Summary
Recent epidemiological studies have revealed a significant association between periodontitis and oral squamous cell carcinoma (OSCC). Furthermore, matrix metalloproteinase 9 (MMP9) is implicated in the invasion and metastasis of tumor cells. We examined the involvement of Porphyromonas gingivalis, a periodontal pathogen, in OSCC invasion through induced expression of proMMP and its activation. proMMP9 was continuously secreted from carcinoma SAS cells, while P. gingivalis infection increased proenzyme expression and subsequently processed it to active MMP9 in culture supernatant, which enhanced cellular invasion. In contrast, Fusobacterium nucleatum, another periodontal organism, failed to demonstrate such activities. The effects of P. gingivalis were observed with highly invasive cells, but not with the low invasive type. P. gingivalis also stimulated proteinase-activated receptor 2 (PAR2) and enhanced proMMP9 expression, which promoted cellular invasion. P. gingivalis mutants deficient in gingipain proteases failed to activate MMP9. Infected SAS cells exhibited activation of ERK1/2, p38, and NF-kB, and their inhibitors diminished both proMMP9-overexpression and cellular invasion. Together, our results show that P. gingivalis activates the ERK1/2-Ets1, p38/HSP27, and PAR2/NFκB pathways to induce proMMP9 expression, after which the proenzyme is activated by gingipains to promote cellular invasion of OSCC cell lines. These findings suggest a novel mechanism of progression and metastasis of OSCC associated with periodontitis.
Keywords: P. gingivalis, squamous cell carcinoma, periodontitis, MAPK
Introduction
Oral squamous cell carcinoma (OSCC) is reported to be the most frequent cancer occurring in the head and neck area (Stewart et al., 2003), with an estimated 263,900 new cases and 128,000 deaths from OSCC occurring worldwide in 2008 (Jemal et al., 2011). These tumors demonstrate invasiveness and frequent lymph node metastasis, and a high rate of early recurrence (da Silva SD et al., 2011). Furthermore, OSCC is considered to be a multi-causal disease influenced by the interrelationships among various etiologic factors, including smoking, alcohol consumption, dietary micronutrient deficiency, occupational activity, external agent exposure, genetic susceptibility, and infection (da Silva SD et al., 2011; Petti, 2009).
Various forms of cancer are linked to microorganisms such as human papilloma virus, hepatitis B and C virus, Epstein Barr virus, human herpes virus type 8, and Helicobacter pylori (Jemal et al., 2011). Chronic marginal periodontitis (henceforth referred to simply as periodontitis) is among the most common bacterial infections (Aas et al., 2005), and recent epidemiological studies have demonstrated a significant relationship between periodontitis and OSCC (Tezal et al., 2007; Tezal et al., 2009; Fitzpatrick and Katz, 2010; Maruyama et al., 2012), with one study reporting an odds ratio of 4.52 (Tezal et al., 2009). Periodontitis was also shown to be predictive of a 5.23-fold increase in risk of tongue cancer (Tezal et al., 2007). Furthermore, a very recent epidemiological survey (Ahn et al., 2012) showed that periodontitis was markedly associated with orodigestive cancer mortality, while Porphyromonas gingivalis, a major periodontal pathogen, was identified as a specific and potentially independent microbial factor to increase the risk of orodigestive cancer death.
The oral cavity is comprised of various boundary surfaces such as the gingiva, buccal mucosa, tongue dorsum, lateral side of the tongue, and hard and soft plate, which are all colonized with a diverse microbiota (Aas et al., 2005). Thus, P. gingivalis can continually interact with OSCC cells in these sites, and this pathogen has been shown to occur frequently in the mouth of oral cancer patients (Mager et al., 2005; Lanzós et al., 2011). Furthermore, antigens of P. gingivalis have also been detected in gingival squamous carcinoma tumors (Katz et al., 2011). B7-H1 expression in renal cell carcinoma and lung cancer is related with pathologic features such as lymph node metastasis, tumor size, higher nuclear grade, degree of differentiation, and lymph node metastasis (Thompson et al., 2007; Mu et al., 2011). As for tongue carcinoma cells, P. gingivalis infection induces expression of the B7-H1 receptor, suggesting involvement of the pathogen in distant metastasis and advanced nuclear grade of tumor cells (Groeger et al., 2011). Collectively, those findings indicate the possibility that interactions of P. gingivalis with OSCC cells are involved in cancer progression and metastasis. However, scant information is available regarding a molecular basis for this causal relationship.
Matrix metalloproteinases (MMPs) have a key role in degradation of basement membranes and extracellular matrix, which promotes carcinoma cell migration and invasion, which is defined as penetration of basement membrane and interstitial stroma by malignant cells. Migration and invasion allow carcinoma cells to enter the lymphatic system and blood vessels for dissemination into the circulation, and then undergo metastatic growth in distant organs (Sternlicht and Wer, 2001; Friedl and Wolf, 2003). MMPs are secreted as inactive pro-enzymes by mammalian cells, with the pro-forms processed into active forms by trypsin-like enzymes (Lijnen, 2001). Among the MMP family members, MMP2 and MMP9 have been shown to be strongly involved in carcinoma cell invasion, which is an essential factor for cancer progression and metastasis (Krüger et al., 2005). MMP9 is reportedly activated by Burkholderia cenocepacia, Brucella abortus, H. pylori, Salmonella enterica, and Streptococcus pyogenes bacterial infection in gastric carcinoma cells, monocytes, lung epithelial cells, and synoviocytes (Tamura et al., 2004; Oliveira et al., 2006; Ramu et al., 2008; Wright et al., 2011; Scian et al., 2011), and migration and invasion of infected cells were accelerated. Also, cellular production of proMMP9 was shown to be induced in vitro by P. gingivalis as well as by stimulation with its lipopolysaccharide (LPS) (Andrian et al., 2007; Jotwani et al., 2010). In addition, culture supernatants of P. gingivalis promoted the production and activation of monocyte proMMP9 (Zhou et al., 2012). On the other hand, butyric acid, an extracellular metabolite of P. gingivalis, had no clear effect on migration of several cancer cell lines (Miyazaki et al., 2010).
In the present study, we examined the effects of P. gingivalis on the expression and maturation of MMP2 and 9 by OSCC cells in order to evaluate a possible molecular basis linking periodontal pathogens to OSCC. We found that P. gingivalis activated the ERK1/2-Ets1, p38/HSP27, and proteinase-activated receptor 2 (PAR2)/NFκB pathways to induce proMMP9 production. Subsequently, the proenzyme was activated by the P. gingivalis gingipain proteases, which promoted the cellular invasion of the OSCC cell lines. These findings suggest a novel mechanism involved in progression and metastasis of OSCC associated with periodontitis.
Results
P. gingivalis induces cell invasion in OSCC cell lines
ProMMPs 2 and 9 are converted to active forms during cancer cell invasion (Ramos-DeSimone et al., 1999; Cheung et al., 2006). Thus, we first examined the effects of P. gingivalis on activation of proMMP2 and 9 in SAS cells. Highly invasive SAS cells were incubated with or without P. gingivalis at a multiplicity of infection (MOI) of 1 for various time periods. proMMPs were continuously secreted from the cells in time dependent manner. Additionally, P. gingivalis increased proMMP9 amounts and processed the proenzyme to the active form of MMP9 (Fig. 1A). Next, P. gingivalis was incubated in culture supernatant from SAS cells without infection and activated MMP9 was clearly detected, indicating that extracellular bacteria processed the proenzyme (Fig. 1B). Incubation with P. gingivalis significantly enhanced cellular invasion into matrigel, while that was significantly prevented by a specific inhibitor of MMP9 (Fig. 1C). P. gingivalis did not process proMMP2, which is localized to the cell surface and activated by extracellular MMP14 (Barbolina and Stack, 2008). However, MMP14 was poorly expressed in cells incubated with or without P. gingivalis (Fig. 1D), thus the possible involvement of MMP2 in OSCC invasion was not further examined.
Figure 1. P. gingivalis induces proMMP9 activation and cell invasion in SAS cells.
(A) Highly invasive SAS cells were incubated with P. gingivalis at an MOI of 1 for the indicated times. Culture supernatant samples from SAS cells were collected and analyzed for proMMP9 activation using gelatin zymography. Enzyme activities are expressed from densitometric analyses with arbitrary units. Data are means ± SD of three independent experiments and were analyzed with a t test. (B) Following growth of SAS cells, P. gingivalis (1×106 cells/ml) was added to the culture supernatant and incubated for 24 hours. proMMP9 activation was examined using gelatin zymography. Fresh medium was used as a control. (C) SAS cell invasion through matrigel-coated transwell membranes was assessed at 24 hours after P. gingivalis infection. When necessary, a specific inhibitor of MMP9 was added to the culture medium 24 hours prior to infection. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test. (D) MMP14 protein was detected using western blotting, and proMMP2 activation in culture supernatant was detected using gelatin zymography in the presence or absence of concanavalin A (ConA, an inducer of MMP14 expression and proMMP2 activation; 20 µg/ml). β-actin was used as a loading control. (E) proMMP activation in SAS cells incubated with F. nucleatum at MOIs of 1 and 10. (F) SAS cell invasion through matrigel-coated transwells at 24 hours after stimulation with P. gingivalis and F. nucleatum. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test.
We also examined the effects of Fusobacterium nucleatum, a periodontal organism suggested to be involved in lymph node metastasis in colorectal carcinoma (Castellarin et al., 2012), on proMMP9 activation and cellular invasion. However, neither proMMP2 or 9 were activated by this pathogen at an MOI of 1 or 10 (Fig. 1E). Cellular invasion of F. nucleatum-stimulated SAS cells was significantly lower than that of those stimulated by P. gingivalis (Fig. 1F). These results indicate the specific involvement of P. gingivalis on MMP9 activation and invasion of OSCC.
OSCC often shows active invasion and lymph node metastasis (Chandler et al., 2011). OSCC cell lines are classified into 2 types based on invasiveness, with highly invasive SAS cells and low invasive for which Ca9-22 cells are representative (Shindoh et al., 1996). proMMP2 was abundantly detected in culture supernatant of Ca9-22 cells, whereas proMMP9 levels were scant and unaffected by the presence of P. gingivalis (Fig. 2A). Furthermore, the effects on cellular invasiveness were negligible (Fig. 2B). We also examined the involvement of MMP9 in cellular invasion. Ca9-22 cells incubated in the culture supernatant of SAS cells infected with P. gingivalis showed significant invasive ability (Fig. 1S). These results also suggests that P. gingivalis promotes cellular invasion by MMP9 activation.
Figure 2. P. gingivalis negligibly induces proMMP9 production and cell invasion of Ca9-22 cells.
(A) Low invasive Ca9-22 cells were incubated with P. gingivalis at an MOI of 1 for the indicated times. Culture supernatant samples were collected and analyzed for proMMP9 activation using gelatin zymography. (B) Ca9-22 cell invasion through matrigel-coated transwell membranes at 24 hours after P. gingivalis stimulation. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test.
PAR2 receptor mediates proMMP9 over-expression by P. gingivalis
The expression of proMMP9 is reportedly mediated via PAR2 signaling pathways in prostate cancer cell lines (Wilson et al., 2004). Therefore, we examined the expression profile of PAR1-4 mRNA in SAS cells. PAR2 transcript was expressed, while the expression levels of the other PARs were negligible or non-detectable (Fig. 3A). We also found that expression levels of PAR2 were elevated following P. gingivalis infection (Fig. 3B), thus we transcriptionally silenced PAR2 expression with siRNA, in order to examine its involvement in proMMP9 expression (Fig. 3C). PAR2 knockdown suppressed proMMP9 expression at 24 hours after P. gingivalis infection (Fig. 3D), while cellular invasion was also inhibited by that infection (Fig. 3E).
Figure 3. PAR2 expression is upregulated in SAS cells incubated with P. gingivalis.
(A) Expression profiles of mRNA of PAR1 to 4 in SAS cells by RT-PCR analysis. β-actin was included as a loading control. (B) Time course of expression level of PAR2 mRNA in SAS cells incubated with P. gingivalis by real-time PCR. SAS cells were incubated with P. gingivalis at an MOI of 1 for the indicated times. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test. (C) SAS cells were transfected with siRNA targeting PAR2 or with nontarget control (siNT). Gene knockdown efficiency after 48 hours was evaluated by real-time PCR. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test. (D) siRNA knockdown cells were incubated with P. gingivalis at an MOI of 1 for 24 hours, then proMMP9 activation was determined using gelatin zymography. Enzyme activities are expressed from densitometric analyses with arbitrary units. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test. (E) Invasion by siRNA knockdown cells through matrigel-coated transwell membranes was assessed at 24 hours after bacterial stimulation. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test.
P. gingivalis induces MAPK phosphorylation and NF-kB translocation
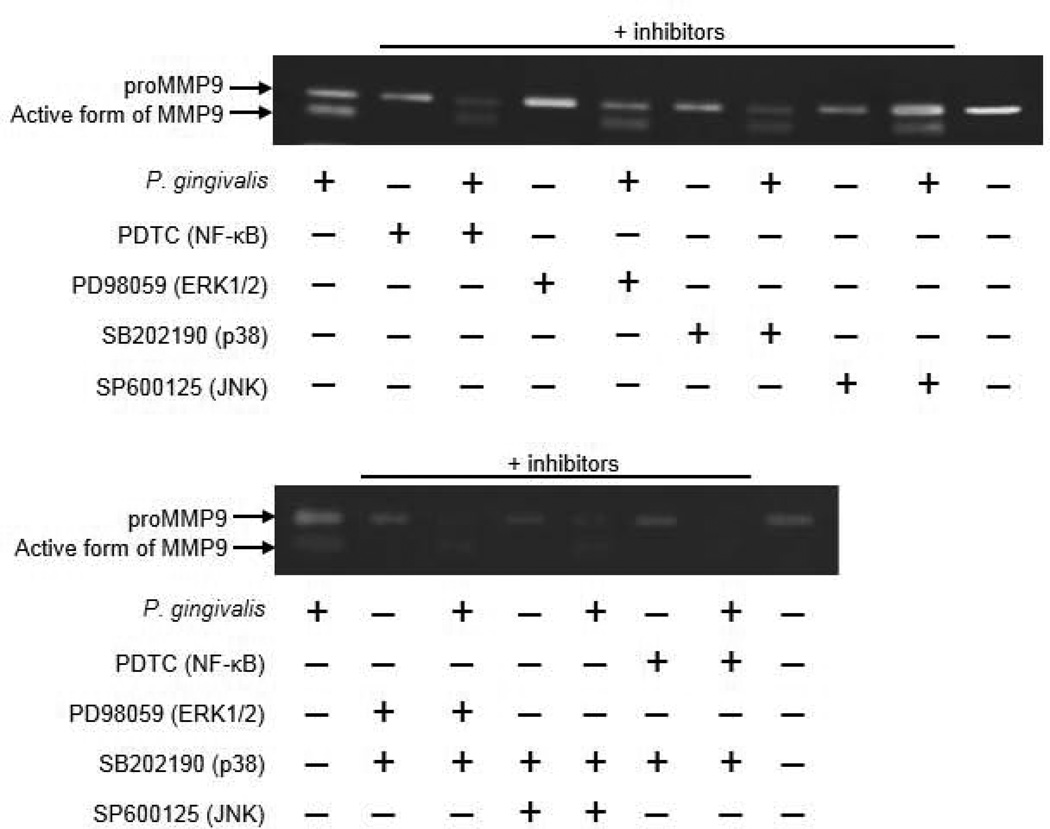
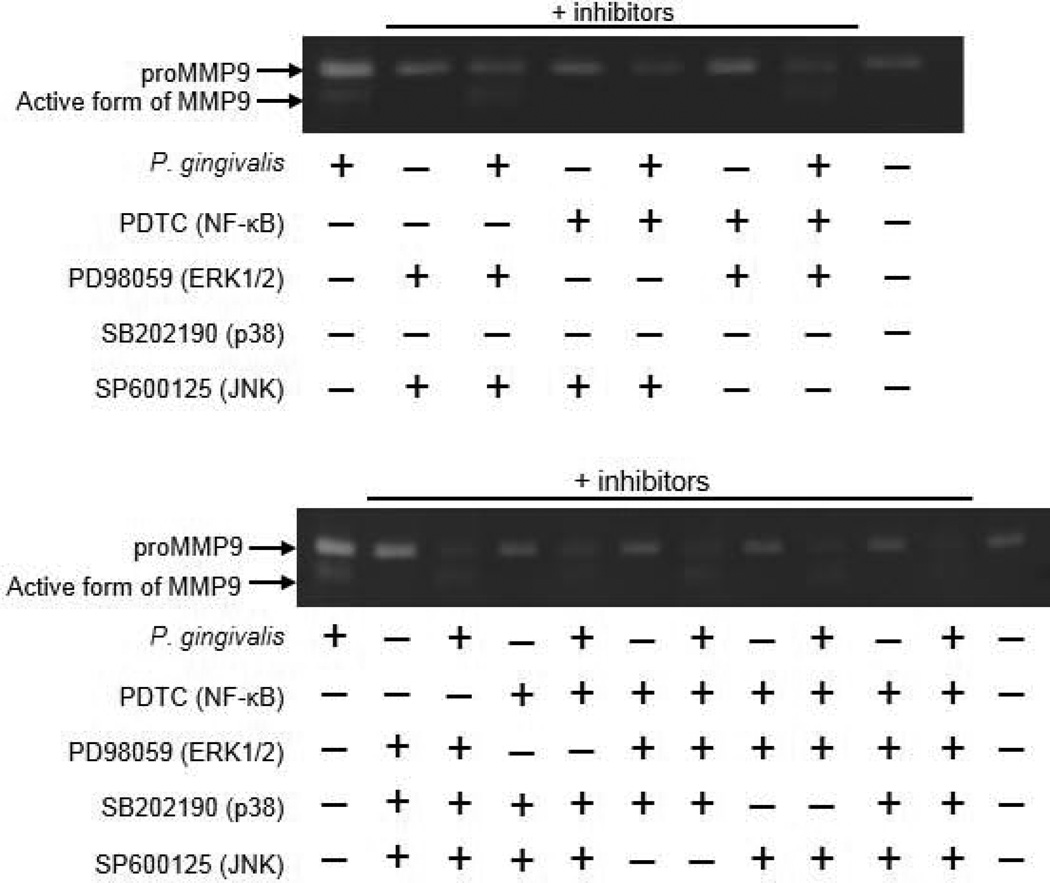
PAR2 activates multiple kinase pathways, such as ERK, p38MAPK, JNK, and NF-kB, in a cell-type specific manner (Kanke et al., 2005; Seits et al., 2007; Kida et al., 2007). Therefore, we examined the signaling pathway that mediates proMMP9 expression in SAS cells. Following P. gingivalis stimulation, phosphorylation of ERK1/2, p38, and JNK was induced (Fig. 4A). Activation and translocation of NF-kB occurs following signal-induced phosphorylation of IkBα, which leads to its proteolysis via the ubiquitin pathway (Wasserman, 1993). We found that the Ser32/36-phosphorylated form of IkBα was increased after P. gingivalis infection (Fig. 4B) with a subsequent increase in translocation of NF-kB into the nuclear fraction (Fig. 4C). Furthermore, pretreatment of SAS cells with PDTC (NF-kB inhibitor), PD98059 (ERK1/2 inhibitor), or SB202190 (p38 inhibitor) prevented proMMP9 production induced by P. gingivalis, which resulted in a decreased amount of activated MMP9 protein in culture supernatant (Fig. 5). Additionally, combinations of these inhibitors enhanced their effects. In contrast, SP600125 (JNK inhibitor) showed a negligible effect on MMP9 production. The inhibitors used were confirmed to have no cellular cytotoxicity (Fig. 2S). These results indicate that proMMP9 overexpression in response to P. gingivalis is mediated via multiple pathways, including those involving p38, ERK1/2, and NF-kB. We also examined whether these pathways are regulated by PAR2. Nuclear translocation of NF-kB was reduced in PAR2-knockdown cells infected with P. gingivalis, whereas phosphorylation of p38 and ERK1/2 was not inhibited (Fig. 6). These results indicate that PAR2 mediates proMMP9 over-expression by P. gingivalis via a pathway involving NF-κB.
Figure 4. Activation of MAPK and NF-κB pathways in SAS cells incubated with P. gingivalis.
SAS cells were incubated with P. gingivalis at an MOI of 1 for the indicated times, then the lysates were subjected to immunoblotting. Blots showing phosphorylation and total proteins are expressed from a densitometric analysis with arbitrary units. β-actin was included as a loading control for whole cell lysates. Nucleolin was included as a loading control for the nuclei. Tubulin was included as a loading control for cytoplasm. Data shown are representative of three independent experiments. (A) MAPK pathways, (B) IκBα, and (C) NFκB in nuclear and cytoplasmic fractions. Densitometric analysis of blots showing phosphorylation and total proteins, expressed in arbitrary units. β-actin was included as a loading control. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test.
Figure 5. Effects of MAPK and NF-kB inhibitors on proMMP9 production.
SAS cells were incubated with P. gingivalis at an MOI of 1 for 24 hours. Inhibitors were added 24 hours prior to P. gingivalis infection. proMMP9 activation in culture supernatant was analyzed using gelatin zymography.
Figure 6. Activation of MAPK and NF-κB pathways in PAR2 knockdown SAS cells incubated with P. gingivalis.
PAR2 cells were subjected to siRNA knockdown and incubated with P. gingivalis at an MOI of 1 for 24 hours. The expression profiles of ERK1/2, p38, and NF-κB were examined by immunoblotting. β-actin was included as a loading control for whole cell lysates. Nucleolin was included as a loading control for the nuclei. Tubulin was included as a loading control for cytoplasm.
Ets1 involved in proMMP9 over-expression by P. gingivalis
The ERK-dependent signaling pathway and transcriptional factor Ets1 reportedly regulate proMMP9 production (Dittmer, 2003). Therefore, we examined the effects of P. gingivalis on the Ets1 and Ets2 pathways. SAS cells infected with P. gingivalis demonstrated significantly increased Ets1 expression, whereas levels of Ets2 were not altered (Fig. 7A). Furthermore, Ets1-knockdown clearly inhibited proMMP9 production induced by P. gingivalis (Fig. 7B and C).
Figure 7. Ets1 expression is upregulated in SAS cells stimulated with P. gingivalis and regulates proMMP9 production.
(A) SAS cells were incubated with P. gingivalis at an MOI of 1 for the indicated times. Cell lysates were immunoblotted with antibodies to Ets1 or Ets2. β-actin was included as a loading control. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test. (B) SAS cells were transfected with siRNA targeting Ets1 or nontarget control (siNT). Immunoblotting was performed with Ets1 or β-actin antibodies at 48 hours after transfection. (C) siRNA knockdown cells were stimulated with P. gingivalis at an MOI of 1 for 24 hours. proMMP9 activation was examined using gelatin zymography.
Phosphorylation of HSP27 by P. gingivalis controls proMMP-9 production
In a study of human hepatocellular carcinoma cell lines, PKC-β/ERK1/2 and PKC-β/p38 MAPKs regulated HSP27 phosphorylation (Guo et al., 2008), which induced proMMP9 expression and promoted cancer invasion (Kumar et al., 2010). The p38/c-Jun/MMP9 pathway has also been reported in human cancer cells (Loesch et al., 2010). On the other hand, c-Jun activated by JNK phosphorylation regulates multiple target genes that contain AP1-binding sites related with cell cycle, apoptosis and MMPs expression (Wagner and Nebreda, 2009). Therefore, we examined the effects of P. gingivalis on phosphorylation of HSP27 and c-Jun by SAS cells. P. gingivalis induced Ser82 phosphorylation of HSP27, whereas a negligible effect was observed on c-Jun phosphorylation (Fig. 8A). HSP27-knockdown inhibited proMMP9 production by P. gingivalis at 24 hours as compared to the controls transfected with a non-target siRNA, indicating the involvement of HSP27 in that production (Fig. 8B and C).
Figure 8. HSP27 is phosphorylated in SAS cells stimulated with P. gingivalis and regulates proMMP9 production.
(A) SAS cells were incubated with P. gingivalis at an MOI of 1 for the indicated times. Cell lysates were immunoblotted with antibodies to phosphorylated or total JNK or HSP. Blots were analyzed by scanning densitometry, and ratios of phospho-JNK or HSP27 to each total protein were determined relative to the zero time point. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test. (B) SAS cells were transfected with siRNA targeting HSP27 or nontarget control (siNT). Immunoblotting was performed with anti-HSP27 or β-actin antibodies at 48 h after transfection, with β-actin as a loading control. (C) siRNA knockdown cells were stimulated with P. gingivalis at an MOI of 1 for 24 hours. proMMP9 activation was examined using gelatin zymography.
Involvement of gingipains in proMMP9 activation and cell invasion
P. gingivalis expresses gingipains comprised of arginine-X [Arg-gingipain A and B (RgpA and RgpB)]- and lysine-X [Lys-gingipain (Kgp)]-specific cysteine proteinases (Guo et al., 2010). We examined the involvement of these enzymes in activation of proMMP9 and cellular invasion. The gingipain-deficient mutants KDP129 (Δkgp), KDP133 (ΔrgpArgpB), and KDP136 (ΔrgpArgpBkgp) failed to activate proMMP9 proteins, whereas the increase in pro-MMP9 production was similar between KDP129 and KDP133 strains. Namely, proMMP9 production requires either Rgp or Kgp. On the other hand, both gingipains be considered essential to proMMP9 activation. (Fig. 9A). Similarly, invasion of SAS cells was not enhanced by any of the mutant strains (Fig. 9B). These results suggest that P. gingivalis gingipains promote proMMP9 activation based on proteolytic activity, resulting in cellular invasion.
Figure 9. P. gingivalis gingipains induce proMMP9 activation and cell invasion in SAS cells.
(A) SAS cells were incubated with P. gingivalis parent and mutants KDP129 (Δkgp), KDP133 (ΔrgpAΔrgpB), and KDP136 (ΔkgpΔrgpAΔrgpB). Culture supernatant samples were analyzed for proMMP9 activation using gelatin zymography. Enzyme activities are expressed from densitometric analyses with arbitrary units. Data are means ± SD of three independent experiments and were analyzed with a t test. (B) SAS cell invasion through matrigel-coated transwells after 24 hours of incubation with P. gingivalis mutants. Data are shown as the mean ± SD of three independent experiments and were analyzed with a t test.
Discussion
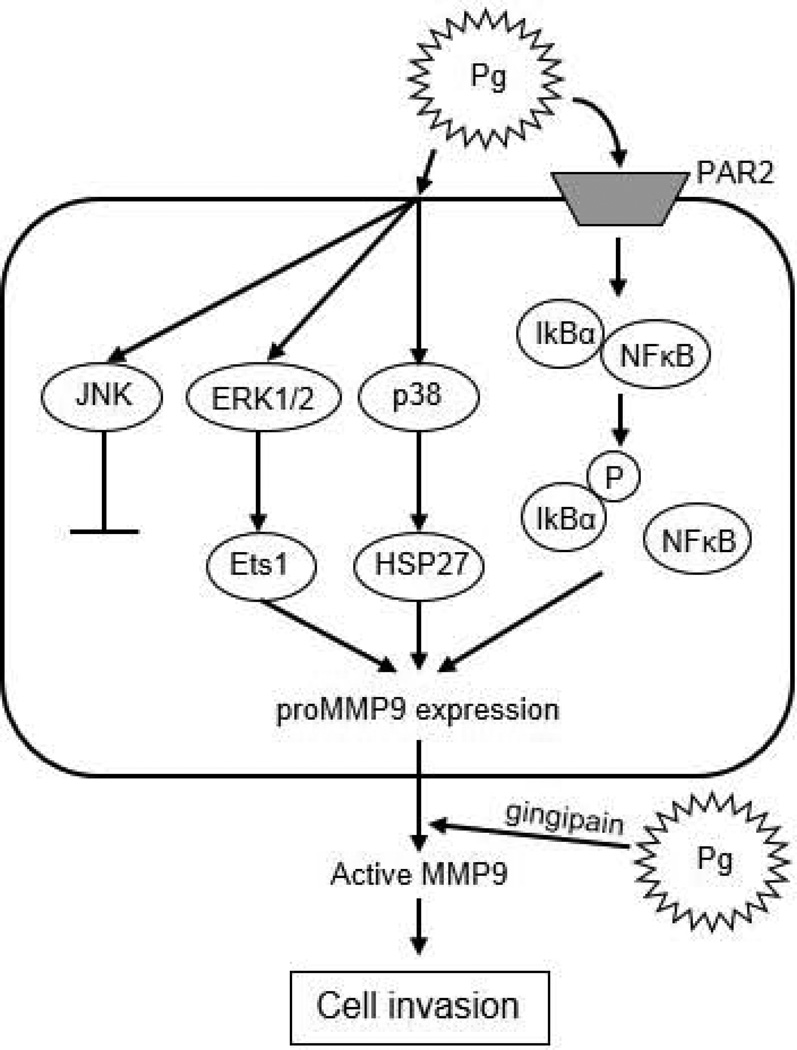
Epidemiological and in vitro studies have provided evidence for a possible link of periodontitis and periodontal pathogens, such as P. gingivalis, with oral cancer progression and metastasis. However, the molecular basis for a causal relationship is unclear. In the present study, we found a novel molecular mechanism by which a major periodontal pathogen promotes invasion of cancer cells, thus linking periodontitis to OSCC (Fig. 10). P. gingivalis induces proMMP9 production via multiple signaling cascades, including the ERK1/2-Ets1, p38/HSP27, and PAR2/NFκB pathways, after which the proenzyme is activated by bacterial gingipains, leading to enhanced invasion of SAS cells.
Figure 10. Proposed schematic model for proMMP9 secretion and activation in SAS cells infected with P. gingivalis.
P. gingivalis activates ERK1/2-Ets1, p38/HSP27, and PAR2/NFκB pathways to induce proMMP9 production. Subsequently, the proenzyme is secreted to the extracellular milieu and activated by gingipains, which promotes cellular invasion of OSCC cell lines.
Cell signaling is part of a complex system of communication that governs basic cellular activities and coordinates cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis for human development, tissue homeostasis, and immunity. Thus, disruption of normal cell signaling can lead to disease development including cancer. MMP9 activity can facilitate tumor progression by degrading collagen IV from basement membranes and extracellular matrix (Westermarck and Kähäri, 1999; Raffetto and Khalil, 2008), and P. gingivalis has been reported to increase proMMP9 expression (Andrian et al., 2007; Jotwani et al., 2010; Zhou et al., 2012) in oral mucosa, dendritic cells, and monocytes. Moreover, monocyte migration is promoted by P. gingivalis via proMMP9 activation, whereas P. gingivalis LPS has no effect on the cell migration and proMMP9 activation (Zhou et al., 2012). However, little is known regarding the signaling cascades that mediate proMMP9 expression induced by the organisms. PAR2 modulates inflammatory responses and acts as a sensor for proteolytic enzymes generated during infection, and has also been implicated in cell migration and metastasis in malignant tumor cell lines, such as breast carcinoma, melanoma, prostate carcinoma, and lymphoma (Wilson et al., 2004; Hjortoe et al., 2004; Shi et al., 2004; Dejean et al., 2012). In addition, PAR2 expression was shown to be related to the progression of gastric cancer and nasopharyngeal carcinoma in clinical samples (Caruso et al., 2006; Li et al., 2009). PAR2 activation promotes cellular proliferation and/or migration in colorectal adenocarcinoma and glioblastoma via intracellular signaling pathways involving ERK1/2 or NFκB (Guo et al., 2011; Svensson et al., 2011). Also, several reports have noted induction of proMMP9 expression via the PAR2 cascade. In experiments with keratinocytes, a Propionibacterium acnes serine protease activated PAR2 and induced MMP9 mRNA (Lee et al., 2010). Furthermore, proMMP9 was released into the extracellular environment by airway epithelial cells via PAR2 activation (Vliagoftis et al., 2000). In the present study, PAR2 activation induced by P. gingivalis promoted phosphorylation of IκB and nuclear translocation of NF-κB leading to proMMP9 overexpression. Previous studies have also demonstrated that P. gingivalis RgpB stimulated PAR2, which then activated p38 and ERK1/2, as well as the AP-1 pathway (Chandler et al., 2011), whereas NF-kB was not activated by PAR2 in another study (Fyfe et al., 2005). In the current study, PAR2 activation in SAS cells did not lead to activation of p38 and ERK1/2. Collectively, these suggest that PAR2 activation is important to cellular responses through activation of multiple kinase pathways in a cell-type specific manner. Further, PAR2 activation by P. gingivalis is likely due to the proteolytic activity of gingipains.
Ets1 has been shown to play a role in regulating invasiveness and metastasis in several cancer cell lines via modulation of genes responsible for migration and invasion (Dittmer, 2003; Hahne et al., 2005; Seth et al., 2005; Ghosh et al., 2012). In addition, HSP27 affects cellular invasion through regulation of MMP9 expression (Hansen et al., 2001). In the present study, we found that SAS cells stimulated with P. gingivalis exhibited sustained activation of Ets1 and phosphorylation of HSP27, while proMMP9 production was inhibited by knockdown of Ets1 or HSP27. Our findings are consistent with several previous reports, as proMMP9 expression was shown to be regulated by ERK1/2-Ets1 and p38/HSP27 in a number of cell lines (Ghosh et al., 2012; Hansen et al., 2001; Liu et al., 2005), and knockdown of ERK1/2 and Ets1 decreased proMMP9 mRNA expression in response to TGFβ1 (Liu et al., 2005). Moreover, cancer cell migration and invasion were reported to be inhibited by knockdown of Ets1 and HSP27 (Shin et al., 2005; Hahne et al., 2005), and p38 deficiency inhibited HSP27 and MMP9 expression (Kumar et al., 2010). Collectively, these results suggest that P. gingivalis exploits the ERK1/2-Ets1 and p38/HSP27 signaling pathways, as well as the PAR2/NFκB pathway to upregulated the production of proMMP9.
JNK1 phosphorylation is a positive regulator of c-Jun expression and cellular proliferation and JNK2 is a negative regulator of c-Jun expression and proliferation in mouse embryo fibroblasts (Jaeschke et al., 2006). On the other hand, JNK2 and c-Jun but not JNK1 are highly activated in human SCC clinical samples and are essential for the invasive human epidermal neoplasia (Zhang and Selim, 2012). In addition, p38 can negatively regulate JNK activity in several contexts (Wagner and Nebreda, 2009). We found that JNK2 was not phosphorylated while JNK1 and p38 phosphorylation were increased in SAS cells following P. gingivalis infection. Our results suggest that p38 phosphorylation may prevent activation of JNK2/c-Jun pathway in P. gingivalis-infected SAS cells.
In the present study, proMMP was produced by highly invasive SAS cells but not by Ca9-22 cells, which have a lower level of invasiveness. On the other hand, Ca9-22 cells incubated in culture supernatant obtained from infected SAS cells showed significant invasive ability (Fig. 1S), suggesting that P. gingivalis promotes cellular invasion by MMP9 activation. In addition, proMMP9 proteins were not activated by any of the gingipain-mutants of P. gingivalis and consequently cellular invasion was not enhanced. The MMP9 proenzyme (92KD) is activated by cleavage on the C-terminal side of arginine and lysine by trypsin, thus generating active mature MMP-9 (82KD) (Duncan et al., 1998). Therefore, both Arg- and Lys-gingipains are likely required for activation of proMMP9. These results suggest that gingipains are critical virulence factors for progression and metastasis of OSCC.
A previous report noted that the invasive potential of cancer cells is linked to proMMP9 activation (Ramos-DeSimone et al., 1999; Cheung et al., 2006). In our study, non-infected SAS cells also revealed invasive ability even in the absence of activated MMP9 (Fig. 1C). Cancer cell invasion and metastasis are affected by urokinase plasminogen activators (uPA) and MMP7, in addition to MMP2 and MMP9 (Crawford et al., 2002; Baker et al., 2007). uPA protein was shown to be expressed in SAS cells (Lu et al., 2011), while MMP7 expression was not detected (data not shown). In addition to MMP9, other factors such as uPA are likely involved in cellular invasion.
In conclusion, P. gingivalis exploits the ERK1/2-Ets1, p38/HSP27, and PAR2/NFκB pathways to induce proMMP9 production, while MMP9 activated by gingipains promotes cellular invasion of OSCC cell lines. Thus, P. gingivalis is suggested to be involved in various systemic diseases and the present findings provide a mechanistic basis for its pathogenicity in OSCC progression and metastasis.
Experimental procedures
Bacterial and cell cultures
The bacterial strains used were F. nucleatum ATCC 25586 and P. gingivalis ATCC 33277 along with isogenic KDP129 (Δkgp) (Okamoto et al., 1998), KDP133 (ΔrgpAΔrgpB) (Nakayama et al., 1995), and KDP136 (ΔrgpAΔrgpBΔkgp) (Shi et al., 1999). Bacteria were grown in trypticase soy broth supplemented with yeast extract (1 mg/ml), menadione (1 µg/ml), and hemin (5 µg/ml), as previously described (Inaba et al., 2009). SAS and Ca9-22 cells from an oral squamous cell carcinoma cell line were obtained from the Japanese Collection of Research Bioresources (Tokyo, Japan), and cultured in RPMI 1640 medium (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2.
Zymography
The activities of MMP2 and MMP9 in SAS cells culture supernatant, harvested from the cultures and as a source of MMPs, was determined using gelatin zymography. Samples were mixed with SDS sample buffer without reducing reagents, then separated on 10% SDS-polyacrylamide gels containing 0.1% gelatin. The gels were incubated at 37°C with 2.5% Triton X-100 for 1 h, followed by 48 h with reaction buffer [20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM CaCl2, 0.02% NaN3]. After staining with 5% Coomassie Brilliant Blue R-250, gelatinolytic activities were visualized as clear bands against a blue background.
Chemicals
Ammonium pyrrolidine dithiocarbamate (PDTC), a NF-kB inhibitor; SB203580, a p38-MAPK specific inhibitor; SP600125, a JNK-MAPK specific inhibitor; PD98059, an ERK1/2-MAPK specific inhibitor; and concanavalin A (ConA, an inducer of MMP14 expression) were purchased from Sigma-Aldrich. A specific inhibitor against MMP2 and MMP9 was purchased from Merck-Calbiochem, (MMP2/9 Inhibitor I; Darmstadt, Germany). The solvents used and final concentrations were as follows: PDTC, 100 µM in H2O; SB203580, 10 µM in dimethyl sulfoxide (DMSO); SP600125, 10 µM in DMSO; ConA, 20 mg/ml in H2O; and MMP2/9 inhibitor I, 20 µM in DMSO. SAS cells were pre-incubated with the inhibitors for 2 hours prior to infection.
Western immunoblotting
SAS cells were solubilized in cell lysis/extraction reagent (Sigma-Aldrich, St Louis, MO) containing a protease and phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL), along with KYT-1 and KYT-36 (Peptide Institute Osaka, Japan). Nuclear proteins were extracted from SAS cells using a Nuclear/Cytosol Fractionation kit (BioVision, Palo Alto, CA), according to the manufacturer’s protocol. Immunoblotting was performed as previously described (Inaba et al., 2009). Blots were probed at 4°C overnight with the following primary antibodies: anti-Phospho-ERK1/2 (Thr202/Tyr 264), 1:1000; anti-ERK1/2, 1:1000; anti-Phospho-p38 (Thr180/Tyr182), 1:1000; anti-p38, 1:1000; anti-Phospho-JNK (Thr183/Tyr185), 1:1000; anti-JNK, 1:1000; anti-Phospho-IkBα (Ser32/36), 1:1000; anti-IkBα, 1:1000, anti-Phospho-p38 (Thr180/Tyr182), 1:1000; anti-Phospho-cJun, 1:1000; anti-Jun, 1:1000; anti-Phospho-HSP27 (Ser82), 1:1000; anti-HSP27, 1:1000; (Cell Signaling Technology, Beverly, MA); anti-Ets1, 1:500; anti-Ets2, 1:500; anti-MMP14, 1:500 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-MDM2, 1:1000 (NOVUS Biologicals, Littleton, CO), and anti-β-actin, 1:1000 (Cell Signaling Technology). Proteins and phosphorylated proteins were detected using a Pierce ECL Substrate (Thermo Scientific).
RNA interference
Small interfering RNA (siRNA) for PAR2, Ets1, and HSP27, and a control siRNA were purchased from Santa Cruz Biotechnology. siRNAs were introduced into SAS cells with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. At 24 hours after transfection, the medium was replaced and cells were incubated for a further 24 hours prior to infection.
Invasion assay
Cell invasion was measured by assessment of the migration rate of OSCC cells using a BD Matrigel™ Invasion Chamber (BD Biosciences, Bedfold, MA). Cells (1 × 105) with or without P. gingivalis were seeded into the upper chambers in serum-free medium, while the lower wells were filled with RPMI1640 containing 10% FBS. After incubation, non-invading cells were removed from the upper wells, and cells that had transferred to the inverse surface of the membrane were subjected to Diff-Quick staining. All experiments were completed in triplicate and six fields/well were counted. Invasion rate is expressed as the percentage of cells that passed through the matrigel membrane relative to migration through the control membrane (100%).
Reverse transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA from SAS cells was isolated using TRIsure (BIOLINE, Luckenwalde, Germany) according to the manufacturer’s instructions, and converted into cDNA using an iscript™ cDNA Synthesis kit (Bio-Rad, Hercules, CA). cDNAs were amplified using a Taq PCR Master Mix kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The amplification reaction was performed with the following primers: for PAR1, 5’-TACACCGGAGTGTTTGTAGT-3’ and 5’-TTGAGGACGAGAGGCACTAC-3’; for PAR2, 5’-GGTAAGGTTGATGGCACATC-3’ and 5’-TGGTCTGCTTCACGACATAC-3’; for PAR3, 5’ATCTCATAGCTTTGTGCCTG-3’ and 5’-CACGCCTGTAATCCAGCACT-3’, for PAR4, 5’-AGTCTGTGCCAATGACAGTG-3’ and 5’-TCATGGCAGAGCACGCGATC-3’ (Tancharoen et al., 2005), and for β-actin, 5’-GATATCGGCCGCGCTCGTCGTCGAC-3’ and 5’-CAGGAAGGAAGGCTGGAAGAGTGC-3’ (Inaba et al., 2004). PCR products were subjected to electrophoresis in 1.5% agarose gel and stained with ethidium bromide.
Real-time PCR
Real-time PCR was performed with the following primers: for PAR2, 5’-ATACATGGCAACAACTGG-3’ and 5’-TTCACGATGACCCAATACCT-3’ (St-Onge et al., 2010); and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5’-GTCTTCACCACCATGGAGAAG-3’ and 5’-GTTGTCATGGATGACCTTGGC-3’ (Kato et al., 2005). GAPDH was used as a housekeeping control and negative RT reactions were included in each assay. Expression values for mRNA were quantified by the ΔΔCt method using GAPDH as the control.
Supplementary Material
Acknowledgments
We thank the members of the Amano and Lamont labs for their helpful discussions. We also thank Mayumi Yoshimori for the technical assistance. This research was supported by grants-in-aid for Scientific Research (23792102 and 25462850 to H.I. and B23390477 to A.A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and DE11111 and DE17921 to R.J.L. from the NIH.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–1058. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrian E, Mostefaoui Y, Rouabhia M, Grenier D. Regulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases by Porphyromonas gingivalis in an engineered human oral mucosa model. J Cell Physiol. 2007;211:56–62. doi: 10.1002/jcp.20894. [DOI] [PubMed] [Google Scholar]
- Baker EA, Leaper DJ, Hayter JP, Dickenson AJ. Plasminogen activator system in oral squamous cell carcinoma. Br J Oral Maxillofac Surg. 2007;45:623–627. doi: 10.1016/j.bjoms.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Barbolina MV, Stack MS. Membrane type-1 matrix metalloproteinase: substrate diversity in pericellular proteolysis. Seminars in cell and developmental biology. Semin Cell Dev Biol. 2008;19:24–33. doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Pallone F, Fina D, Gioia V, Peluso I, Caprioli F, et al. Protease-activated receptor-2 activation in gastric cancer cells promotes epidermal growth factor receptor trans-activation and proliferation. Am J Pathol. 2006;169:268–278. doi: 10.2353/ajpath.2006.050841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Barnes R, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler K, Vance C, Budnick S, Muller S. Muscle invasion in oral tongue squamous cell carcinoma as a predictor of nodal status and local recurrence: just as effective as depth of invasion? Head Neck Pathol. 2011;5:359–363. doi: 10.1007/s12105-011-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LW, Leung PC, Wong AS. Gonadotropin-releasing hormone promotes ovarian cancer cell invasiveness through c-Jun NH2-terminal kinase-mediated activation of matrix metalloproteinase (MMP)-2 and MMP-9. Cancer Res. 2006;66:10902–10910. doi: 10.1158/0008-5472.CAN-06-2217. [DOI] [PubMed] [Google Scholar]
- Crawford HC, Scoggins CR, Washington MK, Matrisian LM, Leach SD. Matrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreas. J Clin Invest. 2002;109:1437–1444. doi: 10.1172/JCI15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean E, Foisseau M, Lagarrigue F, Lamant L, Prade N. ALK+ALCLs induce cutaneous, HMGB-1-dependent IL-8/CXCL8 production by keratinocytes through NF-κB activation. Blood. 2012;119:4698–4707. doi: 10.1182/blood-2011-10-386011. [DOI] [PubMed] [Google Scholar]
- Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. http://www.molecular-cancer.com/content/2/1/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan ME, Richardson JP, Murray GI, Melvin WT, Fothergill JE. Human matrix metalloproteinase-9: activation by limited trypsin treatment and generation of monoclonal antibodies specific for the activated form. Eur J Biochem. 1998;258:37–43. doi: 10.1046/j.1432-1327.1998.2580037.x. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick SG, Katz J. The association between periodontal disease and cancer: a review of the literature. J Dent. 2010;38:83–95. doi: 10.1016/j.jdent.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Fyfe M, Bergström M, Aspengren S, Peterson A. PAR-2 activation in intestinal epithelial cells potentiates interleukin-1beta-induced chemokine secretion via MAP kinase signaling pathways. Cytokine. 2005;31:358–367. doi: 10.1016/j.cyto.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Basu M, Roy SS. ETS-1 protein regulates vascular endothelial growth factor-induced matrix metalloproteinase-9 and matrix metalloproteinase-13 expression in human ovarian carcinoma cell line SKOV-3. J Biol Chem. 2012;287:15001–15015. doi: 10.1074/jbc.M111.284034. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology. 2011;216:1302–1310. doi: 10.1016/j.imbio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Guo D, Zhou H, Wu Y, Zhou F, Xu G, Wen H, Zhang X. Involvement of ERK1/2/NF-κB signal transduction pathway in TF/FVIIa/PAR2-induced proliferation and migration of colon cancer cell SW620. Tumour Biol. 2011;32:921–930. doi: 10.1007/s13277-011-0194-1. [DOI] [PubMed] [Google Scholar]
- Guo K, Liu Y, Zhou H, Dai Z, Zhang J, Sun R, et al. Involvement of protein kinase C beta-extracellular signal-regulating kinase 1/2/p38 mitogen-activated protein kinase-heat shock protein 27 activation in hepatocellular carcinoma cell motility and invasion. Cancer Sci. 2008;99:486–496. doi: 10.1111/j.1349-7006.2007.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Nquyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne JC, Okuducu AF, Kaminski A, Florin A, Soncin F, Wernert N. Ets-1 expression promotes epithelial cell transformation by inducing migration, invasion and anchorage-independent growth. Oncogene. 2005;24:5384–5388. doi: 10.1038/sj.onc.1208761. [DOI] [PubMed] [Google Scholar]
- Hansen RK, Parra I, Hilsenbeck SG, Himelstein B, Fuqua SA. HSP27-induced MMP-9 expression is influenced by the Src tyrosine protein kinase yes. Biochem Biophys Res Commun. 2001;282:186–193. doi: 10.1006/bbrc.2001.4548. [DOI] [PubMed] [Google Scholar]
- Hjortoe GM, Petersen LC, Albrektsen T, Sorensen BB, Norby PL, Mandal SK, et al. Tissue factor-factor VIIa-specific up-regulation of IL-8 expression in MDA-MB-231 cells is mediated by PAR-2 and results in increased cell migration. Blood. 2004;103:3029–3037. doi: 10.1182/blood-2003-10-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H, Kawai S, Nakayama K, Okahashi N, Amano A. Effect of enamel matrix derivative on periodontal ligament cells in vitro is diminished by Porphyromonas gingivalis. J Periodontol. 2004;75:858–865. doi: 10.1902/jop.2004.75.6.858. [DOI] [PubMed] [Google Scholar]
- Inaba H, Kuboniwa M, Bainbridge B, Yilmaz O, Katz J, Shiverick KT, et al. Porphyromonas gingivalis invades human trophoblasts and inhibits proliferation by inducing G1 arrest and apoptosis. Cell Microbiol. 2009;11:1517–1532. doi: 10.1111/j.1462-5822.2009.01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke A, Karasarides M, Ventura JJ, Ehrhardt A, Zhang C, Flavell RA, et al. JNK2 is a positive regulator of the cJun transcription factor. Mol Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jotwani R, Eswaran SV, Moonga S, Cutler CW. MMP-9/TIMP-1 imbalance induced in human dendritic cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2010;58:314–321. doi: 10.1111/j.1574-695X.2009.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke T, Takizawa T, Kabeya M, Kawabata A. Physiology and pathophysiology of proteinase-activated receptors (PARs): PAR-2 as a potential therapeutic target. J Pharmacol Sci. 2005;97:38–42. doi: 10.1254/jphs.fmj04005x7. [DOI] [PubMed] [Google Scholar]
- Kato T, Okahashi N, Kawai S, Kato T, Inaba H, Morisaki I, Amano A. Impaired degradation of matrix collagen in human gingival fibroblasts by the antiepileptic drug phenytoin. J Periodontol. 2005;76:941–950. doi: 10.1902/jop.2005.76.6.941. [DOI] [PubMed] [Google Scholar]
- Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3:209–215. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y, Inoue H, Shimizu T, Kuwano K. Serratia marcescens serralysin induces inflammatory responses through protease-activated receptor 2. Infect Immun. 2007;75:164–174. doi: 10.1128/IAI.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger A, Arlt MJ, Gerg M, Kopitz C, Bernardo MM, Chang M, et al. Antimetastatic activity of a novel mechanism-based gelatinase inhibitor. Cancer Res. 2005;65:3523–3526. doi: 10.1158/0008-5472.CAN-04-3570. [DOI] [PubMed] [Google Scholar]
- Kumar B, Koul S, Petersen J, Khandrika L, Hwa JS, Meacham RB, et al. p38 mitogen-activated protein kinase-driven MAPKAPK2 regulates invasion of bladder cancer by modulation of MMP-2 and MMP-9 activity. Cancer Res. 2010;70:832–841. doi: 10.1158/0008-5472.CAN-09-2918. [DOI] [PubMed] [Google Scholar]
- Lanzós I, Herrera D, Santos S, O'Connor A, Peña C, Lanzós E, Sanz M. Microbiological effects of an antiseptic mouthrinse in irradiated cancer patients. Med Oral Patol Oral Cir Bucal. 2011;16:e1036–e1042. [PubMed] [Google Scholar]
- Lee SE, Kim JM, Jeong SK, Jeon JE, Yoon HJ, Jeong MK, Lee SH. Protease-activated receptor-2 mediates the expression of inflammatory cytokines, antimicrobial peptides, and matrix metalloproteinases in keratinocytes in response to Propionibacterium acnes. Arch Dermatol Res. 2010;302:745–756. doi: 10.1007/s00403-010-1074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bian LJ, Li Y, Liang YJ, Liang HZ. Expression of protease-activated receptor-2 (PAR-2) in patients with nasopharyngeal carcinoma: correlation with clinicopathological features and prognosis. Pathol Res Pract. 2009;205:542–550. doi: 10.1016/j.prp.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Lijnen HR. Plasmin and matrix metalloproteinases in vascular remodeling. Thromb Haemost. 2001;86:324–333. [PubMed] [Google Scholar]
- Liu S, Liang Y, Huang H, Wang L, Li Y, Li J, et al. ERK-dependent signaling pathway and transcriptional factor Ets-1 regulate matrix metalloproteinase-9 production in transforming growth factor-beta1 stimulated glomerular podocytes. Cell Physiol Biochem. 2005;16:207–216. doi: 10.1159/000089846. [DOI] [PubMed] [Google Scholar]
- Loesch M, Zhi HY, Hou SW, Qi XM, Li RS, Basir Z, et al. p38gamma MAPK cooperates with c-Jun in trans-activating matrix metalloproteinase 9. J Biol Chem. 2010;285:15149–15158. doi: 10.1074/jbc.M110.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KW, Chen JC, Lai TY, Yang JS, Weng SW, Ma YS, et al. Gypenosides inhibits migration and invasion of human oral cancer SAS cells through the inhibition of matrix metalloproteinase-2 -9 and urokinase-plasminogen by ERK1/2 and NF-kappa B signaling pathways. Hum Exp Toxicol. 2011;30:406–415. doi: 10.1177/0960327110372405. [DOI] [PubMed] [Google Scholar]
- Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. http://www.translational-medicine.com/content/3/1/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Yamanaka R, Yokoi A, Ekuni D, Tomofuji T, Mizukawa N, et al. Relationship between serum albumin concentration and periodontal condition in patients with head and neck cancer. J Periodontol. 2012;83:1110–1115. doi: 10.1902/jop.2011.110536. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Kikuchi K, González-Alva P, Inoue H, Noguchi Y, Tsuchiya H, et al. Association of Butyric Acid Produced by Periodontopathic Bacteria with Progression of Oral Cancer. J Cancer Sci Ther. 2010;2:26–32. [Google Scholar]
- Mu CY, Huang JA, Chen Y, Chen C, Zhang XG. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol. 2011;28:682–688. doi: 10.1007/s12032-010-9515-2. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- Oliveira MJ, Costa AC, Costa AM, Henriques L, Suriano G, Atherton JC, et al. Helicobacter pylori induces gastric epithelial cell invasion in a c-Met and type IV secretion system-dependent manner. Biol Chem. 2006;281:34888–34896. doi: 10.1074/jbc.M607067200. [DOI] [PubMed] [Google Scholar]
- Petti S. Lifestyle risk factors for oral cancer. Oral Oncology. 2009;45:340–350. doi: 10.1016/j.oraloncology.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Raffetto JD, Khalil RA. Matrix metalloproteinases and their inhibitors in vascular remodeling and vascular disease. Biochem Pharmacol. 2008;75:346–359. doi: 10.1016/j.bcp.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- Ramu P, Lobo LA, Kukkonen M, Bjur E, Suomalainen M, Raukola H, et al. Activation of pro-matrix metalloproteinase-9 and degradation of gelatin by the surface protease PgtE of Salmonella enterica serovar Typhimurium. Int J Med Microbiol. 2008;298:263–278. doi: 10.1016/j.ijmm.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Scian R, Barrionuevo P, Giambartolomei GH, De Simone EA, Vanzulli SI, Fossati CA, et al. Potential role of fibroblast-like synoviocytes in joint damage induced by Brucella abortus infection through production and induction of matrix metalloproteinases. Infect Immun. 2011;79:3619–3632. doi: 10.1128/IAI.05408-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz I, Hess S, Schulz H, Eckl R, Busch G, Montens HP, et al. Membrane-type serine protease-1/matriptase induces interleukin-6 and-8 in endothelial cells by activation of protease-activated Serratia marcescens receptor-2: potential implications in atherosclerosis. Arterioscler Thromb Vasc Biol. 2007;27:769–775. doi: 10.1161/01.ATV.0000258862.61067.14. [DOI] [PubMed] [Google Scholar]
- Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Shi X, Gangadharan B, Brass LF, Ruf W, Mueller BM. Protease-activated receptors (PAR1 and PAR2) contribute to tumor cell motility and metastasis. Mol Cancer Res. 2004;2:395–402. [PubMed] [Google Scholar]
- Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- Shin KD, Lee MY, Shin DS, Lee S, Son KH, Koh S, et al. Blocking tumor cell migration and invasion with biphenyl isoxazole derivative KRIBB3, a synthetic molecule that inhibits Hsp27 phosphorylation. J Biol Chem. 2005;280:41439–41448. doi: 10.1074/jbc.M507209200. [DOI] [PubMed] [Google Scholar]
- Shindoh M, Higashino F, Kaya M, Yasuda M, Funaoka K, Hanzawa M, et al. Correlated expression of matrix metalloproteinases and ets family transcription factor E1A-F in invasive oral squamous-cell-carcinoma-derived cell lines. Am J Pathol. 1996;148:693–700. [PMC free article] [PubMed] [Google Scholar]
- da Silva SD, Ferlito A, Takes RP, Brakenhoff RH, Valentin MD. Advances and applications of oral cancer basic research. Oral Oncol. 2011;47:783–791. doi: 10.1016/j.oraloncology.2011.07.004. [DOI] [PubMed] [Google Scholar]
- St-Onge M, Lagarde S, Laflamme C, Rollet-Labelle E, Marois L, Naccache PH, Pouliot M. Proteinase-activated receptor-2 up-regulation by Fcgamma-receptor activation in human neutrophils. FASEB J. 2010;24:2116–2125. doi: 10.1096/fj.09-146167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Wer Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BW, Greim H, Shuker D, Kauppinen T. Defence of IARC monographs. Lancet. 2003;361:13003–13006. doi: 10.1016/S0140-6736(03)13003-6. [DOI] [PubMed] [Google Scholar]
- Svensson KJ, Kucharzewska P, Christianson HC, Sköld S, Löfstedt T, Johansson MC, et al. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc Natl Acad Sci U S A. 2011;108:13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura F, Nakagawa R, Akuta T, Okamoto S, Hamada S, Maeda H, et al. Proapoptotic effect of proteolytic activation of matrix metalloproteinases by Streptococcus pyogenes thiol proteinase (Streptococcus pyrogenic exotoxin B) Infect Immun. 2004;72:4836–4847. doi: 10.1128/IAI.72.8.4836-4847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancharoen S, Sarker KP, Imamura T, Biswas KK, Matsushita K, Tatsuyama S, et al. Neuropeptide release from dental pulp cells by RgpB via proteinase-activated receptor-2 signaling. J Immunol. 2005;174:5796–5804. doi: 10.4049/jimmunol.174.9.5796. [DOI] [PubMed] [Google Scholar]
- Tezal M, Sullivan MA, Hyland A, Marshall JR, Stoler D, Reid ME, et al. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:2406–2412. doi: 10.1158/1055-9965.EPI-09-0334. [DOI] [PubMed] [Google Scholar]
- Tezal M, Scannapieco FA, Wactawski-Wende J, Hyland A, Marshall JR, Rigual NR, Stoler DL. Chronic periodontitis and the risk of tongue cancer. Arch Otolaryngol Head Neck Surg. 2007;133:450–454. doi: 10.1001/archotol.133.5.450. [DOI] [PubMed] [Google Scholar]
- Thompson RH, Dong H, Kwon ED. Implications of B7-H1 expression in clear cell carcinoma of the kidney for prognostication and therapy. Clin Cancer Res. 2007;13:709s–715s. doi: 10.1158/1078-0432.CCR-06-1868. [DOI] [PubMed] [Google Scholar]
- Vliagoftis H, Schwingshackl A, Milne CD, Duszyk M, Hollenberg MD, Wallace JL, et al. Proteinase-activated receptor-2-mediated matrix metalloproteinase-9 release from airway epithelial cells. J Allergy Clin Immunol. 2000;106:537–545. doi: 10.1067/mai.2000.109058. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Wasserman SA. A conserved signal transduction pathway regulating the activity of the rel-like proteins dorsal and NF-kappa B. Mol Biol Cell. 1993;4:767–771. doi: 10.1091/mbc.4.8.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermarck J, Kähäri VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781–792. [PubMed] [Google Scholar]
- Wilson SR, Gallagher S, Warpeha K, Hawthorne SJ. Amplification of MMP-2 and MMP-9 production by prostate cancer cell lines via activation of protease-activated receptors. Prostate. 2004;60:168–174. doi: 10.1002/pros.20047. [DOI] [PubMed] [Google Scholar]
- Wright C, Pilkington R, Callaghan M, McClean S. Activation of MMP-9 by human lung epithelial cells in response to the cystic fibrosis-associated pathogen Burkholderia cenocepacia reduced wound healing in vitro. Am J Physiol Lung Cell Mol Physiol. 2011;301:L575–L586. doi: 10.1152/ajplung.00226.2010. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Selim MA. The role of the c-Jun N-terminal Kinase signaling pathway in skin cancer. Am J Cancer Res. 2012;2:691–698. [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang J, Chao J. Porphyromonas gingivalis promotes monocyte migration by activating MMP-9. J Periodontal Res. 2012;47:236–242. doi: 10.1111/j.1600-0765.2011.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.