Summary
Given prior evidence for the contribution of rare copy number variations (CNVs) to autism spectrum disorders (ASD), we studied these events in 4,457 individuals from 1,174 simplex families, composed of parents, a proband and, in most kindreds, an unaffected sibling. We find significant association of ASD with de novo duplications of 7q11.23, where the reciprocal deletion causes Williams-Beuren syndrome, featuring a highly social personality. We identify rare recurrent de novo CNVs at five additional regions including two novel ASD loci, 16p13.2 (including the genes USP7 and C16orf72) and Cadherin13, and implement a rigorous new approach to evaluating the statistical significance of these observations. Overall, we find large de novo CNVs carry substantial risk (OR=3.55; CI =2.16-7.46, p=6.9 × 10−6); estimate the presence of 130-234 distinct ASD-related CNV intervals across the genome; and, based on data from multiple studies, present compelling evidence for the association of rare de novo events at 7q11.23, 15q11.2-13.1, 16p11.2, and Neurexin1.
Introduction
Autism spectrum disorders (ASD) are defined by impairments in reciprocal social interaction, communication, and the presence of stereotyped repetitive behaviors and/or highly restricted interests. A genetic contribution is well established from twin studies (Bailey et al., 1995; Lichtenstein et al., 2010) in which the very large difference between the monozygotic and dizygotic concordance rates is consistent with the contribution of de novo mutation and/or complex inheritance. In addition, the over-representation of ASD in monogenic developmental disorders (Klauck et al., 1997; Smalley et al., 1992), gene discovery in families with Mendelian forms of the syndrome (Morrow et al., 2008; Strauss et al., 2006), and long-standing evidence for an increased burden of gross chromosomal abnormalities (Bugge et al., 2000; Veenstra-Vanderweele et al., 2004; Vorstman et al., 2006; Wassink et al., 2001) all point to the importance of genetic risks.
Over the last several years, dramatic advances have emerged from the study of copy number variants (CNVs) such as submicroscopic chromosomal deletions and duplications (Iafrate et al., 2004; Sebat et al., 2004). Sebat et al. (2007) first noted that “large” (mean size of 2.3Mb), rare (<1% frequency in the general population), de novo events were more frequent in ASD probands from families with only a single affected child (i.e. simplex families) than in controls, as well as in comparison to probands from families with more than one affected individual (i.e. multiplex families).
The over-representation of large de novo CNVs in ASD has been replicated in studies ranging from 60 to 393 simplex trios (Itsara et al., 2010; Marshall et al., 2008; Pinto et al., 2010), and two of the three studies (Marshall et al., 2008; Pinto et al., 2010) have confirmed a greater abundance in simplex versus multiplex ASD families. The burden of rare de novo CNVs in simplex probands (i.e. the percentage of individuals carrying ≥1 rare events) has ranged from 5.0-11% (Table S1). Rare structural variants, both transmitted and de novo, also show varying degrees of evidence for association with ASD. These include deletions and/or duplications at specific loci, including: 1q21.1, 15q11.2-13.1, 15q13.2-13.3, 16p11.2, 17q12, and 22q11.2 as well as recurrent structural variations involving one or a small number of genes, including: Neurexin 1 (NRXN1), Contactin 4 (CNTN4), Neuroligin 1 (NLGN1), Astrotactin 2 (ASTN2), and the contiguous genes Patched Domain Containing 1 (PTCHD1) and DEAD box Protein 53 (DDX53) (Bucan et al., 2009; Glessner et al., 2009; Kumar et al., 2008; Marshall et al., 2008; Moreno-De-Luca et al., 2010; Noor et al., 2010; Pinto et al., 2010; Weiss et al., 2008).
To date, the number of definitive replicated findings from these studies has remained small and all evidence has pointed to a highly heterogeneous allelic architecture as no risk variant is present in more than ~1% of affected individuals. In addition, examples of incomplete penetrance (not all mutation carriers have disease), and affected siblings not sharing the same risk variant, have been the rule rather than the exception. Moreover, remarkably diverse outcomes have been identified for apparently identical CNVs. For example, chromosome 16p11.2 deletions and duplications have been found in individuals with ASD and intellectual disability (ID) (Weiss et al., 2008), seizure disorder (Mefford et al., 2009), obesity (Bochukova et al., 2010), macrocephaly, and schizophrenia (McCarthy et al., 2009). These complexities suggest that the use of association strategies to demonstrate an excess of specific de novo CNVs will play an important role in definitively implicating loci in ASD.
We have conducted a genome-wide analysis focusing primarily on the study of rare de novo CNVs in 4,457 individuals comprising 1,174 simplex ASD families from the Simons Simplex Collection (Fischbach and Lord, 2010), nearly three-fold larger than previously reported simplex cohorts. Each family is extensively phenotyped, with a single affected offspring, unaffected parents, and, in the majority of cases, at least one unaffected sibling. This ascertainment strategy is designed to enrich for rare de novo risk variants, the family-based case-control comparisons mitigate a wide range of technical and methodological confounders that have plagued association study designs (Altshuler et al., 2008), and we have developed a new rigorous approach to evaluating the genome-wide significance of recurrent rare de novo events. Consequently, the scale and design of this study provides an extraordinary opportunity to investigate the relative contributions of rare de novo and rare transmitted variants in simplex families, to identify novel ASD risk loci, to evaluate the relationship between rare structural variation and social and intellectual disability (ID), and to place these findings in the context of previous ASD data, particularly with regards rare de novo CNVs.
Results
Simons Simplex Collection summary characteristics
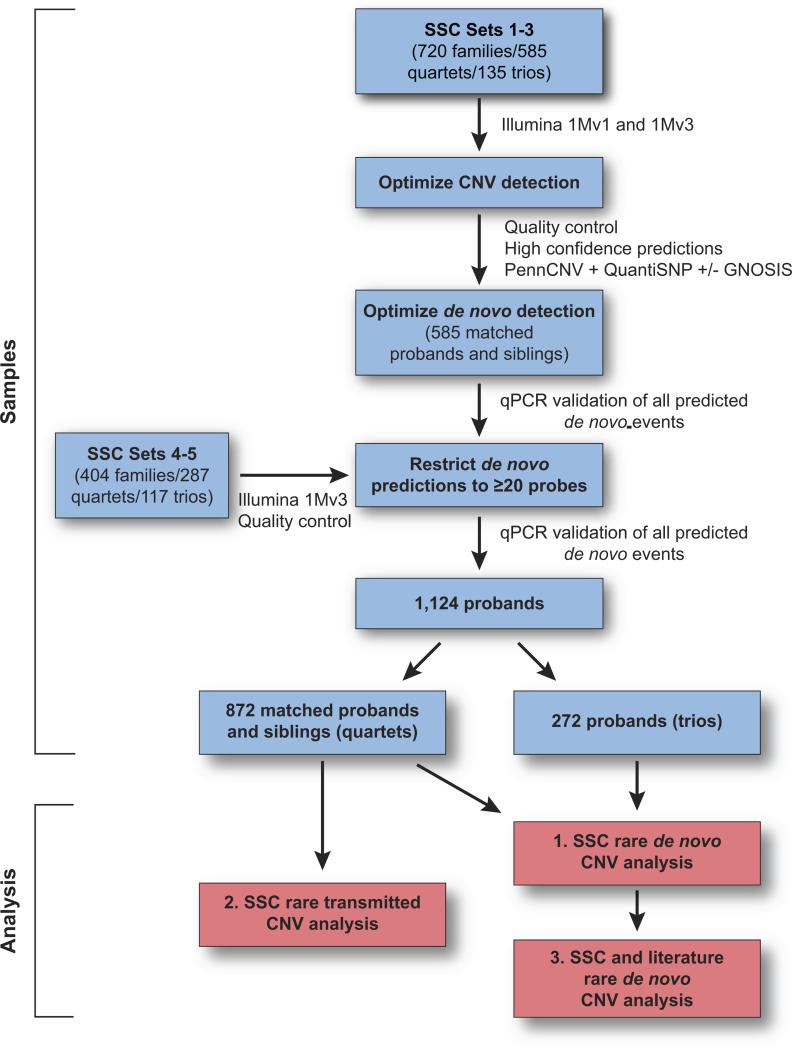
A total of 4,457 individuals from 1,174 families were included in the study. Data from 1,124 families passed all quality control; 872 families were quartets that included two unaffected parents, a proband, and one unaffected sibling; 252 families were trios that included two unaffected parents and a proband (Figure 1).
Figure 1. Flow chart of CNV detection and confirmation in the Simons Simplex Collection (SSC).
CNV detection was optimized by qPCR analysis of 115 predictions (Table S1, Figure S1). Quality control was performed to check for identity error and data quality (supplementary methods). De novo detection was optimized by qPCR analysis of 403 predictions (Figure S1) leading to the threshold of ≥20 probes and refinement of the prediction algorithm. All de novo CNVs reported in the study were confirmed using qPCR with absolute quantification.
The male to female ratio for probands was 6.2:1. All had confirmed ASD diagnoses based on well-accepted research criteria (Risi et al., 2006), including autism: 1,006 (89.5%), Pervasive Developmental Disorder-Not Otherwise Specified: 96 (8.5%), and Asperger Syndrome: 22 (2%). The mean age at inclusion was 9.1 years for probands (4-18 years) and 10.0 years (3.5-26 years) for siblings. The mean (±95% CI) full-scale IQ in probands was 85.1±1.5, however the range was considerable (<20-167, Figure 3); the mean verbal IQ was 81.9±1.7 and the mean non-verbal IQ was 88.4±1.4. Self-reported ancestry was as follows: White, non-hispanic: 74.5%; Mixed: 9.3%; Asian: 4.3%; White, Hispanic: 4.0%; African American: 3.8%; Other: 4.2%. Additional phenotypic data may be found in recent publications (Fischbach and Lord, 2010) and at www.sfari.org/simons-simplex-collection.
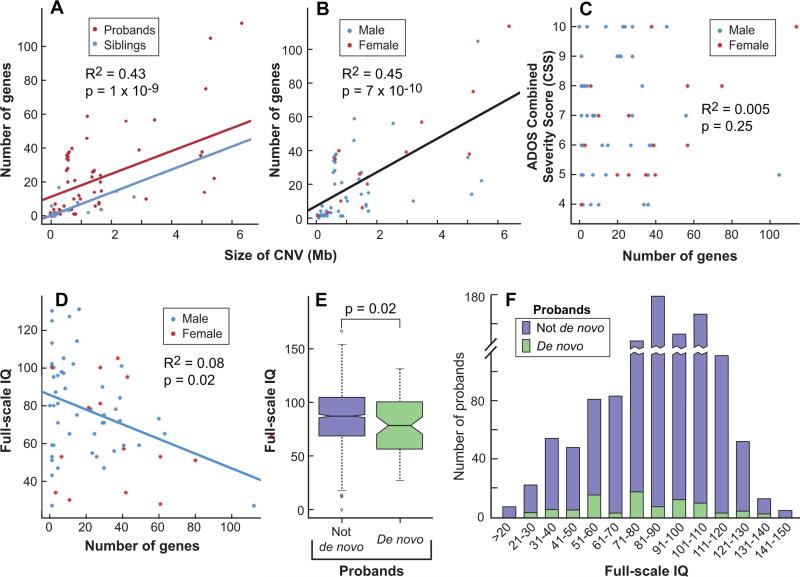
Figure 3. Genotype-phenotype analyses of probands carrying rare de novo CNVs.
A) The number of RefSeq genes within rare de novo CNVs (genes) vs. CNV size (size), with proband (red) vs. sibling (blue). The slope of the lines shows the fitted significant (p=1 × 10−9) relationship between genes and size and the difference between the lines shows the fitted difference for probands and siblings (p=0.025): on average probands have more genes within a rare de novo CNV for any given size. B) genes vs. size, with sex of subject encoded by color as noted (sex). The slope of the line shows the fitted significant (p=7 × 10−10) relationship between genes and size while the presence of only one line reflects the lack of significant difference by sex (p=0.20). C) ADOS Combined Severity Score (CSS), a measure of autism severity, against genes and by sex. The lack of a line indicates the absence of a significant relationship. D) Full-scale IQ (IQ) against genes and by sex. The slope shows that IQ declines as a function of genes in males (p=0.02, Wilcoxon test); there is no significant relationship in females. E) Boxplot with 95% confidence intervals for IQ by presence (yellow) or absence (blue) of a detected rare de novo event in the probands. F) Distribution of IQ in probands with (yellow, N=63) rare de novo CNVs and without (blue, N=1,061).
Illumina 1M arrays accurately detect both rare de novo and transmitted CNVs
DNA samples derived from whole blood (N=4,381), cell lines (N=68), or saliva (N=8) were genotyped on the Illumina IMv1 (334 families) or Illumina IMv3 Duo Bead-arrays (840 families), which share 1,040,853 probes in common. CNV prediction was performed by PennCNV (PN) (Wang et al., 2007), QuantiSNP (QT) (Colella et al., 2007), and GNOSIS (GN), (www.CNVision.org) (Figure 1). 115 predicted rare (≤50% of the span of the event found at >1% in the database of genomic variation (DGV)) CNVs were evaluated by quantitative polymerase chain reaction (qPCR). A higher positive predictive value was observed for CNVs predicted by PN and QT, with or without GN (PPV=97% with GN, PPV=83% without) than for other combinations of algorithms, irrespective of the number of probes mapping within the structural variation (Table S2, Figure S1); these “high-confidence” criteria were subsequently used to identify all rare transmitted CNVs.
However, given the likely importance of de novo variation, and the relative challenge of accurately detecting these CNVs (Lupski, 2007), we focused on this small subset of predictions and further optimized our detection strategy using the first 585 quartets with complete genotyping data (Figure 1). We predicted putative de novo events from the group of rare high-confidence CNVs based on the combination of within-family intensity and genotypic data and used a blinded qPCR confirmation process (Figure S1). 53% of de novo predictions based on ≥20 probes (N=94) were confirmed compared with 2.6% with <20 probes (N=430). 82% of failures were false-positive predictions in offspring, 18% were false-negatives in parents. The data from this experiment were used to refine de novo prediction thresholds (supplementary materials). In addition, given the large number of predictions of small CNVs, and the low yield of true positives in the pilot data set (Figure S1), we elected to restrict all further statistical analysis to rare de novo events encompassing ≥20 probes that were also confirmed by qPCR in whole-blood DNA (Figure S1).
At the conclusion of our study, we were able to evaluate this threshold further via a comparison of confirmed de novo CNVs identified here with those reported in 1,340 overlapping offspring (probands or siblings) using the Nimblegen 2.1M array, as described by Levy and colleagues in this issue (Levy et al., 2011). At a threshold defined by the presence of ≥20 Illumina probes within a genomic interval, a combined total of 58 rare de novo CNVs were identified across both studies, with each array type identifying 95% (n=55) of the total events. This suggests that both arrays have high sensitivity for such events at or above this threshold, and that the combined results are very likely to represent the complete set of large de novo CNVs present in the SSC. This situation is reversed below 20 probes: a total of 31 small rare de novo CNVs were identified between the two groups with approximately twice as many found using the 2.1M Nimblegen array vs. the IM Illumina array (23 CNVs vs. 12 CNVs respectively) as would be expected for the increased probe resolution. Of these 31 events only 13% (n=4) were identified by both groups, suggesting that the sensitivity for small de novo events was low for both arrays and that, as expected, there is a pool of small de novo structural events that were not considered in our analyses.
Analysis of rare de novo CNVs in the Simons Simplex Collection (SSC)
Rare, de novo, genic CNVs are over-represented in simplex probands
In light of strong prior evidence for an increased burden of de novo CNVS in simplex autism (Itsara et al., 2010; Marshall et al., 2008; Pinto et al., 2010; Sebat et al., 2007), we investigated these events in probands versus their unaffected siblings in all 872 quartets in this study (Figure 1). A total of 28,610 rare, high-confidence CNVs were identified; 97 were classified as rare and likely de novo, and 83 events were confirmed to be rare de novo CNVs by qPCR in whole-blood DNA (Table S4).
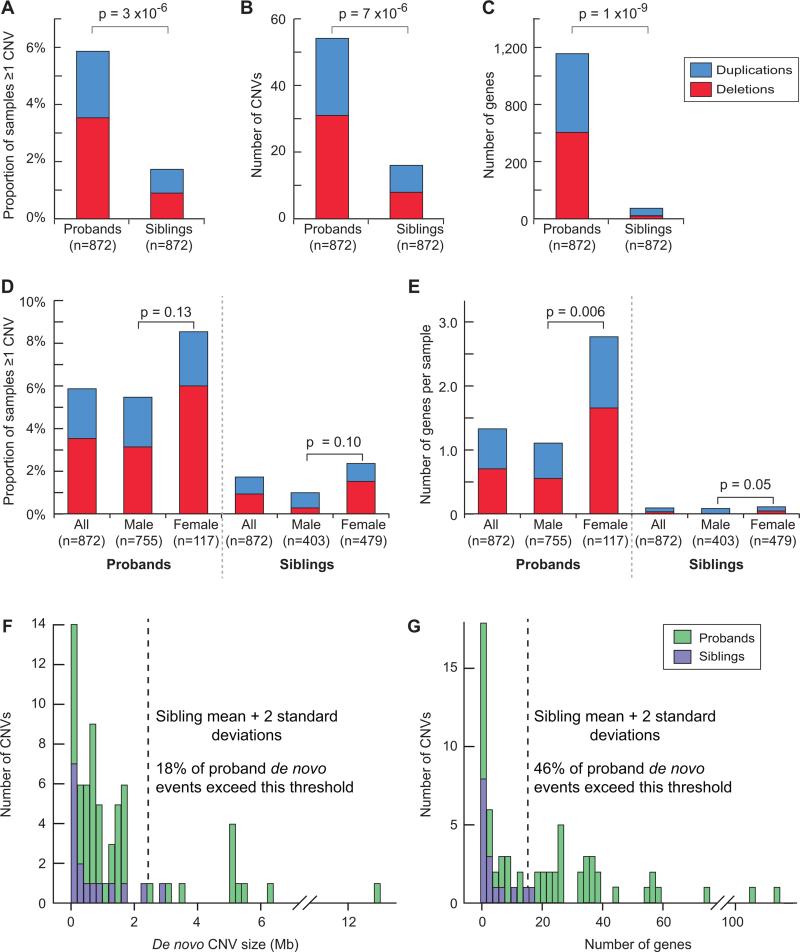
Rare de novo CNVs were significantly more common among probands than siblings. Overall 5.8% of probands (N=51 out of 872) had at least one rare de novo CNV compared with 1.7% of their unaffected siblings (N=15 out of 872) yielding an odds ratio (OR) of 3.55 (CI =2.16-7.46, p=6.9 × 10−6, Fisher's exact test) (Table 1 and Figure 2). When we considered the proportion of individuals carrying at least one rare de novo CNV that also contains known genes (genic CNVs), the OR increased to 4.02 (50 in probands vs. 12 in siblings; CI =1.98-6.36, p=2 × 10−6). These results remained consistent regardless of whether we analyzed total numbers of CNVs as opposed to the proportion of individuals with at least one (Figure 2), or increased the stringency of the threshold for “rarity” (supplementary materials).
Table 1.
Burden of de novo CNVs in probands and siblings
| Category | Analysis | All probands (N=1,124) | Matched probands (N=872) | Matched siblings (N=872) | Ratio (OR) | p-valued |
|---|---|---|---|---|---|---|
| De novo CNVs | CNVs | 67 | 54 | 16 | ||
| Samplesa | 63 | 51 | 15 | |||
| Proportionb | 5.6% | 5.8% | 1.7% | 3.4 (3.5) | 3 × 10−6 | |
| Genesc | 1,417 | 1,153 | 73 | 15.8 | ||
| De novo deletions | CNVs | 35 | 31 | 8 | ||
| Samples | 35 | 31 | 8 | |||
| Proportion | 3.1% | 3.6% | 0.9% | 3.9 (4.0) | 1 × 10−4 | |
| Genes | 638 | 605 | 21 | 728.8 | ||
| De novo duplications | CNVs | 32 | 23 | 8 | ||
| Samples | 29 | 21 | 7 | |||
| Proportion | 2.6% | 2.4% | 0.8% | 3.0 (3.0) | 0.006 | |
| Genes | 779 | 548 | 52 | 10.5 | ||
| De novo genic CNVs | CNVs | 66 | 53 | 13 | ||
| Samples | 62 | 50 | 12 | |||
| Proportion | 5.5% | 5.7% | 1.4% | 4.2 (4.4) | 4 × 16−7 | |
| Genes | 1,417 | 1,153 | 73 | 15.8 | ||
| De novo exonic CNVs | CNVs | 64 | 52 | 11 | ||
| Samples | 60 | 49 | 10 | |||
| Proportion | 5.3% | 5.6% | 1.1% | 4.9 (5.1) | 9 × 10−8 | |
| Genes | 1,415 | 1,152 | 71 | 16.2 | ||
| De novo multigenic CNVs | CNVs | 53 | 44 | 9 | ||
| Samples | 52 | 43 | 8 | |||
| Proportion | 4.6% | 4.9% | 0.9% | 5.4 (5.6) | 2 × 10−7 | |
| Genes | 1,404 | 1,144 | 69 | 16.6 | ||
| De novo autosomal CNVs | CNVs | 66 | 53 | 14 | ||
| Samples | 62 | 50 | 14 | |||
| Proportion | 5.5% | 5.7% | 1.6% | 3.6 (3.7) | 2 × 10−6 | |
| G^nes | 1,416 | 1,152 | 67 | 17.2 | ||
| De novo chrX CNVs | CNVs | 1 (male deletion) | 1 (male deletion) | 2 (female duplications) | ||
| Samples | 1 (male deletion) | 1 (male deletion) | 1 (female duplication) | |||
| Proportion | 0.1% | 0.1% | 0.1% | 1.0 (1.0) | 0.75 | |
| Genes | 1 | 1 | 6 | 0.2 | ||
| Small de novo CNVs (<100kb) | CNVs | 8 | 5 | 3 | ||
| Samples | 8 | 5 | 3 | |||
| Proportion | 0.7% | 0.6% | 0.3% | 1.7 (1.7) | 0.36 | |
| Genes | 8 | 5 | 7 | 0.7 | ||
| Medium de novo CNVs (100-1,000kb) | CNVs | 32 | 26 | 9 | ||
| Samples | 30 | 25 | 8 | |||
| Proportion | 2.7% | 2.9% | 0.9% | 3.1 (3.2) | 0.002 | |
| Genes | 469 | 392 | 34 | 11.5 | ||
| Large de novo CNVs (≥1,000kb) | CNVs | 27 | 23 | 4 | ||
| Samples | 26 | 22 | 4 | |||
| Proportion | 2.3% | 2.5% | 0.5% | 5.5 (5.6) | 2 × 10−4 | |
| Genes | 940 | 756 | 32 | 23.6 | ||
| Single occurrence de novo CNVs | CNVs | 44 | 37 | 14 | ||
| Samples | 40 | 34 | 13 | |||
| Proportion | 3.6% | 3.9% | 1.5% | 2.6 (2.7) | 0.001 | |
| Genes | 862 | 754 | 54 | 14.0 | ||
| Double occurrence de novo CNVs | CNVs | 8 | 8 | 2 | ||
| Samples | 8 | 8 | 2 | |||
| Proportion | 0.7% | 0.9% | 0.2% | 4.0 (4.0) | 0.05 | |
| Genes | 89 | 102 | 19 | 5.4 | ||
| ≥3 occurrence de novo CNVs | CNVs | 15 | 9 | 0 | ||
| Samples | 15 | 9 | 0 | |||
| Proportion | 1.3% | 1.0% | 0.0% | NA (NA) | 0.002 | |
| Genes | 466 | 297 | 0 | NA |
Four individuals have multiple de novo CNVs
% of samples with ≥1 de novo CNV
RefSeq genes within the CNV
Fisher's exact test
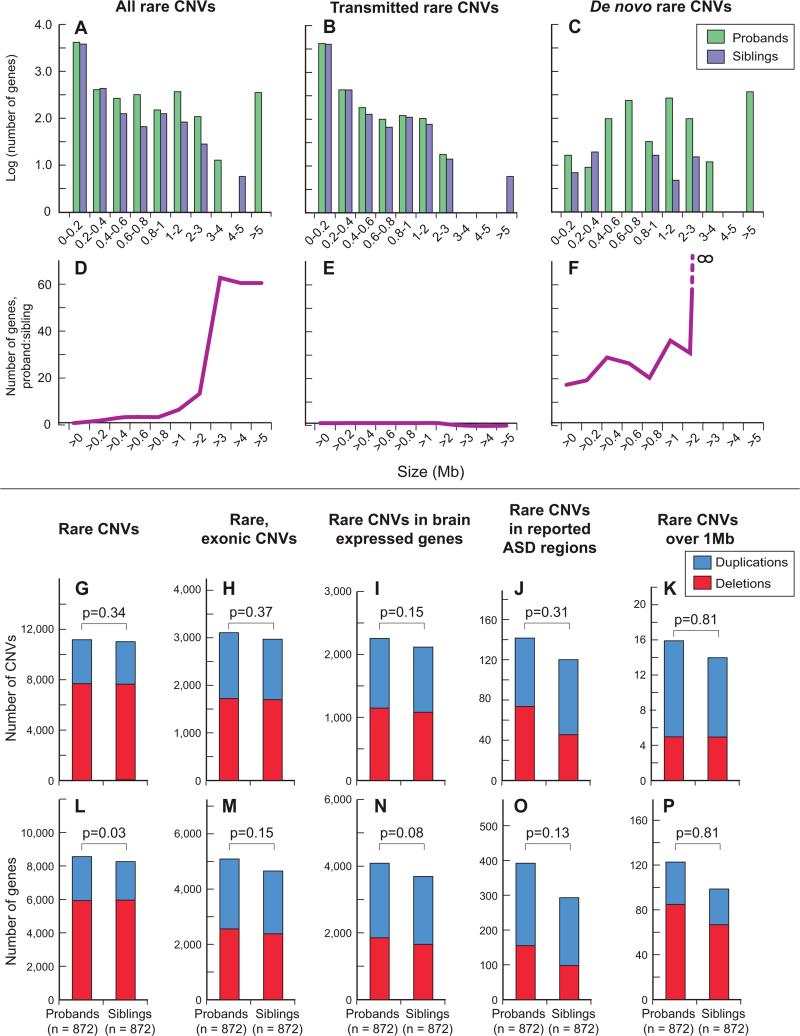
Figure 2. The burden of rare de novo CNVs and genes mapping within them in 872 probands and 872 matched siblings.
A) % of individuals with ≥1 rare de novo CNV in probands vs. siblings. Red = deletions; blue = duplications for A to E. B) Total number of rare de novo CNVs in probands vs. siblings (2 probands and 1 sibling have more than one). C) Number of RefSeq genes (Pruitt et al., 2007) overlapping rare de novo CNVs in probands vs. siblings. D) % of individuals with ≥1 rare de novo CNV (as shown in A) split by sex. Specific comparisons and associated p-values are given. E) Number of RefSeq genes overlapping rare de novo CNVs (as shown in C) split by sex. F) The distribution of rare de novo CNVs by size in probands (green) and siblings (purple). The dashed vertical line represents the mean plus two standard deviations of the sibling events. G) The distribution of rare de novo CNVs by number of RefSeq genes. Statistical significance was calculated using: Fisher's Exact test (A, D), Sign test (B), Wilcoxon paired test (C), and Wilcoxon test (E).
Given the strong male predominance and increased rates of ASD in monogenic X-linked intellectual disability syndromes, we paid particular attention to rare de novo CNVs on the X chromosome but found only 2 events: one genic deletion present in a male at the gene DDX53 and a duplication involving 6 genes in a female sibling (Xq11.1). This small number precluded meaningful group comparisons. Importantly, neither these, nor any subsequent statistical results reported here were substantively altered by the exclusion of 15 confirmed rare de novo CNVs identified during our detection optimization experiments that did not then meet our minimum probe criteria (Table S4). Of note, however, one of these was an exonic deletion of NLGN3 on chromosome X in a male proband. (Table S4)
This burden of rare de novo CNVs in simplex families is remarkably similar to previously published results (Table S1) despite varying CNV discovery approaches and array densities from 85,000 (Sebat et al., 2007) to 1 million probes (Pinto et al., 2010). We reasoned that this was likely due to the particular importance of large de novo events, as their detection would be least sensitive to differences in probe number and distribution. Indeed, we found that rare de novo CNVs in probands tended to be larger than in siblings (mean 1.6Mb vs. 0.7Mb) (Figure 2, Figure S2) and to include a greater number of genes (16-fold increase in probands, and a 29-fold increase considering only deletions).
In fact, we found that de novo CNVs in probands were both larger and contained a greater number of genes when these measures were considered independently. We fit a series of stepwise linear models that increased in complexity from individual predictors to an analysis of covariance model, with size and affected status as predictors, to a three-term model that included the interaction of size and affected status. We confirmed a significant difference between probands and siblings with regard to the number of genes within CNVs (estimated β=11.1 more genes in a proband's de novo CNV; p=0.025) even after accounting for the strong effect of the size of the event (estimated β=6.8 genes per Mb; p=1.1 × 10−9) (Figure 3A). Considering deletions and duplications separately did not alter these findings. In summary, the burden of rare de novo CNVs is greater in probands with regard to number, size, and gene content.
Strong association of rare, recurrent, de novo CNVs
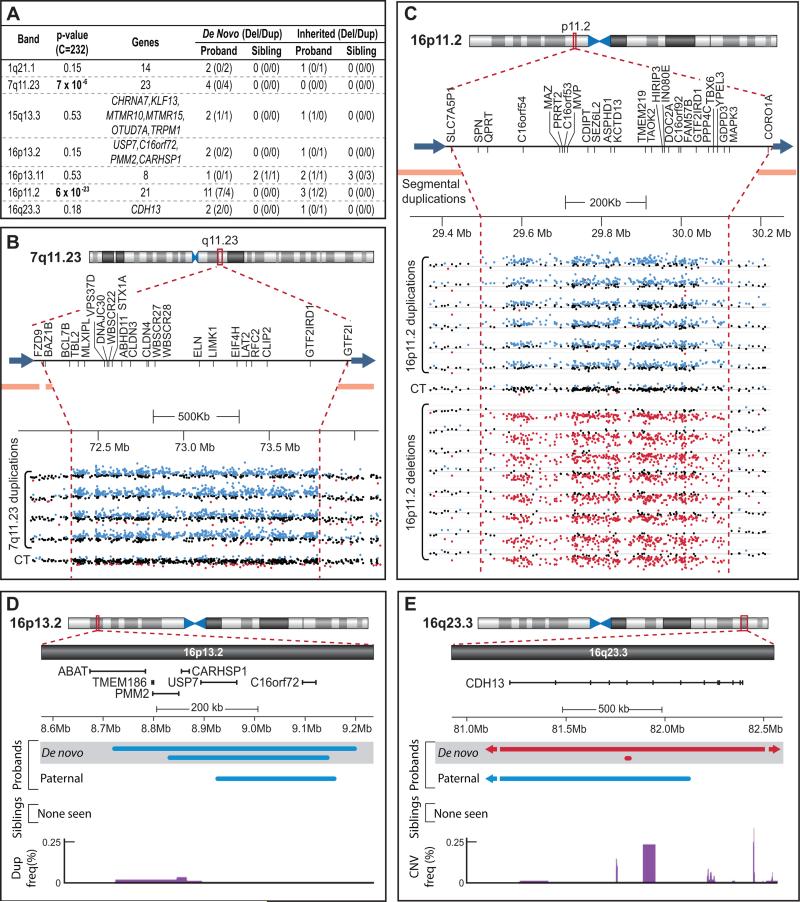
Our interest in identifying specific regions of the genome contributing to ASD led us to next investigate whether multiple overlapping de novo events were present in probands and then to compare these findings to siblings. In total, 23 probands were found to carry recurrent de novo CNVs in 6 separate regions of the genome. Each of these intervals contains from 2 to 11 de novo CNVs in unrelated probands; no de novo CNVs overlapping these regions were found in siblings. In contrast only a single recurrent de novo event was observed in siblings (16p13.11 in 2 unrelated siblings) and one CNV overlapping the region was also found in a proband (Figure 4).
Figure 4. Confirmed recurrent rare de novo CNVs.
A) All recurrent de novo CNVs identified in 1,124 probands and 872 siblings. The gene count is given when >6 RefSeq genes map to an interval; a complete listing of genes is presented in Table S4. The total number of de novo and matching inherited CNVs in probands and siblings are shown for deletions (Del) and duplications (Dup) in parentheses. B) LogR data for 4 de novo duplications and 1 control with no CNV (CT) in the 7q11.23 interval. RefSeq genes within this region are noted below the ideogram; the orange bars represent flanking segmental duplications. NCBI 36 (hg18) genomic coordinates are shown with the scale indicated. The LogR for all probes within the region is shown; LogR values greater than 0.15 are in blue (suggesting a duplication) while LogR values less than −0.15 are in red (suggesting a deletion). B allele frequency data is not shown but supports the presence of a corresponding CNV. The approximate boundaries of the CNVs are shown by the vertical dashed red line and blue arrows. C) LogR data for 6 duplications (4 de novo), 8 deletions (7 de novo), and 1 control with no CNV (CT) in the 16p11.2 interval. The ideogram and intensity plots are as in B. D) Overlapping rare de novo and rare inherited CNVs identified in the 16p13.2 interval. The brackets show the boundaries of RefSeq genes; 2 genes are in common between all 3 duplications: USP7 and C16orf72. The frequency of duplications in the DGV is shown in purple: the majority of the recurrent de novo region is not present in the DGV. E) Overlapping rare de novo and rare inherited CNVs identified in the 16q23.3 interval. A 34kb deletion overlaps a 5Mb deletion over a CDH13 exon (represented by ticks on the gene). The frequency of CNVs observed in the DGV is shown at the bottom in purple.
The 6 regions found in probands included 7 deletions and 4 duplications at chromosome 16p11.2, 4 duplications at 7q11.23 (the Williams-Beuren syndrome region), and 2 CNVs each at 1q21.1 (2 duplications), 15q13.2-q13.3 (1 deletion, 1 duplication), 16p13.2 (2 duplications), and disrupting the gene Cadherin 13 (CDH13) at 16q23.3 (5Mb deletion and an overlapping 34kb exonic deletion).
The presence of multiple regions showing overlapping rare de novo CNVs restricted to probands, and the absence of similar findings in their sibling controls, is striking. However, in contrast to genome-wide common variant association studies, there is no widely accepted statistical approach or threshold to formally evaluate these results. Consequently, we set out to develop a rigorous method to assess the genome-wide significance of de novo events (methods). To do so, we determined the null expectation for recurrent rare de novo CNVs based on our data from unaffected siblings and then used this expectation to evaluate the p-value for finding multiple recurrences in probands.
Using this approach, the probability of finding 2 rare de novo CNVs at the same position in probands is 0.53. However, the observation of 4 recurrent de novo duplications at 7q11.23 (p= 7 × 10−6) and 11 recurrent de novo CNVs at 16p11.2 (p= 6 × 10−23) are both highly significant. In addition, we found that 16p11.2 deletions (N=7; p=2 × 10−14) and duplications (N=4; p=7 × 10−6) are strongly associated with ASD when considered independently (Figure S3).
Prior studies have often found a combination of rare transmitted and de novo CNVs at ASD risk regions. In our data, we observed 8 loci at which rare transmitted CNVs, present only in probands, overlapped one of the 51 regions in probands containing at least one rare de novo CNV. Conversely, in siblings we did not observe any cases in which a rare transmitted CNV, restricted to siblings, overlapped one of the 16 regions showing de novo events. Interestingly, the 8 regions in probands showing overlapping rare de novo and rare transmitted CNVs include 5 of the 6 intervals with recurrent rare de novo variants, 1q21.1, 15q13.3, 16p13.2, 16p11.2, and 16q23.3 (Figure 4), and 3 additional genomic segments with 1 rare de novo event: 2p15, 6p11.2, and 17q12.
While the use of matched sibling controls should preclude any confound of population stratification, we explored whether genotype data from the parents of probands with 16p11.2 or 7q11.23 CNVs suggested unusual ancestral clustering (Crossett et al., 2010; Lee et al., 2009) pointing to a particular haplotype that might increase the frequency of de novo events. We found no evidence for this. In addition, given the very large number of 16p11.2 CNVs in this study and the widespread attention afforded previous findings at this locus, we considered the possibility of ascertainment bias. A review of medical histories obtained at the time of recruitment revealed that parents had prior knowledge of a 16p11.2 CNV in 2 instances (1 de novo duplication, 1 transmitted deletion). Nonetheless, with these events removed, association of both deletions and duplications remains significant (p=3 × 10−19 all de novo events (N=10); p=2 × 10−14 deletions (N=7); p=0.002 duplications (N=3)) (Figure S4).
The distribution of de novo CNVs in probands, supports marked locus heterogeneity
The identification of multiple recurrent de novo events restricted to probands, and the absence of similar observations in siblings, led us to consider what these findings might indicate about the overall number of CNV-mediated ASD risk loci that are present in the genome. Consequently, we used the distribution of 67 de novo CNVs identified in SSC probands to calculate the number of regions likely to be contributing large rare de novo risk variants and estimated 130 loci (methods).
We then evaluated the implications of this likely genomic architecture for the planned second phase of genotyping and CNV analysis in the SSC, which is currently underway. We used the estimated number of predicted ASD loci to guide a simulation experiment (supplementary methods) and found that the most likely outcome of a second SSC cohort of similar composition and size to that reported here will be further confirmation of 7q11.23 and 16p11.2 and the identification of 2-3 additional regions of significant association. These were most likely to emerge at the intervals already identified containing recurrent de novo events in phase 1, namely 1q21.1, 15q13.2-13.3, 16p13.2, and the CDH13 locus.
Genotype-phenotype analyses of probands carrying any rare de novo CNV
Given highly reliable phenotypic data and long standing interest in the role of sex in ASD risk and resilience, we investigated whether males or females carried quantitatively different types of rare de novo events and what impact rare de novo CNVs had on intellectual and social functioning in both groups.
We found little evidence for larger or more gene rich de novo CNVs in males versus females. By fitting a series of stepwise linear models, we evaluated whether the number of genes within a de novo CNV tended to differ after accounting for a critical covariate, CNV size. Neither sex (p=0.20) nor the interaction of size and sex (p=0.06) was a significant predictor of gene number. These results should to be viewed with some caution, given the trend toward significance and a relatively small sample size (Figure 3B).
In contrast, we found that male intellectual functioning was relatively more vulnerable to the effects of rare de novo CNVs. Again using a series of stepwise linear models we evaluated the relationship between intellectual functioning, sex, and the number of genes within rare de novo CNVs. For males, there was a significant relationship between IQ and number of genes (p=0.02), with the model predicting a decrease of 0.42 IQ points for each additional gene. In contrast, for females the estimated effect was ten-fold less and did not approach significance (Figure 3D).
To evaluate whether low IQ predicted if a proband carried a de novo CNV, we fit a logistic regression model with de novo CNV status for probands as the outcome and full-scale IQ as the predictor. We found the accuracy of prediction was quite low (Nagelkerke pseudo R2=0.014). Overall, while the odds of carrying a de novo CNV varied three-fold for those with the lowest versus the highest IQ, the odds were never large (0.111 at IQ=30, 0.063 at IQ=80, and 0.036 at IQ=130). This relationship did not differ significantly by sex (interaction of IQ and sex, p=0.12).
Finally, we investigated the relationship between IQ, sex, and number of genes within rare de novo CNVs to determine if any of the models significantly predicted ASD severity (measured by the ADOS combined severity score (CSS)); of these only full-scale IQ predicted ASD severity (p=0.02).
Overall, the data show a strong effect of large rare genic de novo CNVs on affected status, but do not support either IQ or ASD severity as useful predictors for probands carrying large rare de novo risk variants in the SSC (Figure 3C). We did observe a trend toward more gene rich de novo CNVs in females and found females to be less vulnerable to the reduction in IQ associated with rare de novo CNVs.
Genotype-phenotype analyses of probands carrying 16p11.2 and 7q11.23 CNVs
We next investigate whether individuals with recurrent CNVs at 16p11.2 or 7q11.23 showed distinctive behavioral or cognitive profiles compared with probands who were not carrying rare de novo events. For each proband carrying a de novo CNV at 16p11.2 or 7q11.23, five other probands were selected as controls based on hierarchical matching criteria: first age, then sex, genetic distance, ascertainment site, and whether the sample was from a quartet or trio.
Our primary analysis focused on 4 variables: full-scale IQ, categorical diagnosis, severity of autism, and body mass index (BMI) (Table 2), with the latter motivated by multiple reports that 16p11.2 deletions (Bijlsma et al., 2009; Walters et al., 2010) contribute to obesity and the recent observation that duplications have the opposite impact on weight (Reymond et al., 2010). We then pursued a broader exploratory study of additional phenotypic variables, 10 of which are presented in Table 2 and the remainder in Table S5.
Table 2.
Phenotypic comparisons between subjects with 16p11.2 deletions, 16p11.2 duplications, and 7q11.23 duplications and matched probands.
| Primary | 16p Deletion (N=8) Meana (SD) | Deletion Matches (N=40) Mean (SD) | 16p Duplication (N=6) Mean (SD) | Duplication Matches (N=30) Mean (SD) | Duplications (N=4) Mean (SD) | Duplication Matches (N=20) Mean (SD) |
|---|---|---|---|---|---|---|
| CPEA Diagnosis | ||||||
| • Autism | 75% | 98% | 83% | 97% | 100% | 85% |
| • Autism Spectrum Disorder | 13% | 3% | 0% | 0% | 0% | 10% |
| • Asperger Syndrome | 13% | 0% | 17% | 3% | 0% | 5% |
| ADOS Combined Severity Score (CSS) | 6.5 | 7.0 | 7.2 | 7.4 | 7.0 | 7.6 |
| (1.6) | (1.7) | (2.1) | (1.5) | (2.2) | (1.6) | |
| Full-Scale IQ | 76.9 | 82.5 | 75.7 | 81.0 | 84.0 | 81.3 |
| (17.6) | (27.8) | (23.2) | (26) | (14.9) | (30.5) | |
| Body Mass Index (BMI) | 23.2 | 20.9 | 17.1 | 19.3 | 23.1 | 21.6 |
| (6.4) | (5.3) | (1.4) | (5.2) | (5.9) | (6.4) | |
| Exploratory | ||||||
| ADI-R Social Interaction Total | 17.9 | 21.3 | 21.8 | 19.1 | 20.0 | 19.9 |
| (6.7) | (5.5) | (3.5) | (5.6) | (5.5) | (7.0) | |
| ADOS Social Affect Total | 9.9 | 9.5 | 10.8 | 10.7 | 11.0 | 11.7 |
| (4.0) | (4.2) | (5.3) | (3.2) | (4.8) | (3.3) | |
| ADOS Social and Communication Total | 12.4 | 11.5 | 14.2 | 12.7 | 11.8 | 13.7 |
| (3.9) | (4.2) | (6.0) | (3.1) | (5.7) | (3.6) | |
| ADI-R Restricted and Repetitive Behavior (RRB) Total | 5.4 | 6.8 | 8.5 | 6.3 | 7.3 | 6.3 |
| (2.6) | (2.3) | (1.9) | (2.1) | (1.7) | (2.6) | |
| ADOS RRB Total | 2.0 | 3.7 | 4.3 | 3.6 | 2.0 | 4.1 |
| (1.3) | (1.9) | (2.6) | (1.7) | (1.4) | (2.0) | |
| Aberrant Behavior Checklist (ABC) Total | 46.8 | 42.0 | 68.7 | 42.5 | 66.0 | 47.2 |
| (24.3) | (28.6) | (21.5) | (17.9) | (26.7) | (21.0) | |
| ABC Irritability | 14.9 | 9.6 | 19.5 | 10.5 | 20.0 | 12.0 |
| (9.3) | (9.1) | (6.4) | (7.8) | (13.0) | (7.6) | |
| ABC Hyperactivity | 16.5 | 13.9 | 26.7 | 14.0 | 20.3 | 18.3 |
| (9.1) | (10.3) | (6.6) | (7.9) | (9.7) | (9.1) | |
| ABC Lethargy/Social Withdrawal | 9.5 | 10.0 | 11.5 | 10.3 | 13.8 | 9.8 |
| (6.6) | (7.7) | (8.6) | (7.2) | (8.4) | (6.2) | |
| Age First Concern | 2.8 | 1.7 | 1.7 | 2.0 | 1.8 | 2.1 |
| (1.5) | (0.9) | (0.9) | (0.9) | (1.0) | (1.3) |
Significance is shown in bold (p≤0.05) and italics (0.05<p≤0.1)
We found that probands carrying a 16p11.2 or 7q11.23 de novo CNV were indistinguishable from the larger group with regard to IQ, ASD severity, or categorical autism diagnosis (Table 2). However, we did find a relationship between body weight and 16p11.2 deletions and duplications. When we treated copy number as an ordinal variable (1, 2, and 3 copies), and used the matched controls as the diploid sample, BMI diminished as 16p11.2 copy number increased (estimated β=−3.1kg/m2 for each extra copy, p=0.02).
The extensive phenotypic data available on the SSC sample provides great potential to undertake fine-grained analyses of genotype-phenotype relationships. At present the limiting factor with regard to recurrent de novo CNVs is the small sample size, even for 16p11.2 duplications and deletions in this dataset. However, we undertook an exploratory analysis of a range of phenotypic features and found several that yielded significant p-values. While none would survive correction for multiple comparisons, we report them here (Table 2, Table S5) in the interest of generating hypotheses for future studies. For example, individuals with 16p11.2 duplications had higher hyperactivity scores, compared to matched control probands, while probands carrying 7q11.23 duplications showed significantly more behavioral problems (ABC total), but less severe social and communication impairment during ADOS administration.
Analysis of rare transmitted CNVs in the SSC
Rare, transmitted autosomal CNVs are equally represented in probands and siblings
Given the very strong association of rare de novo CNVs, we were somewhat surprised to find that rare transmitted CNVs were not present in greater numbers in affected individuals or in a greater proportion of probands versus siblings. As prior publications have shown an increased burden of specific subsets of CNVs in neuropsychiatric disorders including autism and schizophrenia, we considered multiple subcategories of rare transmitted events as well, including genic, exonic, brain-expressed, and ASD-related, and did not find a statistically significant result that survived correction for multiple comparisons (Figure 5).
Figure 5. Burden of rare CNVs in 872 probands and 872 matched siblings.
A) Bar graph showing the log (10) number of genes present in all rare CNVs binned by size (in Mb), with probands shown in green and siblings in purple. B) The data from A with confirmed de novo events excluded leaving only CNVs transmitted from a parent to offspring. C) Only confirmed de novo events are shown. D-F) The ratio (y-axis) of number of genes in probands vs. siblings for specific size thresholds (x-axis): D) all rare CNVs (transmitted and de novo); E) transmitted events; F) de novo events only. G-K) The total number of transmitted deletions (red) and duplications (blue) for probands and siblings for varying categories of CNV (shown above the graph). Definitions are described in supplementary methods. P-values (noted above the bars) are calculated using the Sign test and are not corrected for multiple comparisons. L-P) As in G-K, with number of RefSeq genes within the CNVs (y-axis). P-values (noted above the bars) are estimated using a two-tailed paired t-test and are not corrected for approximately 3,000 comparisons.
These findings stood in contrast to a recent rigorous large-scale CNV study undertaken by the Autism Genome Project (AGP) (Pinto et al., 2010). Their sample included both simplex and multiplex families and identified a significantly higher burden of genic and ASD-related CNVs in cases versus unrelated controls. Notably they did not differentiate between transmitted and de novo events in this analysis. We reanalyzed our data using the identical criteria detailed in their manuscript and found similar results (Table S6). However, when we again restricted the analysis of our sample to rare transmitted CNVs, by removing confirmed rare de novo events; there was no significant difference found between proband and siblings, suggesting that the excess burden in the SSC sample was entirely driven by rare de novo events.
We pursued this analysis further given strong evidence that certain rare transmitted CNVs carry ASD risk, as well as reports of particularly significant effects for maternal transmission of rare CNVs to male probands (Zhao et al., 2007). Consequently, we investigated whether mothers were more likely than fathers to transmit a rare CNV to an affected offspring. We also asked whether there were a greater number of maternally transmitted CNVs in probands versus their unaffected siblings. Neither analysis showed a significant result after correction for multiple comparisons despite considering combinations of the following variables: deletions, duplications, size, exonic, brain-expressed, and ASD-related. In addition, based on the possibility that risk might be confined to only the rarest transmitted events, presumably under the strongest purifying selection, we evaluated “singleton” CNVs, i.e. those observed in only one parent and transmitted to only one proband or sibling. In this case, we found a modest, non-significant excess of maternally transmitted CNVs in probands: 344 maternal autosomal singletons are transmitted to probands vs. 303 transmissions to siblings (OR= 1.14; p=0.059 one-sided; p=0.12 two-sided). For fathers, there was no similar trend (OR= 1.03; p=0.37 one-sided).
Rare transmitted X-linked CNVs are equally represented in probands and siblings
We asked similar questions regarding transmission of rare X-linked CNVs from mothers to male probands and obtained similar results. In a group of 353 male probands and 353 matched male siblings, we found, contrary to expectations, that more siblings carried maternally transmitted rare CNVs than probands (14% probands vs. 18% siblings, OR=0.76, p=0.11), though this difference was not significant. The result did not change when we evaluated the various subcategories of rare X-linked CNVs including exonic, deletions, duplications, size, brain-expressed, or ASD-associated.
Rare transmitted CNVs show greater biological coherence in probands versus siblings
We hypothesized that the absence of evidence of association for rare transmitted CNVs might be a consequence of the inability to differentiate functional from neutral variants. Consequently we looked to pathway analyses to help address this question, reasoning that if the specific genic content of CNVs contributed to disease risk, we would find a greater enrichment of biological pathways in probands versus siblings.
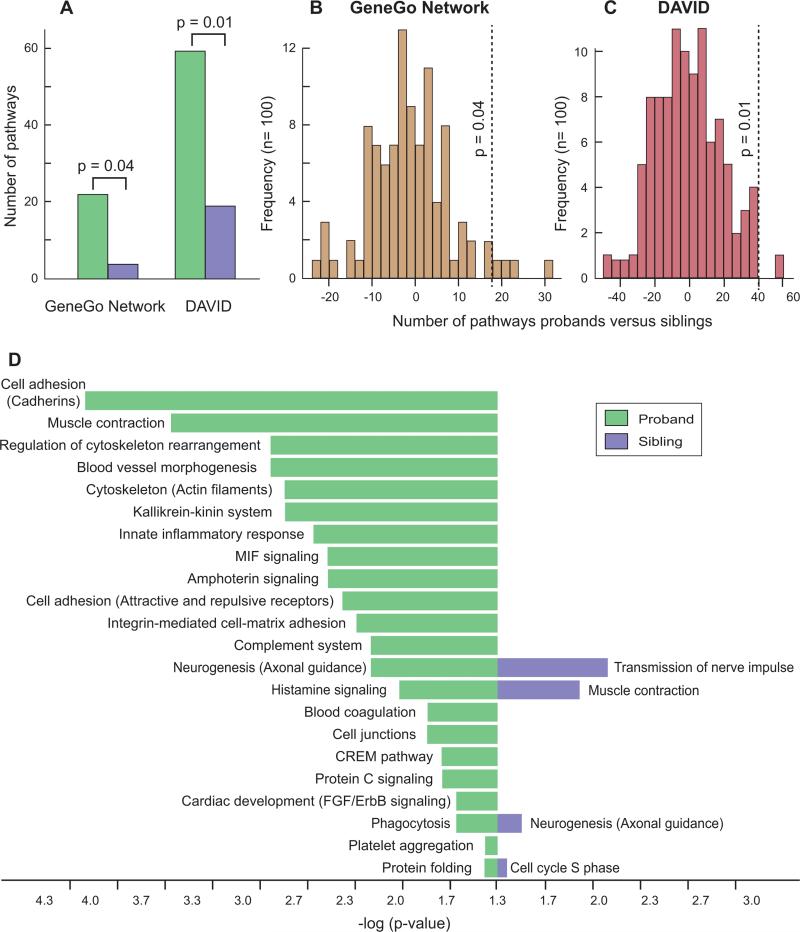
We used two gene ontology and pathway analysis tools, MetaCore from GeneGo Inc. and DAVID (Dennis et al., 2003; Huang et al., 2009), to analyze 1,516 genes within CNVs exclusive to probands and 1,357 genes exclusive to siblings. The total number and size of rare, transmitted CNVs used to determine these gene sets were highly similar in probands and siblings (Figure 5). GeneGo Networks identified 22 pathways showing significant enrichment in probands versus only 4 enriched pathways among siblings. This difference was significant based on 100 permutations of the dataset (p=0.04). DAVID yielded consistent results with 59 pathways enriched in probands and 19 in siblings (p=0.01, permutation analysis) (Figure 6).
Figure 6. Pathway analysis of genes mapping within transmitted rare CNVs.
A) The number of pathways with a corrected p-value ≤0.05 identified in probands (green) and siblings (purple) by the programs Metacore (GeneGo Networks) and DAVID (level 4 terms). The input consisted of 1,516 RefSeq genes found only in transmitted rare CNVs in probands and the 1,357 RefSeq genes found only in transmitted rare CNVs in siblings; p-values are from B and C. B) Permutation analysis to assess significance of the difference between probands and siblings. The 2,873 genes identified in probands or siblings were divided randomly between probands and siblings in the same initial proportions. The lists were submitted to GeneGo Networks and the difference between the number of pathways in probands and siblings was recorded. This process was performed 100 times and the image shows the frequency of the results. Only 4 events showed a difference ≥18 (the difference seen in A, vertical dashed line) yielding a p-value of 0.04. C) Permutation analysis to calculate the significance value with DAVID (level 4 terms) using the same methods as in B. A single result was ≥40 (the difference seen in A, vertical dashed line) giving a p-value of 0.01. D) All pathways with a corrected p-value ≤0.05 identified by GeneGo Networks for probands (green) and siblings (purple). The length of the bar represents the significance value on a logarithmic scale.
For the current analysis, we elected to restrict our evaluation of pathways to the general question described here. A manuscript describing a more extensive pathway analysis is in preparation focusing on both structural and gene expression data from the SSC.
Transmitted autosomal and X chromosome CNVs overlap with previously reported ASD loci
We next examined all rare CNVs in the SSC in light of previously reported findings, comparing our data to the list of ASD regions included in the recent AGP analysis (Pinto et al., 2010). We also considered recent common variant findings, including SEMA5A (Weiss et al., 2009), MACROD2 (Anney et al., 2010), CDH9 and CDH10 (Wang et al., 2009), the MET oncogene (Campbell et al., 2006), EN2 (Gharani et al., 2004), and selected schizophrenia loci (ISC, 2008; McCarthy et al., 2009; Millar et al., 2000; Stefansson et al., 2008; Walsh et al., 2008; Xu et al., 2008) (Table 3). We identified multiple regions in which rare transmitted and/or rare de novo events corresponded to previously characterized loci in both ASD and schizophrenia.
Table 3.
CNVs in genes and regions previously associated with ASD.
| Gene/Regiona | Location (NCBI 36/hg18) | All (de novo)b | Deletions (de novo) | Duplications (de novo) | |||
|---|---|---|---|---|---|---|---|
| Proband | Sibling | Proband | Sibling | Proband | Sibling | ||
| NRXN1 | chr2:50,000,991-51,113,178 | 3 | 1 | 3 | 1 | 0 | 0 |
| CDH10 | chr5:24,522,967-24,680,668 | 0 | 1 | 0 | 1 | 0 | 0 |
| MET | chr7:116,099,695-116,225,676 | 1 | 0 | 0 | 0 | 1 | 0 |
| VPS13B | chr8:100,094,669-100,958,984 | 1 | 0 | 1 | 0 | 0 | 0 |
| CACNA1C | chr12:2,032,676-2,677,376 | 1 | 1 | 0 | 0 | 1 | 1 |
| UBE3A | chr15:23,133,488-23,235,221 | 1 (1) | 0 | 0 | 0 | 1 (1) | 0 |
| NF1 | chr17:26,446,120-26,728,821 | 1 | 0 | 1 | 0 | 0 | 0 |
| MACROD2 | chr20:13,924,146-15,981,841 | 0 | 1 | 0 | 1 | 0 | 0 |
| TBX1 | chr22:18,124,225-18,151,112 | 1 (1) | 0 | 1 (1) | 0 | 0 | 0 |
| ADSL | chr22:39,072,449-39,092,521 | 2 (1) | 2 (0) | 1 (1) | 1 (0) | 1 (0) | 1 (0) |
| NLGN4X | chrX:5,818,082-6,156,706 | 1M | 0 | 0 | 0 | 1M | 0 |
| DMD | chrX:31,047,265-33,267,647 | 1F | 1M,5F | 1F | 1M,5F | 0 | 0 |
| NLGN3 | chrX:70,281,435-70,307,776 | 1M (1M) | 0 | 1M (1M) | 0 | 0 | 0 |
| ATRX | chrX:76,647,011-76,928,375 | 0 | 1F | 0 | 0 | 0 | 1F |
| FMR1 | chrX:146,801,200-146,840,333 | 1F | 1F | 1F | 1F | 0 | 0 |
| RPL10 | chrX:153,279,911-153,285,232 | 1M | 0 | 0 | 0 | 1M | 0 |
| 1q21.1 | chr1:144,022,893-147,496,468 | 3 (2) | 0 | 0 | 0 | 3 (2) | 0 |
| 3q29 | chr3:197,244,288-198,830,238 | 1 (1) | 0 | 1 (1) | 0 | 0 | 0 |
| 4p16.3 | chr4:1-2,043,468 | 1 (1) | 0 | 0 | 0 | 1 (1) | 0 |
| 7q11.23 | chr7:71,970,679-74,254,837 | 4 (4) | 0 | 0 | 0 | 4 (4) | 0 |
| 15q11.2-13.1 | chr15:20,768,955-26,230,781 | 1 (1) | 0 | 0 | 0 | 1 (1) | 0 |
| 15q13.2-13.3 | chr15:28,698,632-30,234,007 | 3 (2) | 0 | 2 (1) | 0 | 1 (1) | 0 |
| 16p13.11 | chr16:15,421,876-16,200,195 | 3 (1) | 5 (2) | 1 (0) | 1 (1) | 2 (1) | 4 (1) |
| 16p11.2 | chr16:29,474,810-30,235,818 | 14 (11) | 0 | 8 (7) | 0 | 6 (4) | 0 |
| 17q12 | chr17:31,893,783-33,277,865 | 2 (1) | 0 | 2 (1) | 0 | 0 | 0 |
| 22q11.21 typical | chr22:17,412,646-19,797,314 | 1 (1) | 0 | 1 (1) | 0 | 0 | 0 |
| 22q11.21 distal | chr22:22,028,923-23,368,015 | 1 | 0 | 0 | 0 | 1 | 0 |
For genes a CNV was included if it overlapped ≥1 exon; for regions CNVs spanning ≥50% of the region and ≥1 exon and included.
De novo CNV count in parentheses; for chromosome X sex is indicated by M for male and F for female.
Rare transmitted CNVs do not show genome-wide association in the SSC
Finally, we looked for evidence of association for any other CNVs in the SSC sample, evaluating all high-confidence autosomal CNVs together with all confirmed de novo CNVs. In this instance, we did not use a frequency cutoff to define a set of rare transmitted events. A total of 3,667 recurrent regions were identified; 6 showed relative enrichment in probands and 5 in siblings. No result reaches significance after correction for multiple comparisons (Table S7, Figure 7C). The region showing the greatest difference in probands compared to siblings was 16p11.2 (p=0.001).
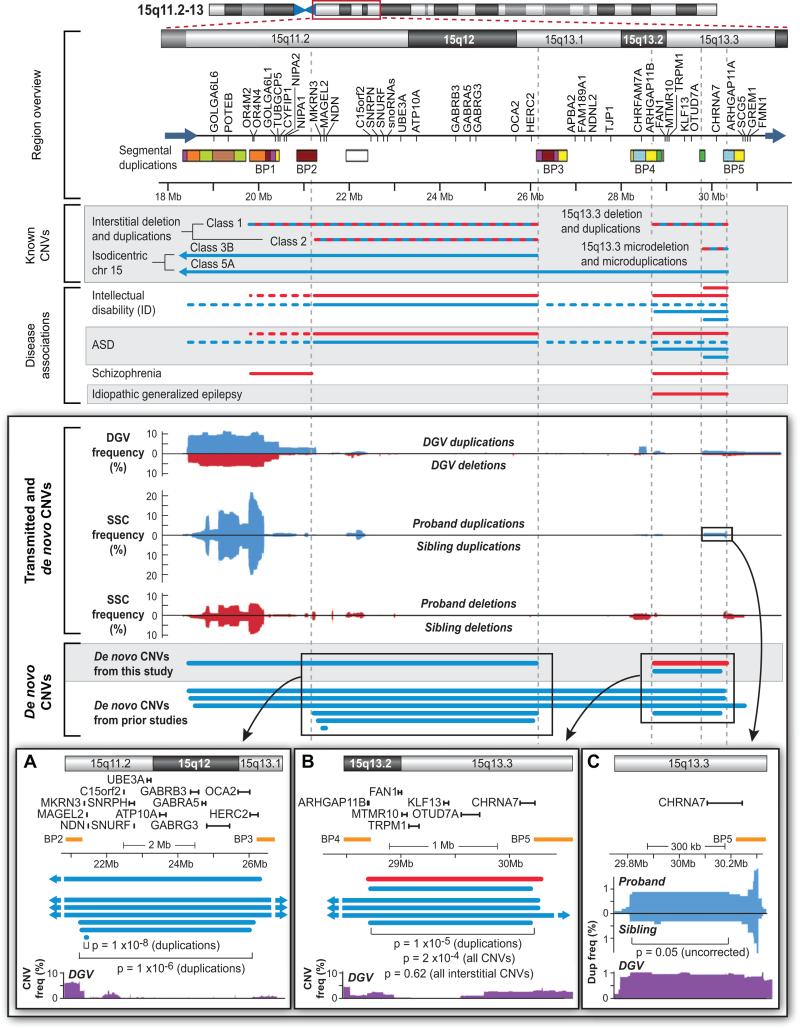
Figure 7. De novo and transmitted CNVs in 15q11.2-13.
A 13Mb region is identified by the red box on the ideogram at the top. The Region overview identifies the RefSeq genes present within the interval and multiple segmental duplications (the colors identify regions of homology (Makoff and Flomen, 2007)). 5 of these segmental duplications are commonly referred to as BP1-BP5. Known CNVs identifies duplications (blue) and deletions (red) that have been reported in the literature; the alternating red and blue colors denote both deletions and duplications. Disease associations identifies regions with reported associations to four developmental and neuropsychiatric conditions (supplementary materials). Of note, BP2-BP3 deletions lead to Prader-Willi or Angelman syndrome. Transmitted and de novo CNVs shows the frequency of duplications (blue) and deletions (red) in the DGV and SSC populations. While CNVs overlying the segmental duplications are common, CNVs between the breakpoints are generally rare. De novo CNVs shows confirmed de novo CNVs in single individuals identified in this study and prior ASD studies (Itsara et al., 2010; Marshall et al., 2008; Pinto et al., 2010; Sebat et al., 2007). A) An enlargement of BP2-3 showing the relationship of de novo CNVs, genes, and common regions in the DGV. A small atypical duplication includes the genes MAGEL2, MKRN3, NDN (Itsara et al., 2010). B) An enlargement of BP4-BP5 showing similar data and methods as A. Removing the three Class 5A isodicentric chr15 events results in a non-significant p-value (p=0.62). C) An enlargement of the CHRNA7 region showing enrichment of duplications in probands (N=10) vs. siblings (N=3). The p-value is p=0.05 (Fisher's exact test); uncorrected for 3,667 comparisons; the rate of duplications in the DGV is similar to that seen in probands (Table S7).
Expanded analysis of rare de novo CNVs across multiple ASD samples
An analysis of de novo CNVs in 3,816 probands from genome-wide studies of idiopathic ASD supports association of 6 genomic intervals
Our approach to assessing the genome-wide significance of rare recurrent de novo CNVs allows for a statistical evaluation of events observed in cases without requiring additional matched control samples. Consequently, we were able to conduct a cumulative analysis across multiple studies in search of additional associated ASD loci. We included 4 other large-scale ASD CNV studies (Itsara et al., 2010; Marshall et al., 2008; Pinto et al., 2010; Sebat et al., 2007) meeting 4 criteria: standardized diagnosis, genome-wide detection, confirmed de novo structural variations, and sufficient information to permit the identification of duplicate samples.
These datasets catalogued 228 confirmed, rare de novo CNVs from a total of 3,816 individuals (Table S1). We found 6 regions that exceeded the threshold for significance (methods). Given prior evidence, and our own data, that reciprocal deletions and duplications at certain loci, both contribute to the ASD phenotype we evaluated significance for combined events at an interval, as well as calculating probabilities for deletions and duplications separately (Table 4, Figure S3).
Table 4.
Recurrent (≥2) de novo regions across this and other studies.
| Type | Band | Location (NCBI 36/hg18) | Size (kb) | Recurrence (del/dup) | Frequency (N=3,816) | p-valuea (C=232) | Studiesb | Genesc (RefSeq) |
|---|---|---|---|---|---|---|---|---|
| Deletions | 2p16.3 | chr2:51,002,576-51,157,742 | 155 | 2 | 0.05% | 0.94 | 3 | NRXN1 d |
| 2q24.2 | chr2:162,212,720-162,311,972 | 99 | 2 | 0.05% | 0.94 | 5 | SLC4A10 (intronic) | |
| 2q37.3 | chr2: 238,217,066-242,701,103 | 4,484 | 2 | 0.05% | 0.94 | 5 | 41 | |
| 3p14.1 | chr3:65,674,445-65,725,692 | 51 | 2 | 0.05% | 0.94 | 2,4 | MAGI1 (intronic) | |
| 3p14.1 | chr3:67,223,272-70,633,200 | 3,410 | 2 | 0.05% | 0.94 | 1,4 | 10 | |
| 5p15.2 | chr5:11,403,359-11,491,117 | 87 | 3 | 0.08% | 0.11 | 1,4 | CTNND2 | |
| 7q31.1-31.31 | chr7:113,335,000-119,223,887 | 5,889 | 2 | 0.05% | 0.94 | 4 | 20 | |
| 7q36.2 | chr7:153,380,710-154,316,928 | 936 | 2 | 0.05% | 0.94 | 3,4 | DPP6 | |
| 9p24.3 | chr9:98,998-334,508 | 235 | 2 | 0.05% | 0.94 | 3 | C9orf66, CBWD1, DOCK8, FOXD4 | |
| 11q13.3 | chr11:70,154,458-70,187,872 | 33 | 2 | 0.05% | 0.94 | 3 | SHANK2 | |
| 14q32.12 | chr14:92,476,815-92,496,373 | 19 | 2 | 0.05% | 0.94 | 2 | ITPK1 | |
| 15q23-24.1 | chr15:69,601,300-71,944,199 | 2,343 | 2 | 0.05% | 0.94 | 1,4 | 22 | |
| 16p11.2 | chr16:29,578,715-30,001,681 | 422 | 14 | 0.37% | 5 × 10−29 | 1,2,3,4,5 | 26 | |
| 16q23.3 | chr16:81,796,275-81,830,296 | 34 | 2 | 0.05% | 0.94 | 1 | CDH13 | |
| 16q23.3 | chr16:82,557,318-82,683,859 | 126 | 2 | 0.05% | 0.94 | 1,2 | MBTPS1, NECAB2, OSGIN1, SLC38A8 | |
| 18q22.1 | chr18:64,812,093-6,484,6196 | 34 | 2 | 0.05% | 0.94 | 2,4 | CCDC102B | |
| 20p12.1 | chr20:14,616,243-14,751,454 | 135 | 2 | 0.05% | 0.94 | 1,3 | MACROD2 (intronic) | |
| 22q11.21 | chr22:17,257,787-19,786,200 | 2,528 | 3 | 0.08% | 0.11 | 1,3,4 | 56 | |
| 22q13.33 | chr22:49,243,247-49,465,883 | 222 | 3 | 0.08% | 0.11 | 4,5 | 16 | |
| Duplications | 1q21.1 | chr1:144,838,175-146,324,832 | 1,487 | 2 | 0.05% | 0.88 | 1 | 14 |
| 2p25.3 | chr2:143,279-196,704 | 53 | 2 | 0.05% | 0.88 | 2 | 0 | |
| 3q21.2 | chr3:125,966,642-127,254,388 | 1,288 | 2 | 0.05% | 0.88 | 2 | 11 | |
| 7q11.23 | chr7:72,411,506-73,782,113 | 1,371 | 4 | 0.09% | 0.003 | 1 | 22 | |
| 8p23.3 | chr8:710,491-1,501,580 | 791 | 2 | 0.05% | 0.88 | 3,4 | DLGAP2 | |
| 10q11.23-21.1 | chr10:52,699,516-54,408,816 | 1,709 | 2 | 0.05% | 0.88 | 1,2,5 | CSTF2T, DKK1, MBL2, PRKG1 | |
| 12q24.31 | chr12:120,628,928-120,862,589 | 233 | 2 | 0.05% | 0.88 | 2,4 | 7 | |
| 15q11.2 | chr15:21,343,866-21,505,342 | 161 | 7 | 0.18% | 4 × 10−9 | 1,2,3,4,5 | MAGEL2, MKRN3, NDN | |
| 15q11.2-13.1 | chr15:21,240,037-26,095,621 | 4,856 | 6 | 0.16% | 4 × 10−4 | 1,2,3,4,5 | 12 | |
| 15q13.2-13.3 | chr15:28,723,577-30,231,488 | 1,508 | 5e | 0.13% | 2 × 10−5 | 1,2,4,5 | CHRNA7, KLF13, MTMR10, MTMR15, OTUD7A, TRPM1 | |
| 16p13.2 | chr16:8,828,382-9,147,487 | 319 | 2 | 0.05% | 0.88 | 1 | PMM2, CARHSP1, USP7, C16orf72 | |
| 16p11.2 | chr16:29,563,365-30,085,308 | 521 | 5 | 0.13% | 2 × 10−5 | 1,3,4 | 26 | |
| 20q13.33 | chr20:60,949,339-61,220,552 | 271 | 2 | 0.05% | 0.88 | 2,4 | 7 | |
| 22q11.21 | chr22:17,265,500-19,791,274 | 2,526 | 2 | 0.05% | 0.88 | 2,4 | 56 | |
| Combinedf | 1q21.1 | chr1:145,013,719-146,293,282 | 1,280 | 3 (1/2) | 0.08% | 0.53 | 1,2 | 14 |
| 2p16.3 | chr2:50,539,877-50,677,835 | 138 | 2 (1/1) | 0.05% | 1.00 | 3 | NRXN1 d | |
| 7q31.1 | chr7:108,242,570-108,393,666 | 151 | 2 (1/1) | 0.05% | 1.00 | 2,4 | C7orf66 | |
| 7q31.1 | chr7:111,065,681-111,454,179 | 388 | 2 (1/1) | 0.05% | 1.00 | 2,4 | DOCK4 | |
| 9p24.3 | chr9:175,632-334,508 | 159 | 3 (2/1) | 0.08% | 0.53 | 1,3 | C9orf66, DOCK8 | |
| 15q13.2-13.3 | chr15:28,723,577-30,231,488 | 1,508 | 6 (1/5)e | 0.16% | 1 × 10−4 | 1,2,4,5 | CHRNA7, KLF13, MTMR10, MTMR15, OTUD7A, TRPM1 | |
| 16p11.2 | chr16:29,578,715-30,001,681 | 521 | 19 (14/5) | 0.50% | 2 × 10−55 | 1,2,3,4,5 | 26 | |
| 16q22.3 | chr16:69,987,425-70,647,241 | 660 | 2 (1/1) | 0.05% | 1.00 | 1,2 | 13 | |
| 20q13.33 | chr20:61,056,624-61,076,763 | 20 | 3 (1/2) | 0.08% | 0.53 | 2,4 | SLC17A9 | |
| 22q11.21 | chr22:17,265,500-19,786,200 | 2,521 | 5 (3/2) | 0.13% | 0.002 | 1,2,3,4 | 56 |
p-values are calculated as described in the methods.
1. This study; 2. Itsara et al., 2010; 3. Pinto et al., 2010; 4: Marshall et al., 2008; 5. Sebat et al., 2007.
Counts are given for CNVs with >6 RefSeq genes mapping to the interval. A complete listing of genes is in Table S4. All genes shown represent exonic overlap unless otherwise indicated.
While only 2 de novo CNVs overlap within NRXN1, there are 5 de novo events overlapping a section of the gene: 1 intronic deletion, 3 exonic deletions, and 1 exonic duplication (p=0.004 combined, p=0.007 for deletions).
3 of the duplications contributing to 15q13.2-13.3 are isodicentric chr15 events; since there is a longstanding associated with ASD and isodicentric chr15, this region is also considered without these events. For interstitial CNVs alone there 2 duplications and 1 deletion (p=0.62 combined; p=0.92 duplications) (Figure 7).
Regions are only listed in the combined category if there are a combination of deletions and duplications resulting in a different p-value when the two types of CNVs are considered together.
The most frequent recurrent de novo CNV identified across all studies was 16p11.2 with 19 identified probands (14 deletions, 5 duplications) showing extremely strong evidence for association with ASD (2 × 10−55 combined; 5 × 10−29 for deletions; 2 × 10−5 for duplications). The proximal long arm of chromosome 15 showed two contiguous intervals: the first corresponds to the region 15q11.2-13.1 or BP2-BP3 (7 duplications; 4 × 10−9) (Figure 7A), long cited as the most common cytogenetic abnormality identified in idiopathic ASD (Cook et al., 1997). We also found evidence of association for the interval mapping to 15q13.2-13.3 or BP4-BP5 (5 duplications and 1 deletion; 1 × 10−4 combined, 2 × 10−5 for duplications) (Figure 7B). Rare deletions and duplications in this region have previously been associated with intellectual disability and ASD, and deletions with schizophrenia and epilepsy (Figure 7). It is important to note, however, that considering only events restricted to 15q13.2-13.3 (i.e. removing 3 overlapping isodicentric chromosome 15 events) we do not find significance (0.53 combined; 0.88 for duplications). This suggests either that the result is an incidental finding due to the proximity to a true ASD risk locus, or, alternatively, that the smaller 15q13.2-13.3 CNVs might point to a minimum region of overlap mapping to one or more ASD-related genes.
Recurrent de novo CNVs exceeding the significance threshold in the combined sample were also present at 7q11.23 (4 duplications; 0.003), 22q11.2 region (3 deletions and 2 duplications; 0.002 combined; 0.11 for deletions; 0.88 for duplications), and at the locus coding for the gene NRXN1. For NRXN1 there were 5 de novo events: 1 intronic deletion, 3 exonic deletions, and 1 exonic duplication (0.002 combined, 0.004 for deletions).
Finally, we used the observed number and distribution of de novo CNVs in the combined proband data set to estimate the likely number of CNV regions contributing to ASD. From the total of 219 confirmed de novo events, we derived an estimate of 234 distinct genomic regions contributing to large ASD-related de novo structural variations (methods).
Discussion
Our results highlight the importance of rare CNVs for simplex ASD. We confirm an over-representation of rare de novo events in probands versus siblings with an odds ratio of 3.55 for all variants and 4.02 for rare de novo genic variants. Using a novel approach to assessing significance specifically for recurrent de novo CNVs, we find very strong evidence for the contribution of duplications at 7q11.23, showing, for the first time., genome-wide association in a case-control study. Moreover, we identify four additional rare recurrent de novo events restricted to probands. Two of these, 16p13.2 and the CDH13 locus, are novel ASD loci and two, 1q21, 15q13.2-13.3, have been previously implicated in neuro-developmental disorders including ASD. Each of these four regions also show rare transmitted CNVs exclusive to probands. Finally, we find compelling evidence confirming the association of both 16p11.2 duplications and deletions.
It is striking that while we replicate findings of elevated rates of rare de novo CNVs in simplex families (5.8% of probands versus 1.7% in siblings), the percentage of the cohort carrying these events is the same magnitude as that seen previously. This is despite the intensive focus on the ascertainment of simplex quartets and the 10-fold increase in probe density since the earliest studies of ASD. We believe these results are best explained by the particular contribution of large genic de novo variants given the results of our analysis of gene number, CNV size, and affected status (Figure 3), and the observation of generally consistent results over time despite steadily increasing detection resolution.
While it may not seem surprising that large de novo events carry the greatest risk for developmental disorders, it is interesting to note that we did not find evidence that ASD diagnosis or severity was mediated by intellectual disability (ID). It has been argued that ASD in the presence of ID may reflect an epiphenomenon, in which a non-specific impairment of brain functioning unmasks and/or exacerbates limitations in an individual's capacity for social reciprocity (Skuse, 2007). It has also been widely held that the detection of large de novo CNVs will be enhanced by the ascertainment of ASD samples with greater intellectual disability. Our data shows that large de novo CNVs confer substantial risk for ASD in the SSC, but they are only modestly correlated with lower IQ and largely independent of ASD severity.
These data suggest both that this study has identified bona fide high-risk variants for autism spectrum disorders and that many of these loci also confer liability to a range of complex neurobehavioral phenotypes. They also suggest a more complex relationship of IQ and large de novo events than is often supposed: for example the relatively high rates of 16p11.2 and 7q11.23 CNVs and low rates of 15q11.2-13.1 duplications seen in this study compared to others may reflect particular subpopulations of rare de novo risk CNVs that are more readily ascertained in cohorts with higher mean IQ.
The results further show that the risk associated with large de novo events is related to their greater genic content, even after controlling for larger size. This observation points to two countervailing hypotheses: first, that the greater gene number is a surrogate for the increased chance of disrupting one particular gene or regulatory region due to the involvement of a larger segment of the coding genome; or second, that it is the contribution of multiple genes and/or regulatory regions simultaneously within these CNVs that increases risk.
Our data do not allow us to resolve this issue. Nonetheless we suspect that if many deletions or duplications encompassing small numbers of genes were as highly penetrant as multigenic events, we would have begun to show more evidence for this either in the form of an overall increased burden for smaller de novo variations and/or association of specific de novo events. However, it is important to note that despite higher resolution than some prior studies, we nonetheless have a clear ascertainment bias for detection of larger CNVs. It is likely that the combination of high-throughput sequencing, larger patient cohorts, and increasingly sophisticated approaches to evaluating combinations of risk variants will begin to shed light on this issue, with regard to both sequence and structural variation.
Our findings with regard to recurrent de novo events in the SSC sample point to 6 putative ASD loci: two of these, 7q11.23 and 16p11.2, show clear evidence for genome-wide association. Moreover, our simulation analysis suggests that the most likely outcome of the ongoing Phase 2 SSC study will be confirmation of 2-3 of the remaining 4 intervals already showing recurrent de novo events, namely 1q21.1, 15q13.2-13.3, 16p13.2, and 16q23.3 (CDH13).
Our findings at 7q11.23 point to extraordinary opportunities to illuminate the molecular mechanisms of social development. Duplications in this interval have previously been described in developmental disorders, including ASD (Berg et al., 2007; Van der Aa et al., 2009), though these have been restricted to case reports or series, with the attendant difficulties in controlling for ascertainment bias. The identification of clear association of duplications in this controlled study of ASD is particularly striking given that the reciprocal deletion results in a developmental syndrome characterized in part by an empathic, gregarious, and highly social personality (Pober, 2010). Moreover, several lines of evidence, including atypical deletions (Antonell et al., 2010), mouse models (Fujiwara et al., 2006; Hoogenraad et al., 2002; Meng et al., 2002; Sakurai et al., 2010), and gene expression x phenotype studies (Gao et al., 2010; Korenberg et al., 2000) have already identified CAPGLY domain containing linker protein 2 (CLIP2), LIM domain kinase 1 (LIMK1), General transcription factor II, i (GTF2i), and Syntaxin 1A (STX1A) as the leading candidates among the 22 genes within the region for involvement in the cognitive and social phenotypes. The characterization of this single region in which opposite changes in copy number contribute to contrasting social phenotypes promises to set the stage for a range of interesting studies of the role of gene dosage within this interval and the genesis of social mechanisms.
The strong replication of findings at 16p11.2 also highlights emerging opportunities for translational neuroscience. Firstly, the region is sufficiently circumscribed to interrogate using molecular biological and model systems. Secondly, though we cannot quantify an odds ratio from our data, given the absence of events in siblings, there is clear evidence from this and prior studies (McCarthy et al., 2009) that 16p11.2 CNVs carry much larger effects than any common variant contributing to complex common disorders. Thirdly, the 1% allele frequency allows for prospective studies of natural history, neuroimaging, and treatment response, as, for example, in the recently launched Simons Variation in Individuals Project (https://sfari.org/simons-vip). Finally, the entirety of the data now strongly supports a role for both duplications and deletions of 16p11.2 in social disability. Together, these suggest that cross-disciplinary approaches can begin to address the means by which a single locus leads to a range of psychiatric outcomes previously conceptualized as distinct and to address the critical role that dosage sensitivity plays in the unfolding of these neuro-developmental phenotypes.
The notion that the 4 remaining recurrent de novo regions represent true ASD variants is supported by multiple lines of additional evidence. For example, they are among only 8 rare de novo CNVs that overlap with rare transmitted events restricted to probands. Moreover, 2 of the remaining 3 loci, 2p15 and 17q12, have been previously implicated in ASD (Liang et al., 2009; Moreno-De-Luca et al., 2010). In addition, rare 1q21.1 and 15q13.2-13.3 CNVs have been identified in developmental and neuropsychiatric syndromes, with deletions found in ASD (Miller et al., 2009; Shen et al., 2010), schizophrenia (ISC, 2008; Stefansson et al., 2008), idiopathic epilepsy (Helbig et al., 2009), and recurrent duplications reported here. CDH13 has not previously been noted to be a risk variant, but the larger family of proteins has been implicated in ASD pathogenesis through CNV studies (Glessner et al., 2009), homozygosity mapping (Morrow et al., 2008), common variant findings (Wang et al., 2009), and our pathway analysis (Figure 6). The 16p13.2 region contains four genes, the most immediately notable of which are C16orf72, coding for a protein of unknown function, recently identified in a schizophrenia CNV study (Levinson et al., 2011), and Ubiquitin Specific Peptidase 7 (USP7), which has been shown to have a role in oxidative stress response, histone modification and regulation of chromatin remodeling (Khoronenkova et al., 2010). Both would represent novel ASD risk genes, however for the latter, these biological processes, and the ubiquitin pathway in particular, have been previously implicated in ASD pathogenesis (Glessner et al., 2009).
The fact that the family-based design used in our study played a key role in allowing us to identify association presents an important contrast to the prevailing wisdom with regard to genome-wide association studies of common variants, in which there is a tendency to rely on unrelated case-control designs, given the relative ease of generating very large sample sizes. It is notable that the statistical power afforded by the low probability of observing multiple recurrent rare de novo events by chance more than compensated for the comparatively small sample reported here. This is particularly striking with respect to 16p11.2. Based on a traditional case-control comparison, the most significant finding in this sample, 14 events in probands and 0 in siblings (p=0.001, Fisher's exact test), did not provide evidence sufficient to withstand correction for multiple comparisons, while the analysis based on the null expectation for de novo recurrence clearly detected association. It is certain that the SSC sample ascertainment process enhanced certain findings and attenuated others. There is little question that restricting the comparison group to matched siblings limited power to identify association of specific rare recurrent transmitted events; our assessment of significance for de novo CNVs was based on conservative assumptions and may have excluded true risk loci; the filtering for rare de novo CNVs and the small sample size curtailed the assessment of multi-hit hypotheses; the generally older parental age may have obscured the relationship between age and de novo variation (Figure S3) and, as noted, poor specificity at the lower bound of detection limited our assessment of small de novo structural variations.
However, despite these limitations, the manner in which the design mitigated important confounds, and preserved sufficient power to detect association of recurrent de novo events, yielded clear benefits, unambiguously replicating prior findings and identifying novel risk loci. Moreover, this report considers less than half of the Simons Simplex Collection: phase 2 of this study is currently underway, as is high-throughput sequencing of the collection, also focusing on de novo events. Together these endeavors promise to further illuminate the genomic architecture of simplex autism and to provide additional critical points of traction for efforts toward elaborating the molecular mechanisms and developmental neurobiology underlying ASD.
Experimental Procedures
Genotyping
All members of each family were analyzed on the same array version: either the Illumina 1Mv1 (334 families) or Illumina 1Mv3 Duo (840 families) Bead-array. These share 1,040,853 probes in common (representing 97% of probes on the IMv1 and 87% of probes on the 1Mv3). 824 or the 872 quartet families (94.5%) had all members hybridized and scanned simultaneously on the Illumina iScan in an effort to minimize batch effects and technical variation.
Identity quality control
Genotyped samples were analyzed using Plink (Purcell et al., 2007) to identify incorrect sex, Mendelian inconsistencies, and cryptic relatedness by assessing inheritance-by-descent (IBD); 11 families were removed as a result.
CNV detection
CNV detection was performed using three algorithms: 1) PennCNV Revision 220, 2) QuantiSNP v1.1, and 3) GNOSIS. PennCNV and QuantiSNP are based on the Hidden Markov Model (HMM). GNOSIS uses a continuous distribution function (CDF) to fit the intensity values from the HapMap data and determine thresholds for significant points in the tails of the distribution that are used to detect copy number changes. Analysis and merging of CNV predictions was performed with CNVision (www.CNVision.org), an in-house script.
CNV Quality Control
Specific genotyping and CNV parameters are detailed in the supplementary methods. 5% of the samples failed and were rerun; 39 families were removed due to repeated failures.
Criteria for a Rare CNVs
A CNV was classified as rare if ≤50% of its length overlapped regions present at >1% frequency in the Database of Genomic Variation (DGV) March 2010.
CNV burden
Burden analyses were performed on the matched set of 872 probands and siblings. Typically, three outcomes were assessed: proportion of individuals with ≥1 CNV matching the criteria (p-value calculated with Fisher's exact test); number of CNVs matching the criteria (p-value calculated with sign test); number of RefSeq genes within or overlapping CNVs matching the criteria (p-value calculated with Wilcoxon paired test). Where burden was assessed for unequal numbers of probands and siblings (e.g. by sex) the sign test and Wilcoxon paired test were replaced with the Wilcoxon test.
Statistical analysis of de novo recurrence
To determine the probability of finding multiple rare de novo CNVs at the same location in probands, we first estimated how many likely positions in the genome were contributing to the observed de novo CNVs in siblings. As there are widely varying mutation rates for structural variation across the genome (Fu et al., 2010), some positions are more likely to result in de novo CNVs observed in our sample than others. Consequently, the likely number of positions is much smaller than the total possible number of positions. We refer to the likely CNV regions as eCNVRs (effective copy number variable regions) and calculate their quantity “C” using the so-called “unseen species problem” which uses the frequency and number of observed CNV types (or species) to infer how many species are present in the population. Based on the observed de novo CNVs in the control sibling group, we apply the formula (Bunge and Fitzpatrick, 1993) C = c/u + g2*d*(1-u)/u, in which c = the total number of distinct species observed; c1= the number of singleton species; d = total number of CNVs observed; g = the coefficient of variation of the fractions of CNVs of each type, and u = 1 – c1/d. (In this calculation, due to the small number of observations, we assume that g equals 1.) For the de novo events in siblings, c1=14, c=15, d=16 and C=232. This calculation is performed in the siblings because the observed rare de novo CNVs in this group are assumed to be predominantly non-risk variants and consequently represent the null distribution.
Next, we calculate the chance that two de novo events match at any one of “C” eCNVRs in probands, using methods from the classic “birthday problem” which assess the likelihood of seeing at least one pair of matching birthdays among a given number of people. Our interest was in seeing >2 matches (m) in probands under the null hypothesis of no association with ASD. This calculation is performed empirically by distributing “d” events at random among “C” eCNVRs and then counting the maximum number of CNVs falling in the same location. Repeating this experiment many times, we obtained an estimate of the probability of finding “≥m” counts for ≥1 eCNVR under the null hypothesis.
Given the importance of the estimate of eCNVRs in unaffected populations for the determination of significance, we re-calculated “C” based on a combined set of confirmed de novo CNVs in controls described in the literature and obtained a highly similar result (C=242) (supplementary materials). Moreover, we determined that the results reported here remain significant under the plausible range of estimates for “C” (supplementary materials).
Estimate of number of de novo CNV regions contributing to ASD risk
The unseen species problem was used to predict the total number of ASD risk-loci based on the distribution of de novo CNVs in probands. This required identification of the de novo CNVs that confer risk; to identify such CNVs we estimated that 75% of de novo CNVs in probands confer risk (67 de novo CNVs in probands – 16 de novo CNVs expected in siblings / 67 de novo CNVs in probands) and assumed that recurrent de novo CNVs were most likely to be associated with risk and should be included within this 75%. The remainder of the 75% is made up of 27 single occurrence de novo CNVs (though we do not identify which ones) leading to an estimate of the total number of risk conferring loci as 130 (c1=27, c=33, d=51). A similar approach was applied to all de novo CNVs in 3,816 probands (count derived from the literature), leading to an estimate of 234 risk conferring loci (c1=59, c=88, d=158).
Stepwise assessment of multiple variables
Predictors were examined in a logical order, e.g. to evaluate the relationship between gene number (G), CNV size (L), and affection status (A, proband vs. sibling), we fit a series of increasingly complex linear models in the following steps: (1) regress response G on predictor L, regress G on A; 2) if ≥1 term was significant, and assuming L had the best predictive power, we regressed G on L and A; (3) assuming L and A were significant jointly, we regressed G on L, A and L*A (L interacting with A). The latter term permits the slope of the relationship between G and L to differ for probands vs. siblings. In each step, we determined if the newest term was significant, given the terms already in the model. We also fit the model using backward elimination, starting with the full model and simplifying it one term at a time.
Population structure of recurrent de novo CNVs
All parents were projected onto a five-dimensional ancestry map using eigenvector decomposition (Crossett et al., 2010; Lee et al., 2009). Euclidean distances were measured for the parent-of-origin. The mean and median distances between these pairs of parents were calculated and were evaluated relative to the remainder of the sample using a bootstrap procedure (supplementary methods).
Genotype-Phenotype analysis
For each sample with a 16p11.2 deletion (8 samples) or duplication (6 samples) or 7q11.23 duplication (4 samples) 5 control probands were selected based on a matching hierarchy: age (100% of control probands matched), sex (100%), genetic distance (91%, based on five-dimensional ancestry map), collecting site (46%), and quartet/trio family (34%). Probands with de novo CNVs or CNVs in regions previously associated with ASD were removed prior to matching; each control proband was only included once.
For continuous variables each stratum of a “case” proband matched to 5 “control” probands was treated as a block and the data analyzed as a randomized block design by using analysis of covariance. Thus mean values were allowed to vary across blocks and to be altered by case-control status. The difference due to the presence of the CNV of interest was assessed with an F-test with N, M degrees-of-freedom (N is the number of CNVs of interest and M is the residual degrees-of-freedom after accounting for model terms). Because IQ is known to affect many behavioral measures associated with ASD, it was treated as a covariate in models for outcomes besides itself and Body Mass Index (BMI). For diagnostic status, matching was taken into account by using a conditional logit model.
Supplementary Material
Acknowledgements
We are most grateful to all of the families at the participating SFARI Simplex Collection (SSC) sites. This work was supported by a grant from the Simons Foundation (SFARI 124827). C.A.W. and R.P.L. are Investigators of the Howard Hughes Medical Institute. We wish to thank: the SSC principal investigators: (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C.A. Walsh, E. Wijsman); the coordinators and staff at the SSC sites; the SFARI staff (M. Greenup and S. Johnson); R. Smith and Z. Galfayan at Microangelo Associates for bioinformatics support; Prometheus Research; the Yale Center of Genomic Analysis (YCGA) staff, in particular S. Umlauf and C. Castaldi; T. Brooks-Boone and M. Wojciechowski for their help in administering the project at Yale; and J. Krystal, G.D. Fischbach, A. Packer, J. Spiro, and M. Benedetti for their suggestions throughout and very helpful comments during the preparation of this manuscript. Approved researchers can obtain the SSC population dataset described in this study by applying at https://base.sfari.org.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonell A, Del Campo M, Magano LF, Kaufmann L, de la Iglesia JM, Gallastegui F, Flores R, Schweigmann U, Fauth C, Kotzot D, Pérez-Jurado LA. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams-Beuren syndrome neurocognitive profile. J Med Genet. 2010;47:312–320. doi: 10.1136/jmg.2009.071712. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, Fong CT, Salamone J, Freedenberg D, Hannig VL, Prock LA, Miller DT, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Pérez Jurado LA, Delle Chiaie B, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O'Rahilly S, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge M, Bruun-Petersen G, Brøndum-Nielsen K, Friedrich U, Hansen J, Jensen G, Jensen PK, Kristoffersson U, Lundsteen C, Niebuhr E, et al. Disease associated balanced chromosome rearrangements: a resource for large scale genotype-phenotype delineation in man. J Med Genet. 2000;37:858–865. doi: 10.1136/jmg.37.11.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge J, Fitzpatrick M. Estimating the Number of Species: A Review. Journal of the American Statistical Association. 1993;88:364–373. [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- Crossett A, Kent BP, Klei L, Ringquist S, Trucco M, Roeder K, Devlin B. Using ancestry matching to combine family-based and unrelated samples for genome-wide association studies. Stat Med. 2010;29:2932–2945. doi: 10.1002/sim.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Fu W, Zhang F, Wang Y, Gu X, Jin L. Identification of copy number variation hotspots in human populations. Am J Hum Genet. 2010;87:494–504. doi: 10.1016/j.ajhg.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Mishima T, Kofuji T, Chiba T, Tanaka K, Yamamoto A, Akagawa K. Analysis of knock-out mice to determine the role of HPC-1/syntaxin 1A in expressing synaptic plasticity. J Neurosci. 2006;26:5767–5776. doi: 10.1523/JNEUROSCI.0289-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao MC, Bellugi U, Dai L, Mills DL, Sobel EM, Lange K, Korenberg JR. Intelligence in Williams Syndrome is related to STX1A, which encodes a component of the presynaptic SNARE complex. PLoS One. 2010;5:e10292. doi: 10.1371/journal.pone.0010292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH. Association of the homeobox transcription factor, ENGRAILED 2, 3, with autism spectrum disorder. Mol Psychiatry. 2004;9:474–484. doi: 10.1038/sj.mp.4001498. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Koekkoek B, Akhmanova A, Krugers H, Dortland B, Miedema M, van Alphen A, Kistler WM, Jaegle M, Koutsourakis M, et al. Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet. 2002;32:116–127. doi: 10.1038/ng954. [DOI] [PubMed] [Google Scholar]
- Huang d.W., Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- ISC ISC. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianova II, Parsons JL, Dianov GL. USP7/HAUSP stimulates repair of oxidative DNA lesions. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck SM, Münstermann E, Bieber-Martig B, Rühl D, Lisch S, Schmötzer G, Poustka A, Poustka F. Molecular genetic analysis of the FMR-1 gene in a large collection of autistic patients. Hum Genet. 1997;100:224–229. doi: 10.1007/s004390050495. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Hirota H, Lai Z, Bellugi U, Burian D, Roe B, Matsuoka R. VI. Genome structure and cognitive map of Williams syndrome. J Cogn Neurosci. 2000;12(Suppl 1):89–107. doi: 10.1162/089892900562002. [DOI] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Lee C, Abdool A, Huang CH. PCA-based population structure inference with generic clustering algorithms. BMC Bioinformatics 10 Suppl. 2009;1:S73. doi: 10.1186/1471-2105-10-S1-S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, et al. Copy Number Variants in Schizophrenia: Confirmation of Five Previous Findings and New Evidence for 3q29 Microdeletions and VIPR2 Duplications. Am J Psychiatry. 2011 doi: 10.1176/appi.ajp.2010.10060876. appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee Y.-h., Leotta A, Kendall J, Marks S, Lakshmi B, Ye K, Buja A, et al. Rare de novo and transmitted copy number variation in autistic spectrum disorders. Neuron. 2011 doi: 10.1016/j.neuron.2011.05.015. (accepted) [DOI] [PubMed] [Google Scholar]
- Liang JS, Shimojima K, Ohno K, Sugiura C, Une Y, Yamamoto T. A newly recognised microdeletion syndrome of 2p15-16.1 manifesting moderate developmental delay, autistic behaviour, short stature, microcephaly, and dysmorphic features: a new patient with 3.2 Mb deletion. J Med Genet. 2009;46:645–647. doi: 10.1136/jmg.2008.059220. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Carlström E, Råstam M, Gillberg C, Anckarsäter H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- Makoff AJ, Flomen RH. Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Cooper GM, Zerr T, Smith JD, Baker C, Shafer N, Thorland EC, Skinner C, Schwartz CE, Nickerson DA, Eichler EE. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, Pakula AT, Eisenhauer NJ, Uhas K, Weik L, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, Orlic-Milacic M, Lionel AC, Sato D, Pinto D, et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci Transl Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR. Williams-Beuren syndrome. N Engl J Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Zufferey F, Harewood L, Kutalik Z, Martinet D, Chrast J, Walters RG, Bouquillon S, Valsesia A, Hippolyte L, et al. Gene dosage at the 16p11.2 locus controls body mass index. American Society of Human Genetics (Washington DC) 2010 [Google Scholar]
- Risi S, Lord C, Gotham K, Corsello C, Chrysler C, Szatmari P, Cook EH, Leventhal BL, Pickles A. Combining information from multiple sources in the diagnosis of autism spectrum disorders. J Am Acad Child Adolesc Psychiatry. 2006;45:1094–1103. doi: 10.1097/01.chi.0000227880.42780.0e. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Dorr NP, Takahashi N, McInnes LA, Elder GA, Buxbaum JD. Haploinsufficiency of Gtf2i, a gene deleted in Williams Syndrome, leads to increases in social interactions. Autism Res. 2010 doi: 10.1002/aur.169. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shen Y, Dies KA, Holm IA, Bridgemohan C, Sobeih MM, Caronna EB, Miller KJ, Frazier JA, Silverstein I, Picker J, et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010;125:e727–735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Smalley SL, Tanguay PE, Smith M, Gutierrez G. Autism and tuberous sclerosis. J Autism Dev Disord. 1992;22:339–355. doi: 10.1007/BF01048239. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, Romano C, Delle Chiaie B, Mortier G, Menten B, et al. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet. 2009;52:94–100. doi: 10.1016/j.ejmg.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Veenstra-Vanderweele J, Christian SL, Cook EH. Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heineman-de Boer JA, Beemer FA, Swaab H, Kahn RS, van Engeland H. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Walters R, Jacquemont S, Valsesia A, de Smith A, Martinet D, Andersson J, Falchi M, Chen F, Andrieux J, Lobbens S, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner J, Abrahams B, Salyakina D, Imielinski M, Bradfield J, Sleiman P, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassink TH, Piven J, Patil SR. Chromosomal abnormalities in a clinic sample of individuals with autistic disorder. Psychiatr Genet. 2001;11:57–63. doi: 10.1097/00041444-200106000-00001. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A, Consortium G.D.P.o.J.H.t.A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.