al., 2009a, 2010; Winkler et al., 2009).
2518 particles (512×512×512 pixels) from 52 tomographic reconstructions were manually selected and visually classified into four classes based on the morphologies of head and tail: DNA-filled capsid, extended tail; DNA-filled capsid, contracted tail; DNA-emptied capsid, contracted tail, and partially DNA-filled capsid, contracted tail. These particles are oriented variably within the tomogram. Their approximate orientation (two of the three Euler angles) can be manually defined based on two coordinates: the center of the capsid and the end of the tail. This turns out to be a critical step, otherwise the full range of the three Euler angles must be scanned, which comes at a higher computational cost and is less reliable.
4×4×4 binned subvolumes (128×128×128 pixels) of the phage particles were generated for the initial rounds of alignment and classification. The subsequent rotational alignment is performed with an orientation search along the long axis of phage particle by maximizing the constrained cross-correlation (Forster et al., 2005). Structures with icosahedral heads and helical tails were apparent after multiple cycles of refinement and classification. Due to the symmetry mismatch between the head and the tail, 5- and 6-fold symmetry was applied to the head and tail regions, respectively. Finally, the original subvolumes (512×512×512) were used for further refinement and 3D averages.
Helical averaging of the tail sheath
The extended tail is flexible (data not shown); therefore only relatively straight tails were selected for 3-D sub-volume averaging. To improve the structures of both extended and contracted tails, the helical reconstruction method (Egelman, 2000) was adapted and applied to the tail sheath. Briefly, fifteen 80×80×200 voxel segments along each contractile tail and twenty-five segments along each extended tail were extracted, respectively. These segments were aligned with each other and were averaged to generate 3-D structures corresponding to the contractile and extended tails respectively. The helical parameters were estimated from the above averaged structures and were then imposed to improve the tail structures.
Map visualization and interpretation
3-D tomograms were visualized using IMOD (Kremer et al., 1996). UCSF Chimera (Pettersen et al., 2004) was utilized for the surface rendering of 3-D averages. Watershed segmentation based on density threshold levels (Volkmann, 2002) was used to initially segment substructures in the intact phage structures. Initial sub-structure assignments were confirmed and refined by comparison with the known structures of other well-determined phages (Fokine et al., 2004; Jiang et al., 2006; Leiman et al., 2004; Morais et al., 2005b; Xiang et al., 2006).
Acknowledgments
We thank Dr. James Stoops for editing and Dr. Michael Sherman for discussion. The project upon which this publication is based was performed pursuant to an agreement with The Methodist Hospital Research Institute which in turn was supported by the Grant DOD/TATRC Alliance for NanoHealth W81XWH-10-2-0125 from the U.S. Department of the Army. This work was supported partially by Welch Foundation Grant AU-1714 (to J.L.). The Polara electron microscope was supported, in part, through the Structural Biology Center at the UT Houston Medical School.
References
- Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, McPartland J, Kaganman I, Bowman VD, Rothman-Denes LB, Rossmann MG. Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J Mol Biol. 2008;378:726–736. doi: 10.1016/j.jmb.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Frutos M, Letellier L, Raspaud E. DNA ejection from bacteriophage T5: analysis of the kinetics and energetics. Biophys J. 2005;88:1364–1370. doi: 10.1529/biophysj.104.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, McKinstry M, Hwang J, Oppenheim AB, Fekete RA, Giulian G, Merril C, Nagashima K, Adhya S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc Natl Acad Sci U S A. 2006;103:4841–4845. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG, Eisenberg DS. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci U S A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG. Cryo-EM study of the pseudomonas bacteriophage [phi]KZ. Structure. 2007;15:1099–1104. doi: 10.1016/j.str.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Forster F, Medalia O, et al. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci U S A. 2005;102 (13):4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P, Han L, Winther T, Phillips R. Real-time observations of single bacteriophage λ DNA ejections in vitro. Proc Natl Acad Sci. 2007;104:14652–14657. doi: 10.1073/pnas.0703274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch A, Pous J, Páraga A, Valpuesta JM, Carrascosa JL, Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage [phi]29 connector particle. J Mol Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- Inamdar MM, Gelbart WM, Phillips R. Dynamics of DNA ejection from bacteriophage. Biophys J. 2006;91:411–420. doi: 10.1529/biophysj.105.070532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–317. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu S, Reedy MC, Winkler H, Lucaveche C, Cheng Y, Reedy MK, Taylor KA. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J Mol Biol. 2006;362:844–860. doi: 10.1016/j.jmb.2006.07.084. [DOI] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C ring assembly flexion. J Bacteriol. 2009;16:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wright ER, et al. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol. 2010;483:267–290. doi: 10.1016/S0076-6879(10)83014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Matias VR, Beveridge TJ. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol. 2005;56:240–251. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Moody MF. Structure of the sheath of bacteriophage T4 : I. Structure of the contracted sheath and polysheath. J Mol Biol. 1967;25:167–174. doi: 10.1016/0022-2836(67)90136-2. [DOI] [PubMed] [Google Scholar]
- Moody MF. Structure of the sheath of bacteriophage T4 : II. Rearrangement of the sheath subunits during contraction. J Mol Biol. 1967b;25:201–202. doi: 10.1016/0022-2836(67)90137-4. [DOI] [PubMed] [Google Scholar]
- Moody MF. Sheath of bacteriophage T4: III. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol. 1973;80:613–620. doi: 10.1016/0022-2836(73)90200-3. [DOI] [PubMed] [Google Scholar]
- Morais MC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage [phi]29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of [phi]29. Mol Cell. 2005a;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005b;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Müler M, Engel A, Aebi U. Structural and physicochemical analysis of the contractile MM phage tail and comparison with the bacteriophage T4 tail. J Struct Biol. 1994;112:11–31. doi: 10.1006/jsbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- Muller DJ, Engel A, Carrascosa JL, Velez M. The bacteriophage [phgr]29 head–tail connector imaged at high resolution with the atomic force microscope in buffer solution. EMBO J. 1997;16:2547–2553. doi: 10.1093/emboj/16.10.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plisson C, White HE, Auzat I, Zafarani A, Sao-Jose C, Lhuillier S, Tavares P, Orlova EV. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007;26:3720–3728. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol. 2004;14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Schofield DA, Westwater C. Phage-mediated bioluminescent detection of Bacillus anthracis. J Appl Microbiol. 2009;107:1468–1478. doi: 10.1111/j.1365-2672.2009.04332.x. [DOI] [PubMed] [Google Scholar]
- Schofield DA, Molineux IJ, Westwater C. Diagnostic bioluminescent phage for detection of Yersinia pestis. J Clin Microbiol. 2009;47:3887–3894. doi: 10.1128/JCM.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The Structure of the Phage T4 DNA Packaging Motor Suggests a Mechanism Dependent on Electrostatic Forces. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Takeda S, Suzuki M, Yamada T, Kageyama M, Arisaka F. Mapping of functional sites on the primary structure of the contractile tail sheath protein of bacteriophage T4 by mutation analysis. Biochim Biophys Acta, Proteins Proteomics. 2004;1699:163–171. doi: 10.1016/j.bbapap.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Tang S, Moayeri M, Chen Z, Harma H, Zhao J, Hu H, Purcell RH, Leppla SH, Hewlett IK. Detection of anthrax toxin by an ultrasensitive immunoassay using Europium nanoparticles. Clin Vaccine Immunol. 2009;16:408–413. doi: 10.1128/CVI.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N. A novel three-dimensional variant of the watershed transform for segmentation of electron density maps. J Struct Biol. 2002;138:123–129. doi: 10.1016/s1047-8477(02)00009-6. [DOI] [PubMed] [Google Scholar]
- Walter MH. Efficacy and durability of Bacillus anthracis bacteriophages used against spores. J Environ Heal. 2003:66. [PubMed] [Google Scholar]
- Walter MH, Baker DD. Three Bacillus anthracis bacteriophages from topsoil. Curr Microbiol. 2003;47:0055–0058. doi: 10.1007/s00284-002-3899-y. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Winkler H, Zhu P, et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165 (2):64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]


