Abstract
Inflammatory bowel disease (IBD) with its two distinct entities, Crohn’s disease and ulcerative colitis, and Type 1 diabetes mellitus (T1D) are autoimmune diseases. The prevalence of these diseases continues to rapidly rise in the industrialized world. Despite the identification of several genetic loci that are associated with both IBD and T1D, thus far, there is a paucity of epidemiological data to support a clinical overlap. In an effort to better understand the underlying pathogenic mechanisms of both IBD and T1D, this review summarizes the literature about these related autoimmune diseases, describes the most recent advances in their etiopathogenesis and emphasizes the genetic and nongenetic factors that exercise a differential influence. Genome-wide association studies have identified genetic loci with a role in immune response regulation that are linked to both IBD (particularly Crohn’s disease) and T1D. Some of these genetic loci (e.g., IL-18RAP) have a divergent role, conferring risk for one disease and protection for the other. Recent evidence highlights an important role of gut microbiota and cellular responses (e.g., endoplasmic reticulum stress) in the pathogenesis of both IBD and T1D.
Keywords: autoimmune diseases, Crohn’s disease, host genetics, IL-18RAP, SNP, Type 1 diabetes mellitus
Inflammatory bowel disease (IBD) comprises two distinct forms: Crohn’s disease (CD), which is a discontinuous, transmural inflammation of the small and/or large bowel, but can affect any part of the GI tract from mouth to anus; and ulcerative colitis (UC), which represents a bowel Inflammatory disorder restricted to the colonic mucosa. Type 1 diabetes mellitus (T1D) is an autoimmune disease (AID) characterized by the destruction of the insulin-producing pancreatic β-cells. In IBD, the important role of the immune response in its pathogenesis is widely accepted. On the other hand, the contribution of autoimmunity in the pathogenesis of IBD remains controversial [1] . CD is mainly a Th1-driven disorder, whereas UC is a predominantly Th2-mediated disease with a strong humoral component. The existence of circulating autoantibodies against colonic epithelial cells in UC patients has been reported in several studies [1–4]. In addition, the autoimmune response is important in the pathogenesis of extraintestinal manifestations present in 25% of IBD patients [1].
The incidence of both IBD and T1D has experienced a dramatic increase in recent decades, especially in the industrialized world [5, 6]. The concordance rate of T1D has been reported to be 20–50% in monozygotic twins and 5% in dizygotic twins [6–8]. CD and UC, which have a higher incidence among patient relatives have a monozygotic concordance of 30% for CD and 15% for UC, compared with 4% concordance in dizygotic twins for each IBD form [5,9] . Thus, although there is no question about the contribution of genetic susceptibility in IBD and T1D, genetics alone does not explain this rapid increase in incidence. Therefore, it appears that additional nongenetic/environmental-related factors contribute to the development of these AIDs [10,11]. Prospective cohort studies for T1D, such as DAISY in the USA, DIPP in Finland and the BABYDIAB in Germany, have focused on the individuals at increased risk to identify potential environmental causes for the disease [12] . Intriguingly, regardless of the identification of several genetic polymorphisms that are associated with both IBD and T1D, there is a lack of epidemiological data to support a clinical concurrence of these AIDs [13] . Plausible explanations would be that both IBD and T1D have common immune pathogenic pathways and that there is a divergent role of genetic and nongenetic factors, including the gut microbiota and endoplasmic reticulum (ER) stress, in the development of IBD and T1D. Here, we review the role of intestinal bacteria and that of enteroviruses in the pathogenesis of CD and T1D. In addition, we discuss the role of ER stress in the pathogenesis of IBD and T1D. ER stress, which comprises three distinct signaling cascades, could play a differential role in IBD and T1D pathogenesis through differential activation or disruption of unfolded protein response (UPR) signaling cascades in a target cell-specific manner (e.g., in intestinal epithelium cells in IBD and in insulin producing β-cells in T1D).
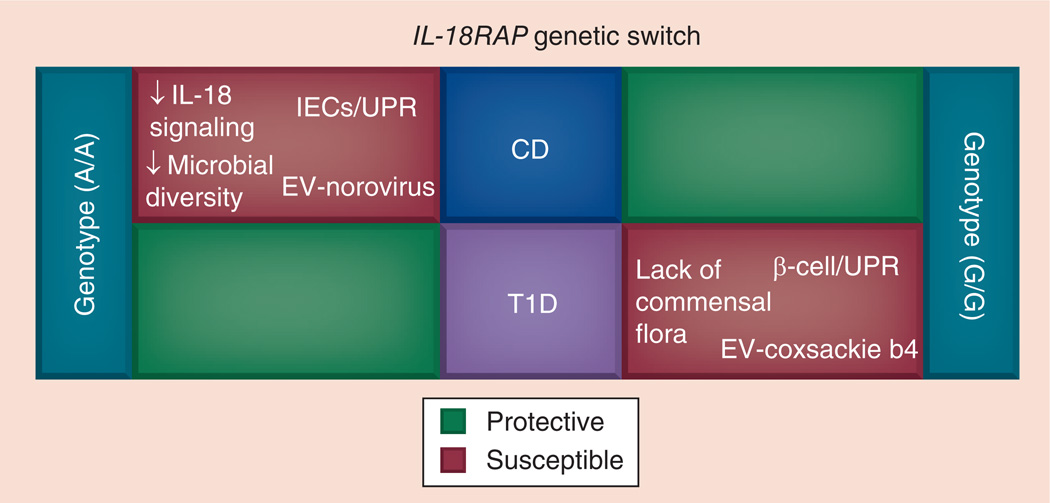
Also, we present an overview of potential interactions among implicated etiopathogenic factors that could play a divergent pathogenic role in IBD and T1D (Figure 1).
Figure 1. Potential interactions that influence the divergent control of IL-18RAP single nucleotide polymorphism in the pathogenesis of Crohn’s disease and Type 1 diabetes mellitus.
The IL-18RAP rs917997 A/A genotype confers risk for CD and protection against T1D, and vice versa is true for the G/G genotype. The overall risk of developing CD or T1D could be determined by the IL-18RAP rs917997 genotype, modulation of the IL-18 signaling pathway, specific changes in gut microbiota, EV (Norovirus for CD and Coxsackie B4 for T1D) infection of the gut mucosa and/or cell-specific dysfunction of UPR signaling. CD: Crohn’s disease; EV: Enterovirus; IEC: Intestinal epithelial cell; T1D: Type 1 diabetes mellitus; UPR: Unfolded protein response.
Host genetics
Genetic associations of AID have been mapped through two main methods: linkage studies and genome-wide association studies (GWAS). Linkage studies identify chromosomal regions that are associated with the disease in affected relatives more frequent than by chance, and are used when the predicted risk factors are rare. GWAS identify disease-associated genetic loci in case–control studies by using high-throughput single nucleotide polymorphism (SNP) genotyping platforms, and are employed when the risk factors are relatively common [14,15]. Large-scale sequencing projects, such as the 1000 Genome Project, with the aim to provide a reference database for human DNA polymorphisms in multiple human populations, are revolutionizing human genetics [16]. In addition, the development of new sequencing technologies, such as second-generation (also called next-generation sequencing) and third-generation sequencing, allows for a variety of complex analysis of whole genomes, exomes and transcriptomes [17,18]. The decrease in cost per base, the development of state-of-the-art sequencing technologies and the building of large genomic databases will make it possible to sequence the entire genome of an individual and compare it with genomic databases, bringing closer the area of personalized medicine. These novel DNA sequencing technologies will provide new avenues to diagnose genetic-based diseases and determine the role of genetics in multifactorial diseases such as IBD and T1D.
HLA genes
The HLA locus is on chromosome 6p. There are two HLA class genes: HLA-A, HLA-B and HLA-C represent class I genes; whereas HLA-DP, HLA–DQ and HLA-DR are class II genes. The HLA-DRB1*0103 is a HLA class II gene that is associated with colonic CD and severe and extensive UC in Caucasians [19–21]. On the other hand, ileal CD is associated with the HLA-DRB1*07 (odds ratio: 1.9; p = 0.006) [22,23]. In addition, the HLA class II genes have a strong association with T1D, conferring 30–50% of the genetic risk. The HLA DR3-DQ2/DR4-DQ8 genotype has the strongest association with T1D. Aly et al. reported that by 15 years of age, 5% of children with this genotype develop anti-islet autoimmunity and T1D, compared with only 0.3% in the general population [24]. T1D is also associated with HLA class I B*39 (involved in antigen presentation to cytotoxic CD8 cells), and HLA-A24 (associated with accelerated islet destruction) [25,26]. Interestingly, Wang et al. reported several strong risk alleles for T1D, located within the HLA complex, that confer protection against CD and UC [27].
Non-HLA genes
GWAS have been very successful in identifying genetic polymorphisms associated with the IBD and/or T1D. Susceptibility genes that are associated with IBD (CD and UC) include IL23R, IL10, SMAD3, ZMIZ1, CARD9, XBP1 and FUT2 [9]. Through a GWAS meta-analysis approach, Franke et al. identified a total of 71 risk loci associated with CD [28]. Wang et al. used these 71 loci in a novel approach involving logic regression to identify genetic interactions that improved CD risk prediction [29]. In a recent study, Doecke et al. identified the association of the rs3024505 SNP, which is located downstream of IL-10, with UC [30]. Several risk genes are specifically associated with CD, such as those that regulate intracellular bacterial processing (NOD2/CARD15) or autophagy (ATG16L1, IRGM and LRRK2) [31–34]. Instead, risk genes that have a role in regulation of epithelial defense functions (ECM1, CDH1, HNF4a and LAMB1) have been associated specifically with UC [20]. Similarly, SNPs of INS and IFIH1 genes have been associated with T1D [35,36]. Huang et al. used a 1000 Genomes-based imputation method to identify genetic polymorphisms associated with T1D (IL-2RA and CUX2) and CD (IL-23R) [37]. Bradfield et al. identified multiple associated loci, including SNPs located within the EFR3B and LMO7 genes, in a genome-wide meta-analysis of six T1D cohorts [38].
Recently, Jostins et al. reported one of the largest genetic experiments to date involving a complex disease [39]. In this study, the number of confirmed IBD susceptibility loci was increased to 163, substantially more than reported for any other complex disease. The authors discovered that 70% (113 out of 163) of the IBD loci are shared with other complex disease or traits, including 20 loci shared with T1D, which, from their calculations, is ten-times the number that would be expected by chance [39]. This overlap of genetic loci, most of which occurs on loci involved in immune response regulation (especially for CD and T1D), strongly suggests that these AIDs share common pathogenic pathways [13,35,40,41]. In comparison to UC that is associated with loci with a role in mucosal barrier function, CD is also associated with many loci involved in the immune response, indicating that pathogenesis of CD relies heavily on immune mechanisms. Furthermore, Wang et al. identified specific SNPs (e.g., PTPN22, IL-10, IL-27 and IL18RAP) that have opposite disease risk effects regarding CD and T1D, indicating that the same modulation of a specific pathogenic immune pathway can confer risk for one disease and protection for the other (Table 1) [27]. PTPN22 (R602W) is broadly associated with AIDs (CD, T1D, rheumatoid arthritis and systemic lupus erythematosus) and it confers risk for T1D and protection for CD [34,42]. Similarly, the rs3024505 SNP, which is located downstream of IL-10, is associated with T1D and IBD (CD and UC) conferring opposite risk effects [30,43,44]. Mutations of the IL-10R and IL-10 genes have been found in infants with severe (Crohn’s-like) colitis, whereas IL-10 knockout mice develop enterocolitis and represent a widely used IBD model [45–50]. In addition, the rs4788084 locus, located in proximity of IL-27 and NURP1 genes, is associated with CD and T1D, having opposite risk effects [27].
Table 1.
Single nucleotide polymorphisms associated with opposite risk effects in Inflammatory bowel disease and Type 1 diabetes
| Gene | SNP | MAF | CD (OR; 95% CI; p-value) | UC (OR; 95% CI; p-value) | T1D (OR; 95% CI; p-value) |
|---|---|---|---|---|---|
| IL-18RAP | rs917997 | 0.23 | 1.23 (1.13–1.34; 2.2 × 10−6)† | 1.08 (0.95–1.23; 0.25) | 0.87 (0.77–0.97; 0.016)† |
| PTPN22 | rs2476601 | 0.09 | 0.72 (0.62–0.83; 6.6 × 10−6)† | 0.89 (0.73–1.09; 0.27) | 1.99 (1.74–2.27; 4.92 × 10−25)† |
| IL-10 | rs3024505 | 0.15 | 1.24 (1.12–1.37; 2.1 × 10−5)† | 1.26 (1.09–1.45; 1.9 × 10−3)† | 0.76 (0.66–0.88; 1.5 × 10−4)† |
| NUPR1/IL27 | rs4788084 | 0.39 | 1.23 (1.14–1.33; 1.4 × 10−7)† | 1.08 (0.96–1.21; 0.19) | 0.88 (0.79–0.97; 9.7 × 10−3)† |
Values that represent significant associations.
CD: Crohn’s disease; MAF: Minor allele frequency; OR: Odds ratio; SNP: Single nucleotide polymorphism; T1D: Type 1 diabetes mellitus; UC: Ulcerative colitis.
Data taken from [27].
Recent GWAS have associated IL-18RAP rs917997 SNP (G > A) with celiac disease, CD and T1D [27,51,52]. The minor allele A of rs917997 SNP confers risk for developing celiac disease and CD, and protection for T1D. The IL-18RAP rs917997 SNP has been shown to have a cis effect on IL-18RAP mRNA expression in the peripheral whole blood of celiac disease patients [53]. A gene–dose effect exercised by the A risk/ rare allele was observed in these subjects. The A/A minor allele homozygotes were associated with the lowest IL-18RAP mRNA levels relative to G/A heterozygotes and G/G wild-type subjects [53]. It is highly likely that IL-18RAP mRNA expression in CD and T1D patients is affected similarly to celiac disease patients. Therefore, it is quite intriguing to decipher how these types of SNPs can exert such opposite disease risk effects. Most likely, AIDs, such as CD or T1D, are multifactorial and are determined by the complex interaction of genetic predisposition, disease specific immune pathways, target cell and/or organ-related mechanisms (e.g., ER stress) and changes in gut microbiota. Figure 1, is a schematic representation of the concept of genetic switch that has emerged as a result of the identification of genetic loci that confer opposite disease risk effects. We use the IL-18RAP rs917997 SNP as an example to describe how the same resulting genotypes (e.g., A/A or G/G), under the differential influence of other genetic and environmental factors, could lead to the development of a particular AID and protection against another. The homozygous rs917997 A/A genotype confers increased genetic risk for CD. A combination of the downregulation of IL-18 signaling (IL-18RAP A allele-associated), a decrease in the diversity of gut microbiota, a gut mucosal infection with an IBD-related Enterovirus (e.g., Norovirus) and/or coexistence of other mutations that could affect the capacity of target intestinal epithelial cells to properly handle ER stress (e.g., the dysfunction of XBP1, shown to be associated with IBD and colitis) could lead to the development of CD. Conversely, in individuals with the IL-18RAP G/G genotype, lack of commensal microbiota, gut mucosal infection with a T1D related Enterovirus (e.g., Coxsackie b4) and/or the disruption of the PKR-like ER kinase (PERK)-mediated arm of UPR, could lead to T1D (Figure 1).
Immune response mechanisms in IBD & T1D
Several overlapping susceptibility loci among IBD and T1D have a regulatory role in the immune response [13]. The shared pathogenesis of IBD and T1D is also indicated by the common features that the two target organs, the bowel and pancreas share: endodermal (one of the three primary germ layers) origin during organogenesis, anatomic proximity and an integrated lymphatic system.
Cytokines (IL-18 & IL-18RAP)
The pathogenic mechanisms of CD and T1D involve aberrant innate and adaptive immune responses in which cytokines play a central role [54,55]. The role of proinflammatory (e.g., IFN-γ, TNF-α, IL-1α and IL-1β, IL-2, IL-6, IL-8, IL-12, IL-17, IL-18, IL-21 and IL-23) and antiinflammatory or Th2 cytokines (IL-4, IL-10 and IL-13) in IBD is reviewed elsewhere by Muzes et al. [54]. The authors emphasize that the inflammation regulatory pathways may not be mutually exclusive, as previously believed, because individual cytokines can have diverse and even opposing functions in various clinical and immunological settings. In depth knowledge about their complex role in the pathogenesis of IBD is of paramount importance in devising targeted biological therapies.
Several cytokines, with well-defined roles in the pathogenesis of IBD have also been reported to play an important role in T1D pathogenesis [56–58]. This group includes TNF-α, IL-1, IL-21 and IL-23. Previous studies identified IL-21 as a central cytokine in β-cell destruction in mouse models. Recent data from a study in NOD mice by Thomas et al. suggest that blocking Inflammatory cytokines, especially IL-1 and TNF-α, may increase the capacity of the immune system to suppress T1D through Tregs [58]. Nepom et al. reviewed the role of several cytokines believed to be directly implicated in the pathogenesis of T1D, suggesting that selective cytokine blockade could be employed as a component of preventive or interventional immunotherapy [57].
Interestingly, even if T1D is thought to be a T-cell-mediated disorder, there is ample evidence of innate immune defects in the pathogenesis of T1D (e.g., serum levels of IL-18 are increased in autoantibody-positive T1D patients) [59]. Also, both serum and mucosal levels of IL-18 are increased in CD patients compared with UC patients who do not show elevated levels [60, 61]. Thus, IL-18 levels could represent a useful and specific clinical marker for CD. An important observation regarding the role of IL-18 in the pathogenesis of bowel inflammation was made in NLRP6 inflammosome knockout mice, which developed spontaneous colitis associated with low levels of IL-18 both in the serum and colonic explants [62].
IL-18, a member of IL-1 family of cytokines, plays a role in both innate and adaptive immune responses. IL-18 is produced by APCs, T and NK cells, and is a stimulator of Th1 cells and an activator of NK cells. The precursor IL-18 protein is cleaved by caspase-1 to produce the mature IL-18 protein [63]. IL-18 binding to the ligand-binding subunit (IL-18 receptor-α) of the IL-18 receptor heterodimeric complex is followed by the recruitment of the signal-transducing subunit, the accesory protein IL-18RAP and the intracellular adaptor protein MyD88. The signaling cascade continues with the phosphorylation of the IL-1 receptor kinases and MAPKs. Next, the NF-κB pathway is activated and the production of proinflammatory cytokines, such as IFN-γ, TNF-α and IL-1β, is elevated [63].
Given the involvement of the IL-18 pathway in the pathogenesis of both T1D and IBD, this signaling cascade could be targeted therapeutically in both disorders. The divergent influence of IL-18RAP rs917997 SNP in CD and T1D indicates a differential role of the IL-18 pathway in their pathogenesis and, consequently, its manipulation for therapeutic purposes will have to be differential and disorder specific. However, in patients that already have developed CD, characterized by a general hyperinflammatory state, and in subjects at high risk for T1D with high serum levels of IL-18, targeting IL-18 with antagonists or blocking antibodies could prove beneficial.
Macrophages & dendritic cells
Intestinal macrophages located in the subepithelial lamina propria represent the largest fraction of tissue macrophages in the human body. Their role is to guard against bacterial pathogens and also to maintain homeostasis of the gut mucosa [64]. Macrophages and dendritic cells (DCs) are key to innate and adaptive immune response polarization [65]. In the intestine, M1 and M2 macrophages play several important roles: antibacterial, proinflammatory (M1 macrophages via TNF-α, IL-12 and IL-23) and anti-inflammatory (M2 macrophages via IL-10 and TGF-β) [66,67]. The important role of macrophages in intestinal inflammation was revealed from IL-10−/− mice that develop spontaneous chronic colitis only in the presence of macrophages, but not when they are depleted [68]. DCs are among the most potent APCs. Intestinal DCs sample antigens from the lumen and, after antigen uptake, migrate to mesenteric lymph nodes. Sakuraba et al. demonstrated that DCs in the mesenteric lymph nodes of CD patients promote Th1 and/or Th17 skewed immune responses [69]. In T1D, during the process of β-islet destruction, there is an infiltration of the islets by macrophages, DCs, T and B cells [70]. In the NOD mouse, which is a model of spontaneous T1D, the inactivation of macrophages protects against T-cell-mediated autoimmune diabetes [71] , whereas DCs prime effector T cells and activate Tregs. This dual role of DCs could be very well exploited in antigen-specific DC-based therapies for T1D [72].
NK cells
NK cells are major players of the antitumoral and antiviral immune responses [73]. In addition, NK cells play a role in immune response regulation and autoimmunity. The regulatory role of NK cells relies on their ability to express both activating (e.g., NKG2D, NKp46, NKp30) and inhibiting (e.g., CD94-NKG2A, KIR2DL1/2) receptors, and their capacity to interact and influence the activation of other immune cells such as DCs, macrophages and T cells. Human NK cells are characterized as CD3−CD56+ cells and are located both in the circulatory system and in the tissues. Blood NK cells are divided into two subtypes: NK CD56bright that are CD16− and secrete cytokines in response to IL-12 and IL-18 stimulation; and CD56dim that express high levels of CD16 and are predominantly cytotoxic, whereas, intestinal NK cells express RORC, CD127 and NKp44 in humans; and RORgt and NKp46 in mice. Several reports have described intestinal NK cell involvement in the pathogenesis of IBD [74]. Steel et al. reported that CD16+ NK cells are enriched in the lamina propria of CD and UC patients compared with unaffected controls and that azathioprine reduced that cell population [75]. In addition, earlier studies have demonstrated that IBD patients had low NK cell function [76].
Regarding the involvement of NK cells in T1D, Dotta et al. reported that β-cell infection with Enterovirus Coxsackie B4 was associated with NK cell infiltration of pancreatic islets in three out of six patients with recent onset T1D [77]. Also, through experiments in the NOD mouse model, Brauner et al. observed that NK cell infiltration of endocrine and exocrine pancreas was present at an early age and preceded T-cell infiltration [78]. These findings denote an immunomodulatory role of NK cells in pancreatic islet inflammation. In addition, Feuerer et al. demonstrated that Treg depletion in NOD mice led to earlier disease onset by a mechanism involving increased NK cell activity within the pancreatic islets [79].
T cells/Tregs
T1D is the result of the autoimmune destruction of insulin secreting β-cells that is mediated by self-reactive T cells, and is controlled by Tregs [80]. CD4+ Tregs are characterized by the expression of the transcription factor FOXP3 [81,82]. Tregs maintain peripheral tolerance to self-antigens by controlling the activation and expansion of self-reactive T cells [80].
Similarly, IBD pathogenesis is mediated by CD4 and CD8 T cells. CD is considered to be mainly a Th1-mediated disease due to Th1 cytokine (IL-12, IFN-γ and TNF-α) involvement. On the other hand, UC is thought to be a Th2 disorder driven primarily by IL-5 and IL-13 [5 4]. The Treg mechanisms of immunosuppression in IBD are mediated by cytokines (IL-10 and TGF-β1) and cell–cell interactions involving perforin/granzyme cytotoxicity. CTLA-4, a Treg-specific receptor, contributes to the Treg suppression function through its high affinity for B7 ligands [83,84].
There is ample evidence that Tregs inhibit NK cell activity in vivo and in vitro [85]. This crosstalk between Tregs and NK cells could be exploited therapeutically in T1D, IBD and cancer [86].
ER stress (XBP1 & PERK)
The ER is a cellular organelle that is responsible for the synthesis, folding and processing of newly synthesized secretory and membrane proteins. The imbalance between protein load and folding capacity of the ER is defined as ER stress, and the coordinated response triggered is known as UPR [87]. The ER resident transmembrane proteins that serve as ER stress sensors include: IRE1, PERK and ATF6. ER-stress triggers the RNase activity of the ER-bound protein IRE1. A precursor XBP1 mRNA is uniquely spliced by IRE1 and ligated by tRNA ligase in response to ER stress. The spliced XBP1 mRNA encodes the protein XBP1 that binds to the UPR target genes [87].
ER stress has been implicated in the pathogenesis of both IBD and T1D. Recent evidence demonstrates that XBP1 links ER stress to intestinal inflammation and human IBD [88]. Kaser et al. used cohorts of IBD (CD and UC) patients to test the association of 20 SNPs across the XBP1 gene region [88]. The authors found that three SNPs rs5997391, rs5762795 and rs35873774 are associated with IBD. The rs35873774 SNP, which is located in intron 4/5 of the XBP1, showed the strongest association (for the C minor allele; odds ratio: 0.74; 95% CI: 0.66–0.84). Xbp1 deletion in intestinal epithelial cells in mice causes ER stress and spontaneous enteritis with histological features of IBD, revealing the ER stress pathway as a genetic contributor to IBD [89]. Conversely, PERK protein signaling appears to be essential in protection against T1D. Perk knockout mice and humans with Wolcott–Rallison syndrome that have mutations in the PERK-encoding EIF2AK3 gene, develop severe diabetes mellitus [90–93]. Thus, this evidence indicates that the differential disruption of UPR signaling in a target cell-specific manner (i.e., in intestinal epithelial cells in IBD and in β-cells in T1D) contributes to the pathogenesis of IBD and T1D.
Gut microbiota
Gut homeostasis
The human GI tract hosts approximately 10 trillion microbes. There is a mutual relationship between the host and gut microbiota. The host provides the habitat and the required nutrients, whereas gut microbiota contributes toward the development of innate and adaptive immunity of the host, maintaining the gut epithelial barrier integrity and protecting against microbial pathogens, and vitamin biosynthesis and nutrient metabolism [94]. An interesting report by Elinav et al. revealed the importance of the host immune system in maintaining gut microbiota homeostasis in mice deficient in NLRP6, ASC, caspase-1 or IL-18 [62]. These NLRP6 inflammosome-deficient mice developed an altered colitogenic microbiota characterized by the expansion of bacterial phyla Bacteroidetes (Prevotellaceae) and TM7, also implicated in the development of human IBD [95,96]. In a recent GWAS, Jostins et al. identified 20 IBD loci shared with primary immunodeficiencies (4.9-fold enrichment) involving skin infections with Staphylococcus and candidiasis, as well as mycobacterial infections [39]. These results focus increasingly closer attention on the interaction between the host mucosal immune system and microbes, both at the epithelial surface level and within the gut lumen. In particular, the results raise the question, in the context of this burden of IBD susceptibility genes, as to what triggers the components of commensal microbiota to switch from a symbiotic to a pathogenic relationship with the host [39].
Gut microbiota in IBD & T1D
The lack of intestinal inflammation in germ-free mice and the reduction of inflammation upon diversion of fecal stream or antibiotic treatment in humans and animal models denote an essential role for gut microbiota in IBD pathogenesis [97,98]. Emerging evidence strongly indicates that the interaction of gut microbiota with mucosal immune cells is responsible for intestinal inflammation and the development of IBD [99]. Studies on fecal and gut mucosa-associated microbiota have demonstrated dysbiosis in CD and UC patients [100,101]. Ott et al. observed a reduction in bacterial diversity in CD and UC patients consisting mainly in a decrease of normal anaerobic species Bacteroides, Eubacterium and Lactobacillus [102]. A metagenomic study by Manichanh et al. used fecal samples to demonstrate that there is a decrease in microbial diversity of the phylum Firmicutes, (especially of Clostridium leptum group) in CD patients compared with healthy control subjects [101].
Regarding microbiota in T1D, data from the DIPP study in Finland revealed a reduction in Firmicutes and an increase in Bacteroidetes in children with T1D compared with age- and HLA-matched healthy controls [11]. The essential role of gut microbiota in T1D was demonstrated in MyD88-deficient NOD mice. Under specific pathogen-free conditions, MyD88 knockout NOD mice did not develop T1D, which is in sharp contrast to germ-free MyD88 knockout NOD mice that developed diabetes [103]. Thus, there is a divergent role of microbiota in IBD versus T1D. The presence of gut microbiota promotes the development of IBD, whereas distinct strains play a protective role against T1D. Furthermore, given the evidence of the involvement of gut microbiota in the pathogenesis of an AID, fecal microbiota transplantation, which has been successfully used for the treatment of recurrent Clostridium difficile infections and IBD (refractory UC and CD), holds great potential for the therapy of other immune-mediated disorders including T1D [104,105].
Enteroviruses in IBD & T1D
Recent evidence shows that T1D is associated with Enterovirus infection in the intestinal mucosa of patients [106,107]. Nevertheless, the debate over the mechanisms involved in β-cell destruction is open and in critical need for specific answers. Potential mechanisms that lead to β-cell destruction by diabetogenic enteroviral strains (e.g., Coxsackie B4) include: triggering of an innate immune response that causes a break in immunological tolerance; molecular mimicry which is autoantibody mediated; and direct β-cell infection with pancreatotropic viruses [108–111]. Conversely, there are limited data on the role of Enteroviruses in IBD. Khan et al. reported that Norovirus, from the family of Caliciviridae, which accounts for the most cases of gastroenteritis worldwide, was detected in the stool samples of pediatric IBD (UC and CD) patients, with exacerbations that presented with bloody diarrhea and required hospitalization [112] . Enteroviruses could trigger IBD or β-cell apoptosis in T1D directly or through the disruption of the barrier function of the intestinal epithelium causing a leaky gut. Deeper access of the bacteria present in the intestinal lumen can cause bowel wall inflammation in IBD and autoantibody-mediated β-cell apoptosis due to bacterial epitope cross-reactivity in T1D [113–115]. Whereas the specific Enteroviruses that could trigger the IBD (e.g., bowel inflammation – Norovirus) or T1D (e.g., diabetogenic–Coxsackie B4) might be different, the Enterovirus infection of the gut mucosa might represent a common phenomenon for both IBD and T1D, indicating that the gut mucosa and Enterovirus infection could be targeted therapeutically in both disorders.
Conclusion
The identification of several genetic polymorphisms that are linked to both IBD and T1D indicate shared pathogenic pathways between these AIDs. The overlap of genetic loci with a role in immune response, especially for CD and T1D, denotes the involvement of immune-related mechanisms, such as those mediated by the IL-18 pathway that could be targeted therapeutically in both diseases. In addition, the divergent role that particular SNPs have in CD versus T1D could be explained by the genetic switch effect of other factors, such as gut microbiota and specific cellular responses (Figure 1).
Future perspective
The discovery of novel genetic and nongenetic factors that influence the development of AID will enable an integrative diagnostic approach (e.g., clinical presentation, serum markers, genotyping and gut microbiota analysis) and the application of personalized medicine. Further understanding of shared immune pathways (e.g., IL-18) and their divergent effects among AIDs, such as IBD and T1D, will contribute toward the identification of novel therapeutic targets and tailoring of more effective treatment strategies.
Executive summary.
The prevalence of inflammatory bowel disease and Type 1 diabetes mellitus (T1D) continues to rapidly rise in the industrialized world.
Susceptibility to inflammatory bowel disease and T1D is multifactorial and includes genetic and nongenetic factors.
Genome-wide association studies have identified single nucleotide polymophisms with a role in the immune response that link Crohn’s disease (CD) and T1D.
IL-18RAP rs917997 SNP confers opposite risk effect for CD and T1D.
Gut microbiota and cell-specific endoplasmic reticulum stress are factors that could influence the divergent control of the pathogenesis of CD and T1D.
Acknowledgments
The authors wish to thank M Mohamadzadeh, J Owen and B Sahay for the critical review of the manuscript.
Footnotes
Financial & competing interests disclosure
This study was supported by a NIH grant (RO1 CA113975-A2) and a Gatorade fund to SC Glover, and a JDRF Career Development Award (2-2012-280) and an American Diabetes Association grant (7–13-BS-022) to TM Brusko. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Das KM, Biancone L. Is IBD an autoimmune disorder? Inflamm. Bowel Dis. 2008;14(Suppl. 2):S97–S101. doi: 10.1002/ibd.20723. [DOI] [PubMed] [Google Scholar]
- 2.Broberger O, Perlmann P. Autoantibodies in human ulcerative colitis. J. Exp. Med. 1959;110:657–674. doi: 10.1084/jem.110.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandtzaeg P. Autoimmunity and ulcerative colitis: can two enigmas make sense together? Gastroenterology. 1995;109:307–312. doi: 10.1016/0016-5085(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 4.Fiocchi C, Roche JK, Michener WM. High prevalence of antibodies to intestinal epithelial antigens in patients with inflammatory bowel disease and their relatives. Ann. Intern. Med. 1989;110:786–794. doi: 10.7326/0003-4819-110-10-786. [DOI] [PubMed] [Google Scholar]
- 5.Brant SR. Promises, delivery, and challenges of inflammatory bowel disease risk gene discovery. Clin. Gastroenterol. Hepatol. 2013;11:22–26. doi: 10.1016/j.cgh.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Dahlquist G. The aetiology of Type 1 diabetes: an epidemiological perspective. Acta Paediatrica. 1998;425:5–10. doi: 10.1111/j.1651-2227.1998.tb01244.x. [DOI] [PubMed] [Google Scholar]
- 7.Tattersall RB, Pyke DA. Diabetes in identical twins. Lancet. 1972;2:1120–1125. doi: 10.1016/s0140-6736(72)92720-1. [DOI] [PubMed] [Google Scholar]
- 8.Kaprio J, Tuomilehto J, Koskenvuo M, et al. Concordance for Type 1 (insulin-dependent) and Type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–1067. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- 9.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704–1712. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd JA. D’oh! Genes and environment cause Crohn’s disease. Cell. 2010;141:1114–1116. doi: 10.1016/j.cell.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for Type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nokoff N, Rewers M. Pathogenesis of Type 1 diabetes: lessons from natural history studies of high-risk individuals. Ann. NY Acad. Sci. 2013;1281:1–15. doi: 10.1111/nyas.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 14.Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn’s disease. Ann. Rev. Genom. Hum. Genet. 2009;10:89–116. doi: 10.1146/annurev-genom-082908-150013. [DOI] [PubMed] [Google Scholar]
- 15.Grant SF, Hakonarson H. Genome-wide association studies in Type 1 diabetes. Curr. Diabetes Reports. 2009;9:157–163. doi: 10.1007/s11892-009-0026-5. [DOI] [PubMed] [Google Scholar]
- 16.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. ▪▪ Reports results of the pilot phase of the 1000 Genomes Project
- 17.Morey M, Fernandez-Marmiesse A, Castineiras D, et al. A glimpse into past, present, and future DNA sequencing. Mol. Genet. Metab. 2013;110:3–24. doi: 10.1016/j.ymgme.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93:105–111. doi: 10.1016/j.ygeno.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Silverberg MS, Mirea L, Bull SB, et al. A population- and family-based study of canadian families reveals association of HLA DRB1*0103 with colonic involvement in inflammatory bowel disease. Inflamm. Bowel Dis. 2003;9:1–9. doi: 10.1097/00054725-200301000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Thompson AI, Lees CW. Genetics of ulcerative colitis. Inflamm. Bowel Dis. 2011;17:831–848. doi: 10.1002/ibd.21375. [DOI] [PubMed] [Google Scholar]
- 21.Lappalainen M, Halme L, Turunen U, et al. Association of IL23R, TNFRSF1A, and HL A-DR B1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm. Bowel Dis. 2008;14:1118–1124. doi: 10.1002/ibd.20431. [DOI] [PubMed] [Google Scholar]
- 22.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman B, Silverberg MS, Gu X, et al. Card15 and HLA DRB1 alleles influence susceptibility and disease localization in Crohn’s disease. Am. J. Gastroenterol. 2004;99:306–315. doi: 10.1111/j.1572-0241.2004.04038.x. [DOI] [PubMed] [Google Scholar]
- 24.Aly TA, Ide A, Humphrey K, et al. Genetic prediction of autoimmunity: initial oligogenic prediction of anti-islet autoimmunity amongst DR3/DR4-DQ8 relatives of patients with Type 1a diabetes. J. Autoimmunity. 2005;25(Suppl.):40–45. doi: 10.1016/j.jaut.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Noble JA, Valdes AM, Varney MD, et al. HLA class I and genetic susceptibility to Type 1 diabetes: results from the Type 1 diabetes genetics consortium. Diabetes. 2010;59:2972–2979. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nejentsev S, Howson JM, Walker NM, et al. Localization of Type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Baldassano R, Zhang H, et al. Comparative genetic analysis of inflammatory bowel disease and Type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. ▪▪ Used genome-wide association studies to find shared genetic loci among Crohn’s disease (CD), ulcerative colitis and Type 1 diabetes (T1D). PTPN22, IL27, IL18RAP and IL10 genetic loci were identified as associated with opposite disease risk effects in CD and T1D.
- 28.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang MH, Fiocchi C, Ripke S, et al. A novel approach to detect cumulative genetic effects and genetic interactions in Crohn’s disease. Inflamm. Bowel Dis. 2013;19(9):1799–1808. doi: 10.1097/MIB.0b013e31828706a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doecke JD, Simms LA, Zhao ZZ, et al. Genetic susceptibility in IBD: overlap between ulcerative colitis and Crohn’s disease. Inflamm. Bowel Dis. 2013;19:240–245. doi: 10.1097/MIB.0b013e3182810041. [DOI] [PubMed] [Google Scholar]
- 31.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 32.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene ATG16L1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Concannon P, Rich SS, Nepom GT. Genetics of Type 1a diabetes. N. Engl. J. Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 36.Smyth DJ, Cooper JD, Bailey R, et al. A genome-wide association study of nonsynonymous snps identifies a Type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome trust case-control consortium Phase 1 data. Eur. J. Hum. Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. ▪▪ Identified genetic polymorphisms associated with T1D and CD using 1000 Genomes-based imputation.
- 38.Bradfield JP, Qu HQ, Wang K, et al. A genome-wide meta-analysis of six Type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 2011;7:e1002293. doi: 10.1371/journal.pgen.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jostins L, Ripke S, Weersma RK, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. ▪▪ Reported the results of one of the largest genetic experiments involving a complex disease; the number of confirmed inflammatory bowel disease (IBD) susceptibility loci was increased to 163, and 20 loci were identified to be shared with T1D.
- 40.Onengut-Gumuscu S, Concannon P. The genetics of Type 1 diabetes: lessons learned and future challenges. J. Autoimmunity. 2005;25(Suppl.):34–39. doi: 10.1016/j.jaut.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Claesson MH, Nicoletti F, Stosic-Grujicic S, Doria A, Zampieri S. Interactions between infections and immune-inflammatory cells in Type 1 diabetes mellitus and inflammatory bowel diseases: evidences from animal models. Clin. Exper. Rheumatol. 2008;26:S8–S11. [PubMed] [Google Scholar]
- 42.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with Type I diabetes. Nat. Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 43.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and metaanalysis find that over 40 loci affect risk of Type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersen V, Ernst A, Christensen J, et al. The polymorphism rs3024505 proximal to IL-10 is associated with risk of ulcerative colitis and Crohns disease in a Danish case-control study. BMC Med. Genet. 2010;11:82. doi: 10.1186/1471-2350-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glocker EO, Kotlarz D, Boztug K, et al. inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glocker EO, Kotlarz D, Klein C, Shah N, Grimbacher B. IL-10 and IL-10 receptor defects in humans. Ann. NY Acad. Sci. 2011;1246:102–107. doi: 10.1111/j.1749-6632.2011.06339.x. [DOI] [PubMed] [Google Scholar]
- 47.Kotlarz D, Beier R, Murugan D, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143:347–355. doi: 10.1053/j.gastro.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 48.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 49.Hagenbaugh A, Sharma S, Dubinett SM, et al. Altered immune responses in interleukin 10 transgenic mice. J. Exper. Med. 1997;185:2101–2110. doi: 10.1084/jem.185.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glocker EO, Frede N, Perro M, et al. Infant colitis – it’s in the genes. Lancet. 2010;376:1272. doi: 10.1016/S0140-6736(10)61008-2. [DOI] [PubMed] [Google Scholar]
- 51.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 2009;10:43–55. doi: 10.1038/nrg2489. ▪▪ Analyzes shared genetics and pathogenesis of 11 immune-related disorders.
- 52.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in Type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hunt KA, Zhernakova A, Turner G, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muzes G, Molnar B, Tulassay Z, Sipos F. Changes of the cytokine profile in inflammatory bowel diseases. World J. Gastroenterol. 2012;18:5848–5861. doi: 10.3748/wjg.v18.i41.5848. ▪▪ Thorough review on the complex role of pro- and anti-inflammatory cytokines involved in the pathogenesis of IBD.
- 55.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol. Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 56.Grunnet LG, Mandrup-Poulsen T. Cytokines and Type 1 diabetes: a numbers game. Diabetes. 2011;60:697–699. doi: 10.2337/db10-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nepom GT, Ehlers M, Mandrup-Poulsen T. Anti-cytokine therapies in T1D: concepts and strategies. Clin. Immunol. 2013 doi: 10.1016/j.clim.2013.02.003. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas HE, Graham KL, Chee J, et al. Proinflammatory cytokines contribute to development and function of regulatory T cells in Type 1 diabetes. Ann. NY Acad. Sci. 2013;1283:81–86. doi: 10.1111/j.1749-6632.2012.06797.x. [DOI] [PubMed] [Google Scholar]
- 59.Hanifi-Moghaddam P, Schloot NC, Kappler S, Seissler J, Kolb H. An association of autoantibody status and serum cytokine levels in Type 1 diabetes. Diabetes. 2003;52:1137–1142. doi: 10.2337/diabetes.52.5.1137. [DOI] [PubMed] [Google Scholar]
- 60.Pizarro TT, Michie MH, Bentz M, et al. IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn’s disease: expression and localization in intestinal mucosal cells. J. Immunol. 1999;162:6829–6835. [PubMed] [Google Scholar]
- 61.Furuya D, Yagihashi A, Komatsu M, et al. Serum interleukin-18 concentrations in patients with inflammatory bowel disease. J. Immunother. 2002;25(Suppl. 1):S65–S67. doi: 10.1097/00002371-200203001-00010. [DOI] [PubMed] [Google Scholar]
- 62.Elinav E, Strowig T, Kau AL, et al. Nlrp6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dinarello CA. Interleukin-18 and the pathogenesis of inflammatory diseases. Semin. Nephrol. 2007;27:98–114. doi: 10.1016/j.semnephrol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Schenk M, Mueller C. Adaptations of intestinal macrophages to an antigen-rich environment. Semin. Immunol. 2007;19:84–93. doi: 10.1016/j.smim.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Hisamatsu T, Kanai T, Mikami Y, et al. Immune aspects of the pathogenesis of inflammatory bowel disease. Pharmacol. Ther. 2013;137:283–297. doi: 10.1016/j.pharmthera.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mantovani A, Sica A, Sozzani S, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 68.Watanabe N, Ikuta K, Okazaki K, et al. Elimination of local macrophages in intestine prevents chronic colitis in interleukin-10-deficient mice. Dig. Dis. Sci. 2003;48:408–414. doi: 10.1023/a:1021960401290. [DOI] [PubMed] [Google Scholar]
- 69.Sakuraba A, Sato T, Kamada N, et al. Th1/Th17 immune response is induced by mesenteric lymph node dendritic cells in Crohn’s disease. Gastroenterology. 2009;137:1736–1745. doi: 10.1053/j.gastro.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 70.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol. Rev. 2011;91:79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 71.Jun HS, Santamaria P, Lim HW, Zhang ML, Yoon JW. Absolute requirement of macrophages for the development and activation of beta-cell cytotoxic CD8+ T-cells in T-cell receptor transgenic NOD mice. Diabetes. 1999;48:34–42. doi: 10.2337/diabetes.48.1.34. [DOI] [PubMed] [Google Scholar]
- 72.Han S, Donelan W, Reeves W, Yang L-J. Autoantigen-specific immunotherapy. In: D Wagner., editor. Type 1 Diabetes – Pathogenesis, Genetics and Immunotherapy. Croatia: InTech, Rijeka; 2011. [Google Scholar]
- 73.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142:847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takayama T, Kamada N, Chinen H, et al. Imbalance of NKP44(+)NKP46(−) and NKP44(−)NKP46(+) natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 2010;139:882–892. doi: 10.1053/j.gastro.2010.05.040. 892. e881-e883. [DOI] [PubMed] [Google Scholar]
- 75.Steel AW, Mela CM, Lindsay JO, Gazzard BG, Goodier MR. Increased proportion of CD16(+) nk cells in the colonic lamina propria of inflammatory bowel disease patients, but not after azathioprine treatment. Aliment. Pharmacol. Ther. 2011;33:115–126. doi: 10.1111/j.1365-2036.2010.04499.x. [DOI] [PubMed] [Google Scholar]
- 76.Ginsburg CH, Dambrauskas JT, Ault KA, Falchuk ZM. Impaired natural killer cell activity in patients with inflammatory bowel disease: evidence for a qualitative defect. Gastroenterology. 1983;85:846–851. [PubMed] [Google Scholar]
- 77.Dotta F, Censini S, van Halteren AG, et al. Coxsackie b4 virus infection of beta cells and natural killer cell insulitis in recent-onset Type 1 diabetic patients. Proc. Natl Acad. Sci. USA. 2007;104:5115–5120. doi: 10.1073/pnas.0700442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brauner H, Elemans M, Lemos S, et al. Distinct phenotype and function of NK cells in the pancreas of nonobese diabetic mice. J. Immunol. 2010;184:2272–2280. doi: 10.4049/jimmunol.0804358. [DOI] [PubMed] [Google Scholar]
- 79.Feuerer M, Shen Y, Littman DR, Benoist C, Mathis D. How punctual ablation of regulatory T cells unleashes an autoimmune lesion within the pancreatic islets. Immunity. 2009;31:654–664. doi: 10.1016/j.immuni.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brusko TM, Putnam AL, Bluestone JA. Human regulatory T cells: role in autoimmune disease and therapeutic opportunities. Immunol. Rev. 2008;223:371–390. doi: 10.1111/j.1600-065X.2008.00637.x. [DOI] [PubMed] [Google Scholar]
- 81.Kornete M, Mason ES, Piccirillo CA. Immune regulation in T1D and T2D: prospective role of FoxP3+ Treg cells in disease pathogenesis and treatment. Front. Endocrinol. 2013;4:76. doi: 10.3389/fendo.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Ann. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boden EK, Snapper SB. Regulatory T cells in inflammatory bowel disease. Curr. Opin. Gastroenterol. 2008;24:733–741. doi: 10.1097/mog.0b013e328311f26e. [DOI] [PubMed] [Google Scholar]
- 84.Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J. Exper. Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghiringhelli F, Ménard C, Martin F, Zitvogel L. The role of regulatory T cells in the control of natural killer cells: relevance during tumor progression. Immunol. Rev. 2006;214:229–238. doi: 10.1111/j.1600-065X.2006.00445.x. [DOI] [PubMed] [Google Scholar]
- 86.Kerdiles Y, Ugolini S, Vivier E. T cell regulation of natural killer cells. J. Exper. Med. 2013;210:1065–1068. doi: 10.1084/jem.20130960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaser A, Martinez-Naves E, Blumberg RS. Endoplasmic reticulum stress: implications for Inflammatory bowel disease pathogenesis. Curr. Opin. Gastroenterol. 2010;26:318–326. doi: 10.1097/MOG.0b013e32833a9ff1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. ▪▪ Established that XBP1 is the link between endoplasmic reticulum stress and intestinal inflammation/IBD.
- 90.Harding HP, Zeng H, Zhang Y, et al. Diabetes mellitus and exocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in secretory cell surviva l. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 91.Delepine M, Nicolino M, Barrett T, et al. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 92.Wolcott CD, Rallison ML. Infancy-onset diabetes mellitus and multiple epiphyseal dysplasia. J. Pediatrics. 1972;80:292–297. doi: 10.1016/s0022-3476(72)80596-1. [DOI] [PubMed] [Google Scholar]
- 93.Thornton CM, Carson DJ, Stewart FJ. Autopsy findings in the Wolcott-Rallison syndrome. Pediatr. Pathol. Lab. Med. 1997;17:487–496. [PubMed] [Google Scholar]
- 94.Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Transl. Med. 2012;160:246–257. doi: 10.1016/j.trsl.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 95.Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand. J. Gastroenterol. 2002;37:1034–1041. doi: 10.1080/003655202320378220. [DOI] [PubMed] [Google Scholar]
- 96.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 97.D’Haens GR, Geboes K, Peeters M, et al. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–267. doi: 10.1016/s0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- 98.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 99.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 101.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. ▪▪ Showed that there is a decrease in microbial diversity in CD patients.
- 102.Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aroniadis OC, Brandt LJ. Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol. 2013;29:79–84. doi: 10.1097/MOG.0b013e32835a4b3e. [DOI] [PubMed] [Google Scholar]
- 105.Borody TJ, Warren EF, Leis SM, et al. Bacteriotherapy using fecal flora: toying with human motions. J. Clin. Gastroenterol. 2004;38:475–483. doi: 10.1097/01.mcg.0000128988.13808.dc. [DOI] [PubMed] [Google Scholar]
- 106.Oikarinen M, Tauriainen S, Oikarinen S, et al. Type 1 diabetes is associated with enterovirus infection in gut mucosa. Diabetes. 2012;61:687–691. doi: 10.2337/db11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vaarala O. Is the origin of Type 1 diabetes in the gut? Immunol. Cell Biol. 2012;90:271–276. doi: 10.1038/icb.2011.115. [DOI] [PubMed] [Google Scholar]
- 108.Bason C, Lorini R, Lunardi C, et al. In Type 1 diabetes a subset of anti-Coxsackievirus b4 antibodies recognize autoantigens and induce apoptosis of pancreatic beta cells. PLoS ONE. 2013;8:e57729. doi: 10.1371/journal.pone.0057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Serreze DV, Wasserfall C, Ottendorfer EW, et al. Diabetes acceleration or prevention by a Coxsackievirus b4 infection: critical requirements for both interleukin-4 and gamma interferon. J. Virol. 2005;79:1045–1052. doi: 10.1128/JVI.79.2.1045-1052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cinek O, Tapia G, Witso E, et al. Enterovirus rna in peripheral blood may be associated with the variants of rs1990760, a common Type 1 diabetes associated polymorphism in IFIH1. PLoS ONE. 2012;7:e48409. doi: 10.1371/journal.pone.0048409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Coppieters KT, Wiberg A, Tracy SM, von Herrath MG. Immunology in the clinic review series: focus on Type 1 diabetes and viruses: the role of viruses in Type 1 diabetes: a difficult dilemma. Clin. Exper. Immunol. 2012;168:5–11. doi: 10.1111/j.1365-2249.2011.04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khan RR, Lawson AD, Minnich LL, et al. Gastrointestinal norovirus infection associated with exacerbation of inflammatory bowel disease. J. Pediatric Gastroenterol. Nutr. 2009;48:328–333. doi: 10.1097/mpg.0b013e31818255cc. [DOI] [PubMed] [Google Scholar]
- 113.Christen U, Bender C, von Herrath MG. Infection as a cause of Type 1 diabetes? Curr. Opin. Rheumatol. 2012;24:417–423. doi: 10.1097/BOR.0b013e3283533719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hara N, Alkanani AK, Ir D, et al. The role of the intestinal microbiota in Type 1 diabetes. Clin. Immunol. 2013;146:112–119. doi: 10.1016/j.clim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 115.Knosel T, Schewe C, Petersen N, Dietel M, Petersen I. Prevalence of infectious pathogens in Crohn’s disease. Pathol. Res. Prac. 2009;205:223–230. doi: 10.1016/j.prp.2008.04.018. [DOI] [PubMed] [Google Scholar]