Abstract
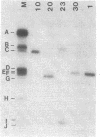
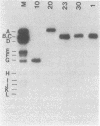
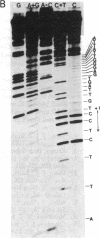
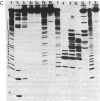
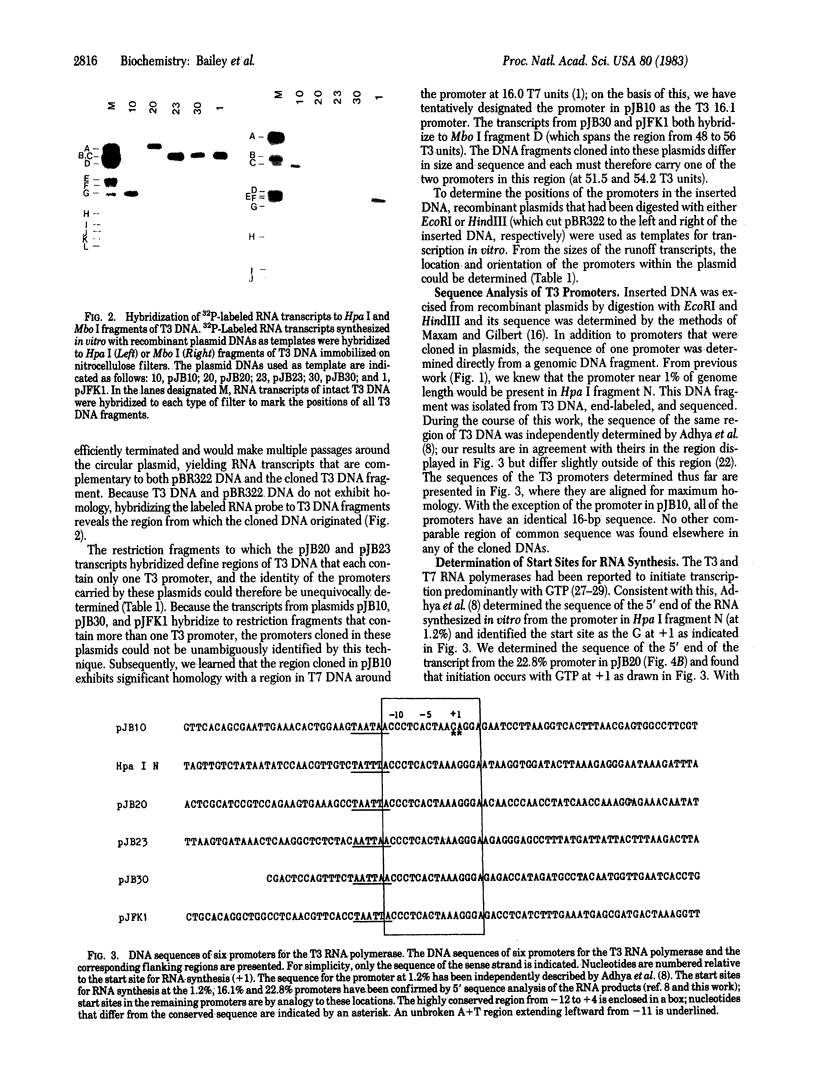
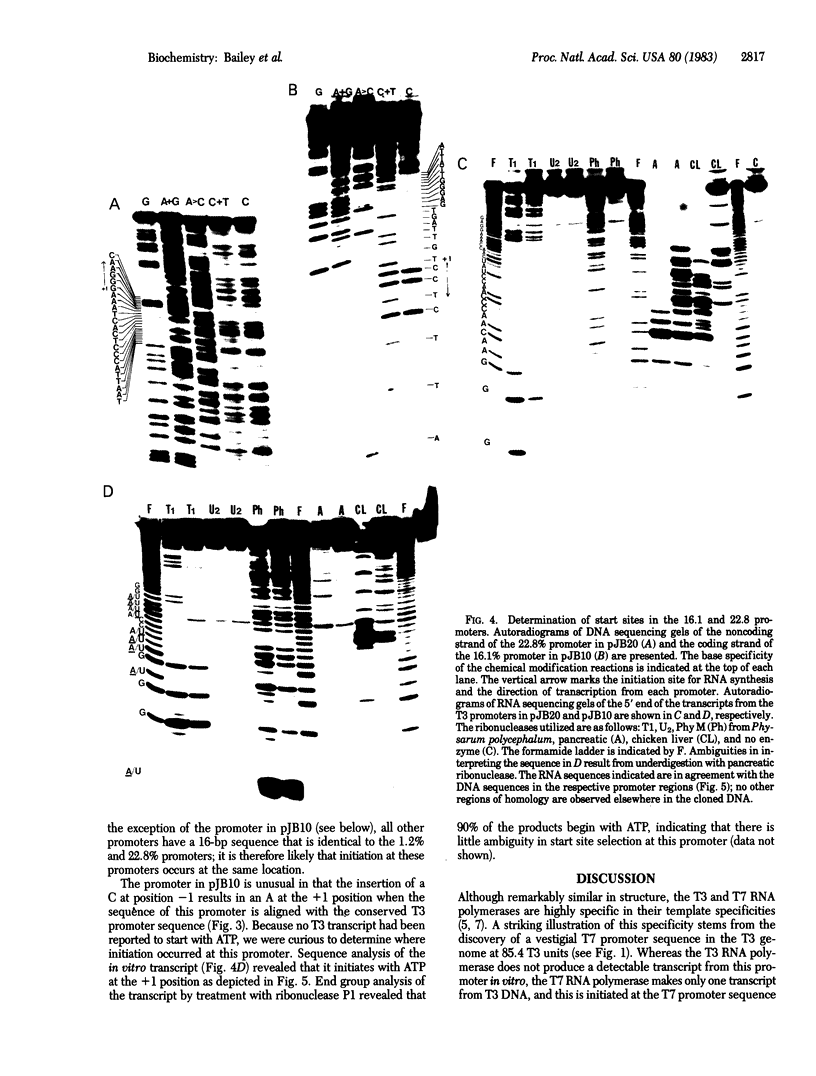
To explore the basis for the template specificities of the bacteriophage T3 and T7 RNA polymerases (EC 2.7.7.6), we determined the nucleotide sequences of six promoters recognized by the T3 RNA polymerase and compared them with the previously determined promoter sequences recognized by the bacteriophage T7 RNA polymerase. Recombinant plasmids containing random Hpa II and Taq I fragments of T3 DNA were screened for T3 promoter activity in vitro in a transcription assay using purified T3 RNA polymerase. Five promoters for the T3 RNA polymerase were identified in this manner and their sequences were determined; the sequence of an additional promoter was determined directly from a genomic DNA fragment. In five of the T3 promoters an identical 16-base-pair sequence (A-C-C-C-T-C-A-C-T-A-A-A-G-G-G-A) extends from -12 to +4 (initiation occurring with GTP at +1); this sequence is preceded by a 6-base-pair A + T region. The remaining promoter contains an inserted C at position -1 and an A at the +1 position. The sequence of the 5' end of the RNA transcript from the latter promoter confirms that transcription is initiated with ATP at the +1 position. Previously, late T3 or T7 transcripts had not been found to initiate with ATP. The highly conserved T3 promoter sequence was compared to the T7 promoter consensus sequence. The fundamental difference between the two kinds of phage promoters is the occurrence of G-A at positions -11 and -10 in the T7 promoter, whereas there is a single C at position -10 in the T3 promoter.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Basu S., Sarkar P., Maitra U. Location, function, and nucleotide sequence of a promoter for bacteriophage T3 RNA polymerase. Proc Natl Acad Sci U S A. 1981 Jan;78(1):147–151. doi: 10.1073/pnas.78.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. N., Dembinski D. R., McAllister W. T. Derivation of a restriction map of bacteriophage T3 DNA and comparison with the map of bacteriophage T7 DNA. J Virol. 1980 Jul;35(1):176–183. doi: 10.1128/jvi.35.1.176-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. N., McAllister W. T. Mapping of promoter sites utilized by T3 RNA polymerase on T3 DNA. Nucleic Acids Res. 1980 Nov 11;8(21):5071–5088. doi: 10.1093/nar/8.21.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H., Hausmann R. Genetic map of bacteriophage T3. J Virol. 1973 Aug;12(2):417–419. doi: 10.1128/jvi.12.2.417-419.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski M. S., Hieter P. A., Levy C. C. Identification of a cytidine-specific ribonuclease from chicken liver. J Biol Chem. 1980 Mar 10;255(5):2160–2163. [PubMed] [Google Scholar]
- Campbell J. L., Richardson C. C., Studier F. W. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc Natl Acad Sci U S A. 1978 May;75(5):2276–2280. doi: 10.1073/pnas.75.5.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Bautz F. A., Bautz E. K. Different template specificities of phage T3 and T7 RNA polymerases. Nat New Biol. 1971 Mar 17;230(11):94–96. doi: 10.1038/newbio230094a0. [DOI] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. J. T7- and T3-specific RNA polymerases: characterization and mapping of the in vitro transcripts read from T3 DNA. J Virol. 1977 Feb;21(2):743–752. doi: 10.1128/jvi.21.2.743-752.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann R. Bacteriophage T7 genetics. Curr Top Microbiol Immunol. 1976;75:77–110. doi: 10.1007/978-3-642-66530-1_3. [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Schroeder C. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol Rev. 1981 Mar;45(1):9–51. doi: 10.1128/mr.45.1.9-51.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra U., Huang H. H. Initiation, release, and reinitiation of RNA chains by bacteriophage-T3-induced polymerase from T3 DNA templates (E. coli-guanosine triphosphate terminus-purified polymerase). Proc Natl Acad Sci U S A. 1972 Jan;69(1):55–59. doi: 10.1073/pnas.69.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., Barrett C. L. Hybridization mapping of restriction fragments from the early region of bacteriophage T7 DNA. Virology. 1977 Oct 15;82(2):275–287. doi: 10.1016/0042-6822(77)90003-4. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., Küpper H., Bautz E. K. Kinetics of transcription by the bacteriophage-T3 RNA polymerase in vitro. Eur J Biochem. 1973 May 2;34(3):489–501. doi: 10.1111/j.1432-1033.1973.tb02785.x. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., Morris C., Rosenberg A. H., Studier F. W. Utilization of bacteriophage T7 late promoters in recombinant plasmids during infection. J Mol Biol. 1981 Dec 15;153(3):527–544. doi: 10.1016/0022-2836(81)90406-x. [DOI] [PubMed] [Google Scholar]
- Rosa M. D., Andrews N. C. Phage T3 DNA contains an exact copy of the 23 base-pair phage T7 RNA polymerase promoter sequence. J Mol Biol. 1981 Mar 25;147(1):41–53. doi: 10.1016/0022-2836(81)90078-4. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. A micromethod for base analysis of 32P-labeled oligoribonulcleotides. Anal Biochem. 1977 Nov;83(1):222–227. doi: 10.1016/0003-2697(77)90530-9. [DOI] [PubMed] [Google Scholar]