Abstract
Traffic through endosomes and lysosomes is controlled by small G-proteins of the Rab5 and Rab7 families. Like humans, Saccharomyces cerevisiae has three Rab5s (Vps21, Ypt52, and Ypt53), and one Rab7 (Ypt7). Here, we elucidate the functional roles and regulation of the yeast Rab5s. Using GFP-tagged cargoes, a novel quantitative multivesicular body (MVB) sorting assay, and electron microscopy, we show that MVB biogenesis and cargo sorting are severely impaired in vps21Δ ypt52Δ double mutants. Ypt53, the third Rab5 paralog, is hardly expressed during normal growth but its transcription is strongly induced by cellular stress through the calcineurin-Crz1 pathway. The requirement for Rab5 activity in stress tolerance facilitated identification of Msb3/Gyp3 as the principal Rab5 GAP (GTPase accelerating protein), and in vitro GAP assays verified that Vps21 is a preferred Gyp3 target. Finally, we demonstrate that Gyp3 spatially restricts active Vps21 to intermediate endosomal compartments by preventing Vps21 accumulation on lysosomal vacuoles. Gyp3 therefore operates as a compartmental insulator that enforces the boundary between penultimate and terminal compartments of the endolysosomal pathway.
Keywords: Rab GTPase, Rab GAP, endosome, vacuole, MVB, LUCID, ESCRT
INTRODUCTION
Membrane traffic through the secretory and endolysosomal systems is highly dynamic. Within these systems small G proteins of the Arf and Rab families simultaneously regulate compartmental identity and control rates of membrane influx and egress from each compartment (Cai et al., 2007; Rivera-Molina & Novick, 2009; Barr and Lambright, 2010; Angers and Merz, 2011). Rabs are anchored to membranes through C-terminal prenyl anchors. In their inactive GDP-bound state, Rabs can be extracted from membranes and relocated by the cytoplasmic chaperone GDI (GDP dissociation inhibitor). Activated GTP-bound Rabs and Arfs bind effector proteins and complexes that execute diverse functions in membrane traffic including vesicle budding, transport, and fusion. Rab signaling is stringently regulated. Like other small G proteins within the Ras superfamily, Rabs bind GDP tenaciously and become activated through catalyzed exchange of GTP for GDP mediated by guanosine nucleotide exchange factors (GEFs). Rabs generally have slow intrinsic GTPase activity and are inactivated by GTPase accelerating proteins (GAPs). All known Rab GAPs contain TBC domains. In ternary Rab-GTP-GAP complexes, the TBC domain generally supplies catalytic Arg and Gln residues which stabilize the hydrolytic transition intermediate (Albert et al, 1999; Pan et al., 2006). Hence, the integrated activities of GEFs, GAPs, and chaperones define the spatial and temporal domains of Rab signaling and activity.
Anterograde transport from early to late endosomes, and then to terminal lysosomes, is principally controlled by members of the Rab5 and Rab7 families, with Rab5 paralogs operating at earlier compartments and Rab7 at later compartments (Huotari & Helenius, 2011). Recent studies also suggest that Rab5 and Rab7 GTPases coordinate retrograde traffic to the Golgi through interactions with the retromer complex (Rojas et al., 2008; Balderhaar et al., 2010). S. cerevisiae, like humans, has three Rab5 paralogs that reside on endosomes (Vps21, Ypt52, and Ypt53), and one Rab7 (Ypt7) found mainly on the vacuolar lysosome (Singer-Krüger et al., 1994 and 1995; Horazdovsky et al., 1994; Wichmann et al., 1992). No clear role for Ypt53 has previously been established, and the relative roles of Vps21, Ypt52, and Ypt53 in MVB biogenesis and cargo sorting have not been fully characterized. During maturation of the late endosome Rab5 is rather abruptly replaced by Rab7 in a process termed endosomal Rab conversion (Rink et al., 2005). The acquisition of Rab7 is thought to confer competence for fusion with terminal endolysosomal compartments.
An important waypoint within the endolysosomal system is the late endosomal multivesicular body (MVB). At the MVB, ESCRTs (endosomal sorting complexes required for transport) package specific cargo into intralumenal vesicles (ILVs) that sequester cargo away from the cytoplasm (Hurley and Hanson, 2010; Wollert and Hurley, 2010). When mature MVBs fuse with lysosomes, ILVs and their cargos are dumped into the lysosome lumen and destroyed, while lipids and transmembrane proteins not packaged into ILVs are delivered to the lysosomal limiting membrane.
The relative timing and regulatory dependencies of ESCRT activity, Rab7 activation, and Rab5 inactivation are not fully understood. Activation of Rab7 requires the activities of the Vps-C complexes HOPS and CORVET (Rink et al., 2005; also reviewed in Nickerson et al., 2009), and the Rab7 GEF complex Ccz1-Mon1 (Kucharczyk et al., 2000; Nordmann et al., 2011). It is unclear when and where GAPs negatively regulate Rab5 in the endolysosomal Rab cascade. Multiple GAPs have been reported to target Vps21 in vitro (Albert et al., 1999; Albert and Gallwitz, 1999), but it has been unclear which (if any) of the yeast GAPs target the Rab5 paralogs in vivo. Here, we employ genetic, biochemical, and cell biological analyses to elucidate the roles and clarify the regulation of the three S. cerevisiae Rab5 paralogs. We find that Rab5 signaling (by either Vps21 or Ypt52) is essential for MVB cargo sorting and biogenesis, and show that Ypt53 is a stress-induced functional homolog of Vps21. We exploit a general requirement for Rab5 signaling during ionic stress to identify Msb3/Gyp3 as the principal in vivo GAP of Vps21. We demonstrate that Gyp3 (Gyp, GAP for Ypt/Rab proteins) has a pivotal role in the endosomal Rab cascade: it spatially restricts Vps21 signaling to prevacuolar endosomal compartments by preventing accumulation of active Vps21 on the terminal vacuole.
RESULTS
Roles of yeast Rab5 paralogs in Golgi-endosome traffic
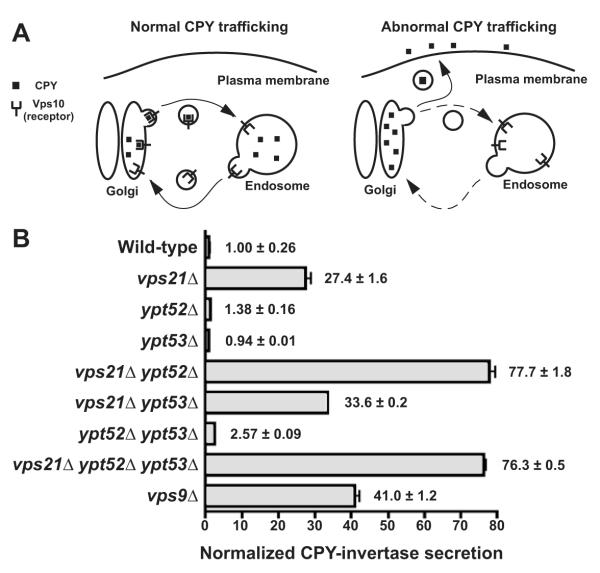
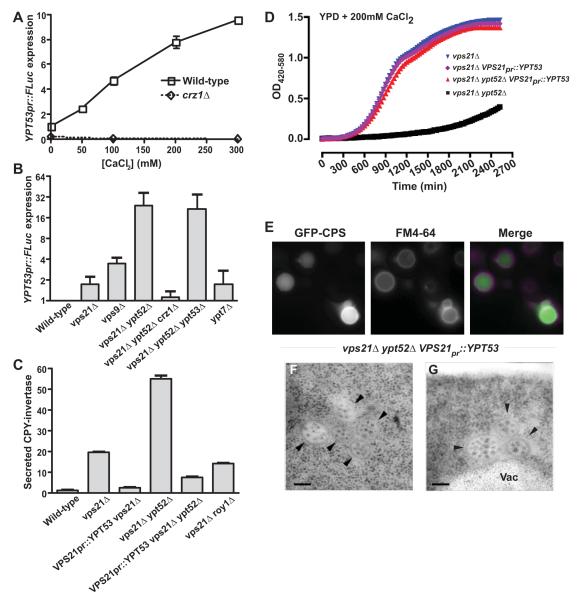
Correct targeting of the soluble vacuolar hydrolase CPY (Fig 1A) requires cycling of the CPY receptor Vps10 between late Golgi and endosomal compartments (Marcusson et al., 1994; Seaman et al., 1998). Vps10 or CPY missorting results in CPY mistargeting into the extracellular space (Rothman and Stevens, 1986). Of the three S. cerevisiae Rab5 GTPase paralogs, Vps21 is known to function in CPY traffic (Horazdovsky et al., 1994), as is its GEF Vps9 (Burd et al., 1996). Conversely, the loss of two other Rab5 paralogs, Ypt52 and Ypt53, did not strongly impair CPY sorting (Singer-Kruger et al., 1994). Using a chimeric CPY-invertase fusion to quantify CPY secretion (Fig 1B), we verified the sorting defects of vps21Δ and vps9Δ single mutants and observed a strong synthetic defect in a vps21Δ ypt52Δ double mutant. Vps21 and Ypt52 therefore have both distinct and complementary activities (see also Singer-Kruger et al., 1994; Singer-Kruger et al., 1995). A vps9Δ mutant exhibited a less severe CPY sorting defect than a vps21Δ ypt52Δ double mutant, suggesting that residual Rab5 signaling occurs in the absence of Vps9-catalyzed nucleotide exchange.
Figure 1.
Relative contributions of three S. cerevisiae Rab5 paralogs in Golgi-endosome traffic. (A) Cartoon depicts how carboxypeptidase Y (CPY) sorting defects depend largely upon Golgi-endosome traffic of the CPY receptor, Vps10. (B) Measurement of extracellular CPY-invertase in cells lacking Rab5 paralogs or Rab5 family GEF, Vps9. Bars indicate mean ± s.d. of triplicate samples from a representative experiment.
Rab5 signaling is required for MVB biogenesis and sorting
Previous work suggested that Vps21 operates upstream of the MVB to accept incoming endosomes and Golgi-derived vesicles (Horazdovsky et al., 1994; Peterson et al., 1999; Tall et al., 1999; Gerrard et al., 2000). In cells lacking the Vps9 GEF, missorting of the endocytic MVB cargo Ste3 to cytoplasmic punctae further suggested a Rab5 requirement in MVB cargo sorting or biogenesis (Prag et al., 2003). Hence, we were surprised to observe correct sorting of both a ubiquitin-independent cargo, Sna3-GFP (Fig 2A) and a ubiquitin-dependent MVB cargo, GFP-CPS (carboxypeptidase S; unpublished results) to the vacuole lumen in vps21Δ or ypt52Δ single mutants. The persistence of cargo sorting in Rab5 single mutants is supported by TEM (transmission electron microscopy) observations, which show that morphologically normal MVBs are present in vps21Δ cells (M. Russell and G. Odorizzi, submitted), albeit at reduced frequency versus the wild-type.
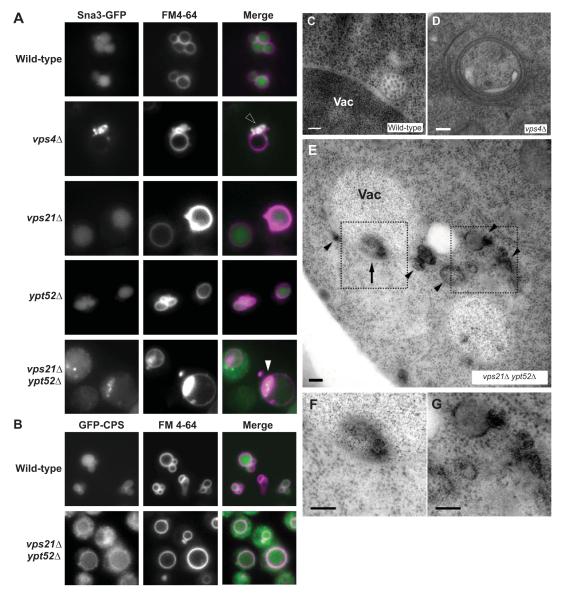
Figure 2.
Effects of Rab5 pathway defects on MVB cargo sorting and biogenesis. (A-B) Fluorescence micrographs show Sna3-GFP (A) or GFP-CPS (B) and FM4-64, an endocytic tracer that marks the vacuole, in cells with genotypes as indicated. Open arrowhead, perivacuolar colocalization of GFP and FM 4-64 at class E compartment puncta. Closed arrowhead, colocalization of GFP and FM 4-64 at puncta that appear to be inside the vacuole limiting membrane. (C-G) Thin-section electron micrographs of wild-type (C), vps4Δ (D) and vps21Δ ypt52Δ (E-G) cells, including electron-dense structures both within (arrows) and outside (arrowheads) the vacuole limiting membrane in vps21Δ ypt52Δ cells. (F-G) Increased magnification of highlighted regions in E. Scale bars span 100 nm. Vac, vacuole.
In contrast to the single mutants, vps21Δ ypt52Δ double mutants strongly missorted both Sna3-GFP (Fig 2A) and GFP-CPS (Fig 2B). In the double mutant cells Sna3-GFP was present throughout the cytoplasm and occasionally (<10% of cells) at large, bright FM 4-64-positive punctae near the vacuole (Fig 2A, closed arrowheads). By light microscopy, this phenotype resembled the class E phenotype of cells with defective ESCRT function (e.g. vps4Δ, Fig 2D). However, by transmission electron microscopy (TEM) vps21Δ ypt52Δ double mutants were devoid of both MVBs (e.g. wild-type, Fig 2C) and stacked class E compartments. Instead, vps21Δ ypt52Δ cells occasionally accumulated unstacked electron-dense structures (Fig 2E-G), both in the cytoplasm (Fig 2G) and within the vacuole limiting membrane (Fig 2F). These electron-dense structures were not observed in wild-type cells or in vps21Δ single mutants. The perivacuolar and intravacuolar locations of the electron-dense structures suggest that they correspond to Sna3-GFP and FM4-64 positive structures observed by light microscopy (Fig. 2A), as described below.
Because ILV cargoes are missorted and ILVs fail to form in vps21Δ ypt52Δ double mutants, we examined ESCRT localization in these cells. ESCRT recruitment to endosomes depends largely on binding of Vps27 (ESCRT-0) to PI(3)P (phosphatidylinositol-3-phosphate) (Katzmann et al., 2003), a lipid marker of endosomes required for efficient ILV formation (Futter et al., 2001). PI(3)P localization was probed with a GFP-FYVE (Fab1, YOTB, Vac1 and EEA1). The endocytic membrane tracer FM 4-64 was used (without a chase step) to label both endocytic organelles and the vacuole (Fig 3A). In wild-type cells GFP-FYVE localized efficiently to FM 4-64-stained perivacuolar punctae as well as to the vacuole limiting membrane, indicating the presence of PI(3)P in cytoplasmic leaflets of both endosomal and vacuolar membranes (Wurmser & Emr, 1998; Gillooly et al., 2000). Cells lacking vps4Δ accumulated PI(3)P at FM 4-64-stained class E compartments. In striking contrast, both vps21Δ and vps21Δ ypt52Δ mutants lacked discrete cytoplasmic GFP-FYVE punctae but showed strong GFP-FYVE signal on the vacuole limiting membrane. The accumulation of GFP-FYVE at the vacuole limiting membrane in vps21Δ and vps21Δ ypt52Δ cells argues against a global defect in PI(3)P synthesis and suggests an endosome-specific defect. Consistent with this finding, mammalian endosomal PI-3-kinase is stimulated by Rab5 signaling (Christofordis et al., 1999). Thus, in vps21Δ and vps21Δ ypt52Δ mutants a key signal for ILV formation is depleted from prevacuolar endosomal compartments.
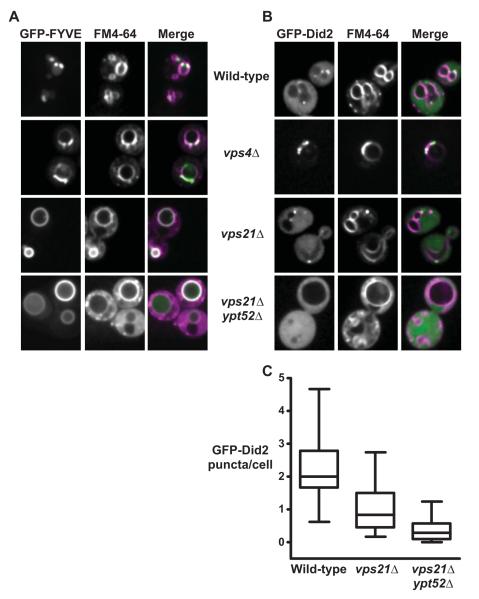
Figure 3.
Rab5 signaling is required for PI(3)P and ESCRT-III localization at endosomes. (A-B) Confocal fluorescence micrographs show PI(3)P probe GFP-EEA1FYVE (‘GFPFYVE’) (A) or ESCRT-III component GFP-Did2 (B) in cells pulsed with endocytic tracer dye FM4-64 for 30 minutes without chase. Scale bars, 2 μm (C) Box plot quantification of observed GFP-Did2 fluorescent foci. 130-150 cell profiles from 3 independent experiments.
ILV scission involves ESCRT-III assembly on endosomal membranes followed by Vps4-catalyzed disassembly (Wollert & Hurley, 2010). We therefore assessed the localization of Did2, perhaps the only ESCRT-III subunit that retains function when fused to GFP (Azmi et al., 2008; Nickerson et al., 2010). Endosomal GFP-Did2 punctae were abundant in wild-type cells, while in vps4Δ cells GFP-Did2 concentrated at large class E punctae (Fig 3B); Azmi et al., 2008; Nickerson et al., 2010). vps21Δ ypt52Δ double mutants had few or no GFP-Did2 punctae (Fig 3B and C) despite increased FM 4-64 stain in the cytoplasm relative to wild-type, indicating delayed FM4-64 traffic through endocytic compartments. vps21Δ single mutants had an intermediate phenotype with fewer GFP-Did2 punctae relative to wild-type (Fig 3C), consistent with the reduced frequency of endosomal lumenal vesicles in vps21Δ cells (Russell et al., submitted). We emphasize that although Rab5 deficiency delays FM 4-64 trafficking to the vacuole the endocytic marker dye does reach the vacuole, indicating that at least a subset of endocytic vesicles retain a vacuolar itinerary. Together, these data support working models in which Rab5-deficient cells have endosomes, but these organelles do not support proper ESCRT activity and fail to form ILVs.
LUCID: Quantitative assessment of Rab5 function in MVB sorting
Existing assays of ILV cargo sorting have significant limitations. Localization of GFP-cargo fusions to the vacuole lumen versus the limiting membrane is not readily quantifiable, and while pulse-chase analyses of cargo maturation resolve subtle differences in rates of transport to the vacuole/lysosome, the proteolytic maturation of MVB cargos does not necessarily result from correct sorting into lumenal vesicles (Odorizzi et al., 2003). To address the requirement for a rapid and quantitative assay of MVB cargo missorting, we constructed a reporter system called LUCID (luciferase reporter of intralumenal deposition).
The LUCID vector has two components. The reporter component is a chimeric fusion of firefly luciferase to the MVB cargo Sna3 (Sna3-FLuc; Fig 4A). Sna3-FLuc is readily quantified using luciferase activity assays. Correct targeting of Sna3-FLuc to ILVs results in sequestration and eventual destruction of the FLuc reporter. Because the FLuc tag faces the cytoplasm, defects in Sna3-FLuc traffic to the MVB, or failure to package Sna3-FLuc into ILVs, result in the accumulation of active Sna3-Fluc. To enable ratiometric correction for variations in cell number, plasmid copy number, protein synthesis rate, or global protein turnover, the LUCID vector’s normalization component encodes a soluble cytoplasmic Renilla luciferase (RLuc; Salas-Marco and Bedwell, 2005; Steffen et al., 2008). Sna3-FLuc and RLuc are sequentially assayed from each cell lysate using a microplate luminometer that incorporates robotic solution injectors. In pilot experiments we found that signal-to-background is optimal when cells are lysed after a 20-30 min. cycloheximide chase. The chase blocks new protein synthesis and allows nascent Sna3-FLuc to reach the vacuole, resulting in lower FLuc activity in wild-type cells, while mutants defective for MVB targeting exhibit relatively higher FLuc activity.
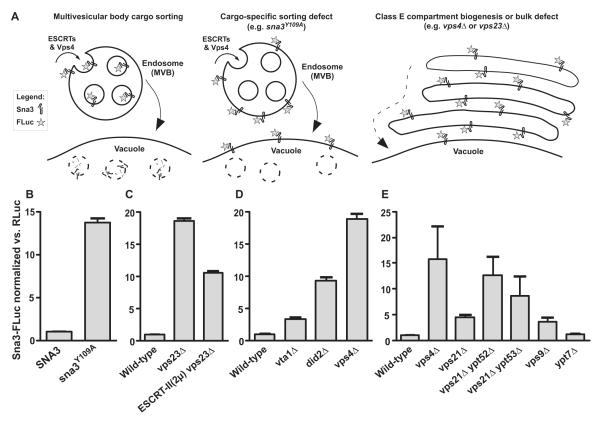
Figure 4.
LUCID assay of MVB cargo missorting in Rab5 mutants. (A) Schematic diagrams show predicted Sna3-FLuc sorting itineraries in wild-type and mutant cells. (B-D) Validation of the LUCID assay. (B)Effect of Sna3Y109A-FLuc mutation (C) Partial rescue of early ESCRT cargo missorting defect by overexpression of ESCRT-II. (D) Mutants with escalating degrees of disrupted ESCRT-III dynamics and cargo missorting display escalating degrees of Sna3-FLuc signal. (E) MVB cargo missorting phenotypes in Rab GTPase-signaling deficient mutants. In panels B-D, bars show mean ± s.d. of triplicate samples from representative experiments. In panel E bars show mean ± s.e.m. of four independent experiments (except ypt7Δ, n=3). RLuc, Renilla luciferase. MVB, multivesicular body. 2μ, yeast high-copy plasmid.
Sna3 is sorted into ILVs through ubiquitin-dependent and –independent mechanisms (Reggiori and Pelham, 2001). To validate the LUCID assay we tested a mutant (Sna3Y109A) that is defective in both mechanisms and inefficiently sorted to ILVs (McNatt et al., 2007). Sna3Y109A-FLuc accumulated to levels at least 10 times higher than the wild-type Sna3-FLuc reporter (Fig 4B). The signal-to-background ratio of LUCID therefore exceeds 10:1. Importantly, LUCID reliably reports intermediate defects in MVB cargo sorting. Deletion of the ESCRT-I subunit Vps23 substantially increased Sna3-Fluc activity, while overproduction of the ESCRT-II holocomplex partially suppressed the vps23Δ defect (Fig 4C), consistent with work using other assays (Babst et al., 2002). Previous work connected defects in disassembly or remodeling of ESCRT-III oligomers to proportional defects in MVB cargo sorting (Azmi et al., 2006; Azmi et al., 2008; Nickerson et al., 2010; Wemmer et al., 2011). We observed a corresponding phenotypic progression of signal intensity (Fig 4D) in mutants with compromised ESCRT-III dynamics, ranging from partial (vta1Δ; did2Δ) to complete (vps4Δ) defects.
LUCID experiments with Rab5 mutant cells (Fig 4E) revealed moderate MVB sorting defects in the vps21Δ and vps9Δ single mutants and severe defects in the vps21Δ ypt52Δ double mutant, with missorting at levels comparable to strong vps class E mutants that fail to form lumenal vesicles (e.g. vps4Δ). Among the Rab5 signaling-deficient mutants, the relative severities of MVB missorting (Fig 4E) correlated with the pattern seen in analyses of CPY secretion (Fig 1B). We conclude that both formation of morphologically normal MVBs (Fig 2) and sorting of MVB cargos to ILVs (Figs 2A and B; Fig 4E) exhibit a strong dependence on Rab5 signaling.
Rab5 signaling is required for efficient autophagy
Cells lacking VPS21 and YPT52 sometimes accumulate endocytic markers at foci near or within the vacuole (Fig 2), and we wondered if these structures might mark attempts to clean up stalled endocytic traffic through autophagy. Atg8 marks nascent and mature autophagic membranes (Cheong & Klionsky, 2008; Nakatogawa et al., 2009). Upon fusion of autophagosomes with the vacuole, some Atg8 is recycled to the cytosol and some is deposited in the vacuole lumen. While intravacuolar membrane accumulations appeared infrequently in vps21Δ ypt52Δ mutants (est. <10%), we observed colocalization of GFP-Atg8 with intravacuolar foci of both FM 4-64 (Fig S2A) and the MVB cargo mCherry-CPS (Fig S2B), in cells both with and without diffuse GFP-Atg8 signal in the vacuole lumen. In contrast, GFP-Atg8 did not colocalize with FM 4-64-stained class E compartments in vps4Δ cells (Fig S2A). Thus, a key autophagic component colocalizes with aberrant endosome-derived structures in vps21Δ ypt52Δ cells.
Recent genetic studies indicate that traffic through the Golgi-endosome system is crucial for CVT vesicle and autophagosome assembly (Ohashi & Munro, 2010). We therefore asked whether autophagy is globally up-regulated or defective in Rab5-deficient cells. Proteolytic maturation of vacuolar aminopetidase I (Ape1) is a hallmark of constitutive cytosol-to-vacuole transport (CVT) and macroautophagy (Cheong & Klionsky, 2008). Unstressed vps4Δ and vps21Δ cells contained precursor and mature Ape1 at approximately wild-type levels (Fig S2C, top panel). In marked contrast, unstressed vps21Δ ypt52Δ cells showed little or no maturation of Ape1, indicating a penetrant defect in CVT transport. Rapamycin stimulates autophagy by inhibiting TOR (Target of rapamycin) kinase, resulting in dephosphorylation of the TOR substrate Atg13 (Kamada et al., 2010). Wild-type, vps4Δ, vps21Δ, and vps21Δ ypt52Δ cells all contained similar levels of slow-migrating phospho-Atg13, and in all cases rapamycin treatment depleted phospho-Atg13 with comparable kinetics (Fig S2D). Thus, TORC1 signaling to the autophagy machinery is not markedly perturbed in Rab5-deficient cells. Nevertheless, rapamycin treatment only partially restored Ape1 maturation in vps21Δ ypt52Δ cells (Fig S2C, bottom panel). It is important to note that vacuoles in vps21Δ ypt52Δ cells are proteolytically competent (Singer-Krüger et al., 1994). The accumulation of precursor Ape1 in vps21Δ ypt52Δ mutants therefore indicates significant defects in CVT and macroautophagy.
Rab5 signaling required for tolerance of ionic stress
Many endolysosomal trafficking mutants are sensitive to salt and osmotic stress, and numerous endocytic trafficking genes were shown to be genetic modifiers of Ca2+ influx (Martin et al., 2011). We therefore tested mutants defective in Rab5 signaling for sensitivity to extracellular Ca2+. Mutants lacking any single Rab5 ortholog grew as well as the wild-type at up to 400 mM Ca2+ (Fig S3A and B; Table II). vps21Δ ypt53Δ double mutant cells had strong synthetic sensitivity to high Ca2+ and were somewhat more Ca2+--sensitive than cells lacking the Vps9 GEF. vps21Δ ypt52Δ double mutants were hypersensitive to extracellular Ca2+, as were cells lacking the Rab5 effectors Vps3 and Vac1. Deletion of a third effector, Vps8, had little effect on Ca2+ tolerance. Triple mutants lacking all three Rab5 orthologs (vps21Δ ypt52Δ ypt53Δ) were hypersensitive to Ca2+, even more so than cells lacking Crz1, a key calcineurin-responsive transcription factor (Yoshimoto et al., 2002). The triple mutants grew poorly even in low extracellular [Ca2+] (Fig S3A; Table II), again indicating a functional role for Ypt53 in stress tolerance.
Table II.
Growth rates of Rab5 family mutants under calcium stress. Inflection doubling time (min), mean ± SD, n=5 each.
| Genotype | YPD | YPD + 200 mM CaCl2 |
|---|---|---|
| Wild-type | 90.3 ± 7.2 | 117.0 ± 5.2 |
| vps21 Δ | 83.3 ± 4.7 | 114.0 ± 2.8 |
| ypt52 Δ | 83.1 ± 3.2 | 107.5 ± 5.6 |
| ypt53 Δ | 92.7 ± 2.7 | 114.1 ± 4.2 |
| vps21Δ ypt52Δ | 89.9 ± 7.0 | 255.0 ± 22.6 |
| vps21Δ ypt53Δ | 82.6 ± 6.8 | 170.3 ± 11.6 |
| ypt52Δ ypt53Δ | 99.4 ± 4.5 | 122.9 ± 5.5 |
| vps21Δ ypt52Δ ypt53Δ | 122.1 ± 9.4 | 277.1 ± 50.2 |
| vps9 Δ | 94.7 ± 3.4 | 152.9 ± 10.8 |
Ypt53 is a stress-induced functional homolog of Vps21
vpt53Δ single mutants lack obvious growth or trafficking phenotypes (Singer-Kruger et al., 1994) and YPT53 has evaded identification in forward genetic screens. YPT53 is transcribed at low levels in unstressed cells, while VPS21 and YPT52 transcripts are considerably more abundant (MacKay et al., 2004). However, YPT53 was identified in microarray experiments as a target of the calcineurin stress response pathway (Yoshimoto et al., 2002). Using a luciferase reporter to measure transcription from the YPT53 promoter, we verified that YPT53 exhibits dose-dependent induction in elevated extracellular Ca2+ (Fig 5A). Moreover, YPT53 transcription was strongly induced in Rab5-deficient mutants (Fig 5B). YPT53 induction by either extracellular Ca2+ or Rab5 deficiency was abolished in mutants lacking the calcineurin-regulated transcription factor Crz1, indicating that responses to both stresses converge on the calcineurin-Crz1 pathway (Fig 5A and B).
Figure 5.
Ypt53 is a stress-induced Vps21 paralog. (A) Transcription from the YPT53 promoter was monitored using FLuc transcriptional fusions in wild-type or crz1Δ cells grown in medium supplemented with different concentrations of Ca2+. Bars shown mean ±s.d. of duplicate samples from a representative experiment. (B) YPT53 expression is induced in cells deficient in Rab5 signaling. Bars show mean ± s.e.m. from independent experiments (n=5, 5, 4, 5, 3, 4, and 4 respectively). (C-G) YPT53 driven by the VPS21 promoter rescues CPY trafficking (C), calcium growth tolerance (D), ILV cargo trafficking (E) and ILV biogenesis (F-G) in vps21Δ ypt52Δ mutants. (C) Assay of extracellular CPY-invertase, normalized to the wild type. Bars indicate mean ± s.d. of triplicate samples from a representative experiment. (D) Growth curves in liquid culture, 30°C. Mean, n=4. (E) Fluorescence microscopy of FM 4-64-stained cells expressing GFP-CPS (carboxypeptidase S). (F-G) Thin-section electron micrographs show restored MVB morphology. Vac, vacuole. Arrowheads, individual MVBs. Bars span 100 nm.
Synthetic phenotypes observed in vps21Δ ypt53Δ double mutants (Fig S3; Table II) raised the possibility that Vps21 and Ypt53 are functional homologs, but differentially expressed. To test this hypothesis we expressed YPT53 from the VPS21 promoter. VPS21PR-YPT53 suppressed the loss of Vps21 alone or Vps21 and Ypt52 together (Fig 5C-E). It restored sorting of CPY and of MVB cargo (Fig 5C, E) and almost completely restored growth in the presence of high extracellular Ca2+ (Fig 5D). Roy1 (repressor of ypt52) was recently reported to oppose Ypt52 signaling (Liu et al., 2011). VPS21pr::YPT53 suppressed CPY-invertase secretion in vps21Δ cells more completely than did loss of Roy1 (Fig 5C). VPS21pr::YPT53 also restored characteristic MVB ultrastructure to vps21Δ ypt52Δ cells (Fig 5 F and G). However, MVBs were now more abundant than in wild-type cells (0.82 versus 0.37 MVB per cell; n=50 cell profiles each), and more often appeared tethered or clustered than in wild-type cells (71% clustered, n=41 MVBs versus 18% clustered, n=40 MVBs). Thus, when expressed from the same VPS21 promoter, Vps21 and Ypt53 are largely but not totally interchangeable.
Gyp3 opposes Rab5 signaling
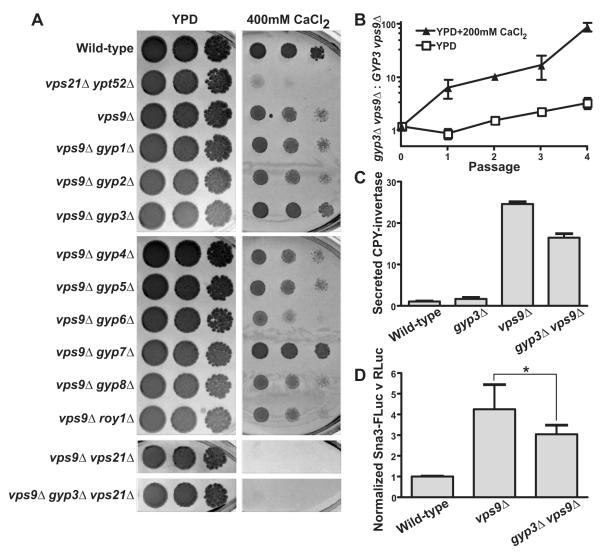
Cells lacking the Rab5 GEF Vps9 had less severe trafficking and growth defects than vps21Δ ypt52Δ double mutants, implying that vps9Δ cells contain a reduced but functionally significant pool of active Rab5. We reasoned that deletion of Rab5 inhibitors might enrich the pool of activated Rab5 and thereby suppress the defects of vps9Δ mutants. To test this idea, we deleted each of the known S. cerevisiae Rab GAPs in a vps9Δ genetic background and tested the resulting double mutants for growth on high Ca2+ media. The loss of two GAPs, Msb3/Gyp3 and Gyp7, partially suppressed the Ca2+ sensitivity of vps9Δ cells (Fig 6A).
Figure 6.
Rab GAP Msb3/Gyp3 opposes endosomal Rab5 signaling. (A) Limiting dilution plate growth assay, 30° C on YPD ± 200 mM CaCl2. (B) vps9Δ gyp3Δ cells outcompete vps9Δ cells in mixed cultures. Bars indicate mean ± s.d., n=2. (C) Assay of extracellular CPY-invertase, normalized to the wild-type. Mean ± s.d., of 4 samples per point from a representative experiment. (D) LUCID assay of ILV sorting. Bars indicate mean ± s.e.m. from 6 independent experiments;*p=0.024, two-tailed, paired t-test.
Gyp7 is the major in vivo GAP of Ypt7, the yeast Rab7 ortholog (Brett et al., 2008). Suppression of vps9Δ by gyp7Δ is probably not a direct effect of Gyp7 on Rab5 signaling, as discussed below. At the plasma membrane Gyp3 functions redundantly with Msb4/Gyp4 to regulate delivery of secretory vesicles by targeting the Sec4 Rab (Gao et al., 2003). As reported previously (Albert & Gallwitz, 1999) and as we confirm here, Gyp3 has strong GAP activity against Vps21. To verify suppression of the vps9Δ growth defect by gyp3Δ, we performed a competition experiment. Double mutant vps9Δ gyp3Δ cells had increased competitive fitness versus vps9Δ cells in Ca2+-rich media (Fig 6B), but not under unstressed conditions. We also found that gyp4Δ deletion, in contrast to gyp3Δ, did not suppress the vps9Δ mutant’s Ca2+ sensitivity (Fig 6A). Thus, GYP3 and GYP4 function redundantly with respect to Sec4 (Gao et al., 2003), but only GYP3 interacts genetically with VPS9 and, presumably, Rab5 signaling.
To test whether gyp3Δ suppression of the vps9Δ mutant’s Ca2+-sensitivity depends on Vps21, we constructed a vps9Δ gyp3Δ vps21Δ triple mutant. The inability of this mutant to grow in the presence of elevated extracellular [Ca2+] demonstrated that gyp3Δ suppression of the vps9Δ growth defect requires an intact copy of VPS21. To test whether gyp3Δ suppresses trafficking defects caused by the vps9Δ mutation, we assayed cargo sorting. vps9Δ gyp3Δ double mutants secreted less CPY-invertase than vps9Δ single mutants (Fig 6C) and in LUCID assays the double mutants accumulated significantly less of the MVB cargo Sna3-FLuc (Fig 6D). We conclude that GYP3 functionally opposes Rab5 signaling in vivo.
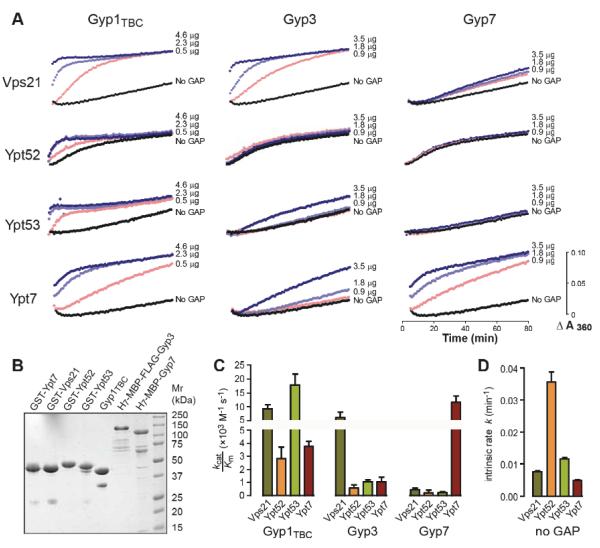
Vps21 is a preferred target of the Gyp3 GAP
Using purified proteins, we assayed the GAP activities of the Gyp1TBC catalytic core, full-length Gyp3, and full-length Gyp7 against a panel of endolysosomal Rabs (Fig 7). Single-turnover kinetics of GTP hydrolysis were measured using a real-time coupled assay of inorganic phosphate evolution, with each GAP assayed over a range of concentrations. The results were analyzed by nonlinear fitting of a pseudo first-order Michaelis-Menten model, which allowed us to estimate the intrinsic hydrolysis rate for each Rab and the specificity constant (Kcat/KM) for each Rab-GAP combination (Fig 7C). As reported previously Gyp1TBC is relatively promiscuous and efficiently accelerated GTP hydrolysis by all four Rabs tested (Albert et al., 1999). Compared to Gyp1TBC, Gyp7 and Gyp3 were more selective. Consistent with previous in vitro assays (Albert et al., 1999), cell-free fusion experiments (Eitzen et al., 2000; Brett et al., 2008) and multiple lines of in vivo evidence (Brett et al., 2008), Gyp7 strongly accelerated hydrolysis on Ypt7 but had low or undetectable activity against the Rab5 paralogs. Gyp3 had robust GAP activity against Vps21 and low but reproducible activity against Ypt53 and Ypt7. These results verify and extend previous work (Albert and Gallwitz, 1999; Albert et al., 1999; Eitzen et al., 2000; Brett et al., 2008) and, together with our genetic experiments, support the hypothesis that Gyp3 operates in vivo as the major Vps21 GAP. Moreover, the observed GAP activities of Gyp3 and Gyp7 against Ypt7 are consistent with in vivo fragmentation of vacuoles upon GYP3 or GYP7 overexpression (Brett et al., 2008).
Figure 7.
Msb3/Gyp3 is a Vps21 GAP in vitro. (A) Time course of solution-based GTP hydrolysis by Vps21, Ypt52, Ypt53 or Ypt7 in the presence of increasing concentrations of Gyp3, Gyp7 or Gyp1TBC. The data shown are representative of three independent experiments. (B) Purified proteins were separated by polyacrylamide gel electrophoresis (2.5 μg per lane) and stained with Coomassie brilliant blue. (C & D) Kinetics curves were fitted to a pseudo-first-order Michaelis-Menten model function to obtain Kcat/KM of a particular Rab-GAP pairing and the intrinsic GTP hydrolysis rate of each Rab. The plots represent the fitted mean ± S.E.M. rate constants obtained from duplicate experiments.
Gyp3 enforces the endolysosomal boundary
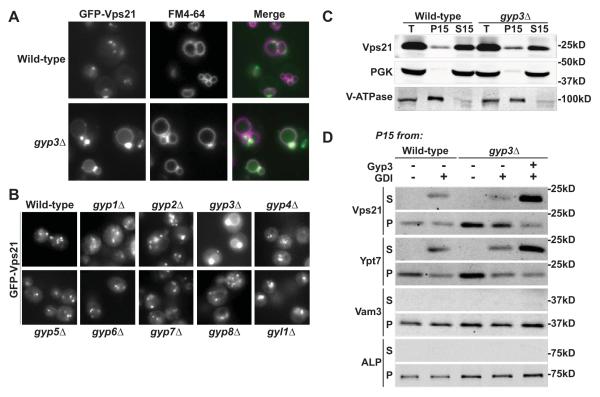
Rab GAPs are thought to regulate the timing and levels of Rab GTPase signaling, and also to police the spatial boundaries of Rab activity (Rivera-Molina and Novick, 2009; Barr and Lambright, 2010). During endolyosomal Rab conversion in mammals, Rab5 dissociates from late endosomes and is replaced by Rab7, a process that requires Mon1/SAND and the Vps3 ortholog hVam6 (Rink et al., 2005; Nordmann et al., 2010; Poteryaev et al. 2010; Nickerson et al., 2009). The Rab5 paralog Vps21 typically appears on cytoplasmic puncta, often adjacent to FM 4-64-stained vacuoles (Fig 8A), while downstream vacuoles are marked by Ypt7. In gyp3Δ mutant cells we observed a dramatic mislocalization of Vps21 to the vacuole limiting membrane. In mutants lacking each of the other known Rab GAPs, Vps21 remained on endosomal punctate structures and was excluded from the vacuole (Fig 8B), indicating a highly selective role for Gyp3 in Vps21 inactivation. These results suggested a working model in which Vps21 is first activated at the endosome by Vps9 and then inactivated by Gyp3 during late endocytic Rab conversion. In the absence of Gyp3, Vps21 remains active through endosome fusion at the vacuole, resulting in steady-state Vps21 accumulation on the vacuole.
Figure 8.
Msb3/Gyp3 inactivates Vps21 and spatially constrains Vps21 activity. (A) GFP-Vps21 contaminates the vacuole (marked by the fluorescent tracer FM4-64) in gyp3Δ mutants. (B) Mislocalization of GFP-Vps21 to the vacuole occurs in cells lacking Gyp3, but not in cells lacking other yeast Rab GAPs. (C) Subcellular fractionation of wild-type and gyp3Δ cells. PGK and V-ATPase (Vph1) were used as fiduciary markers for cytosol and endosolysosomal membrane fractions. T, total lysate after 1000 × g spin. P15, membrane-associated 15,000 × g pellet fraction. S15, cytosolic 15,000 × g soluble fraction. (D) Extraction of Rab GTPases Vps21 and Ypt7 by RabGDI. P15 membrane pellets prepared from wild-type or gyp3Δ cells were resuspended in lysis buffer or buffer supplemented with 3 μM His7-MBP-Gyp3, and incubated for 15 min on ice. Indicated samples received 9 μM recombinant GDI immediately prior to another 15,000 × g spin in which all samples were again separated to membrane-bound (P) and soluble fractions (S). Alkaline phosphatase (ALP) and the vacuolar SNARE Vam3 served as fiduciary markers for sedimentation of vacuoles.
Two GFP-tagged Vps21 effectors, the CORVET subunits Vps3 and Vps8 (Fig S4) exhibited apparently normal localization to prevacuolar endosomes in gyp3Δ cells, indicating that CORVET localization is specified not only by the location of Vps21 but also by other as-yet unidentified cues (see Discussion). In gyp3Δ cells, the GFP-Vps21 perivacuolar puncta appeared brighter than in wild-type cells, somewhat reminiscent of class E compartments or clustered endosomes when viewed by fluorescence microscopy. Ultrastructural examination of gyp3Δ cells by TEM revealed morphologically normal, unclustered MVBs (Fig S1), indicating that the Vps21 mislocalization in gyp3Δ cells does not have obvious morphological consequences. Similarly, gyp3Δ mutants exhibited near-normal sorting of the Golgi-endosome-vacuole cargo CPY (Fig 6C).
The mislocalization of Vps21 in gyp3Δ cells was verified and further characterized in biochemical fractionation experiments. Loss of a Rab’s cognate GAP is predicted to result in enrichment of Rab-GTP relative to Rab-GDP, rendering the Rab resistant to membrane extraction by Rab GDI and increasing the steady-state level of membrane-associated Rab (Brett et al., 2008). About 50% of the Vps21 in the vacuole-enriched P13 membrane fraction from wild-type cells was vulnerable to extraction in incubations with purified GDI; in contrast, most of the Vps21 in a comparable fraction from gyp3Δ cells resisted GDI extraction (Fig 8D, compare lanes 2 and 4). We further predicted that purified Gyp3 should target the GDI-resistant (and presumably GTP-bound) population of Vps21 in membranes from gyp3Δ cells. To test this prediction we isolated membranes from gyp3Δ cells and incubated them with recombinant purified Gyp3 prior to GDI extraction. Gyp3 rapidly converted almost all membrane-bound Vps21 to a GDI-extractable and presumably GDP-bound form (Fig 8D, compare lanes 4 and 5). The most straightforward interpretation of this result is that Gyp3 inactivates Vps21 on intact endolysosomal membranes, rendering Vps21 susceptible to GDI extraction. Gyp3 also enhanced GDI extraction of the vacuolar Rab Ypt7, consistent with its ability to slightly stimulate GTP hydrolysis on Ypt7 in vitro (Fig 7) and its ability to cause vacuole fragmentation when overexpressed in vivo (Brett et al., 2008). In sum, our experiments show that Gyp3 acts to limit the total pool of activated Vps21 in vivo, and that it specifically targets Vps21 at the terminal vacuolar lysosome for inactivation. We conclude that Gyp3 restricts the territory of Vps21 activity to pre-vacuolar endosomal compartments, thereby enforcing a key aspect of compartmental identity.
DISCUSSION
Our experiments show that Rab5 signaling is required not only for endosomal cargo sorting, but for normal MVB biogenesis and for cellular stress tolerance. The requirement for Rab5 signaling in survival of severe Ca2+ stress facilitated a suppression analysis that identified GYP3 as a negative regulator of Rab5 signaling. Further experimentation demonstrated that Gyp3 is an strong and relatively selective Vps21 GAP, and revealed that Gyp3 functions to spatially restrict Vps21 signaling to pre-vacuolar endosomal compartments. In parallel work, Ungermann, Barr, and colleagues performed a biochemical survey of Rab GAP selectivity, independently identifying Gyp3 as a Vps21 GAP, and similarly observed mislocalization of Vps21 to the vacuole in gyp3Δ cells (C. Ungermann, personal communication).
The CORVET subunits Vps3 and Vps8 bind Vps21-GTP and are released from membranes in vps21Δ cells (Horazdovsky et al., 1996, Peplowska et al., 2007, Ostrowicz et al., 2010, Plemel et al., 2011). However, in gyp3Δ cells Vps3 and Vps8 expressed from native promoters did not follow Vps21 to the vacuole (Fig S4). However, overexpression of Vps8 in gyp3Δ cells resulted in partial mislocalization of Vps8 to vacuole membranes (C. Ungermann, personal communication). Taken together, these results suggest that the presence of Rab5 is necessary but insufficient for CORVET localization. We speculate that additional determinants including phosphoinositides and SNAREs operate in concert with Rab5 to recruit CORVET, and indeed we have recently shown that CORVET directly binds the endosomal Qa-SNARE Pep12 (B. Lobingier and AJM, unpublished results).
Rab 5 signaling was required not only for ILV cargo sorting (Fig 2; Fig 4), but for formation of morphologically normal MVBs (Fig 2). ESCRT-0, I, II, and III drive these processes, and both ESCRT recruitment (Katzmann et al., 2003) and ILV formation (Futter et al., 2001) are strongly promoted by PI(3)P. In mammals, endosomal PI-3-kinase is stimulated by Rab5 (Christoforidis et al. 1999). Consistent with these findings, we found that both PI(3)P and ESCRTs are depleted from endosomes in Rab5-deficient yeast cells. The data suggest a working model in which loss of Rab5 signaling causes loss of PI(3)P from endosomes, preventing efficient recruitment and activity of ESCRTs. In addition, both Rab5 and PI(3)P have important roles in endosomal membrane fusion. A mature MVB contains on the order of 20 times more total membrane than a typical endocytic vesicle or post-Golgi carrier. Thus, the biogenesis of a single mature MVB may require dozens of preceding homotypic and heterotypic membrane docking and fusion events orchestrated by Rab5 and PI(3)P signaling. Reduced membrane fusion in vps21Δ ypt52Δ double mutant cells might reduce endosome size and directly limit the quantity of membrane available to form ILVs.
Although the three yeast Rab5 proteins are partially redundant, they exhibit some biochemical and functional specialization. In recent work characterizing an intrinsic membrane tethering activity of yeast endosomal Rabs, we detected robust tethering mediated by Vps21 or Ypt53, but none with Ypt52 (Lo et al., 2012). Similarly, although Gyp3 had robust GAP activity against Vps21 in our assays, Gyp3 activity against Ypt52 and Ypt53 was lower (Fig 7). These results are in good qualitative and quantitative agreement with previous work (Albert and Gallwitz, 1999; Albert et al., 1999). In contrast, an exhaustive assessment of all yeast Rab-GAP pairs in an accompanying study (C. Ungermann, personal communication) found relatively strong stimulation of Ypt52 GTP hydrolysis by Gyp3, and also reported anemic stimulation of Ypt7 GTP hydrolysis by Gyp7, where we and others (Albert et al., 1999) saw robust stimulation. These differences in apparent in vitro GAP selectivity may reflect divergent assay conditions or the measurement of time courses rather than single endpoints. We emphasize that every study to date has demonstrated strong activity of Gyp3 against Vps21.
Ypt52 is notable for its fast intrinsic hydrolysis rate, which results in hydrolysis of ≥90% of bound GTP within 60 min. without assistance from a GAP (Fig. 7; Albert and Gallwitz, 1999). In vivo, Ypt52 (but not Vps21 or Ypt53) is selectively bound and inhibited by the F-box containing inhibitor Roy1 (Liu et al., 2011; Fig. 5C; and AJM and R. Plemel, unpublished results). YPT53 is a stress-inducible gene that encodes a functional homolog of Vps21 (Fig 5). The relative immunity of Ypt53 to Gyp3 may account for increased tethering of MVBs when YPT53 is expressed from the VPS21 promoter in a vps21Δ ypt52Δ genetic background (Fig 5F,G). Given its immunity to Gyp3, Ypt53 might also extend the spatial domain of active Rab5 signaling to the vacuole. Consistent with this hypothesis, overproduced Ypt53 localizes to both endosome and vacuole membranes (Buvelot-Frei et al., 2006).
In the Rab countercurrent model of GAP recruitment (Rivera-Molina and Novick, 2009), an activated Rab recruits the GAP for the preceding upstream Rab. In this way, activation of the downstream Rab triggers inactivation of the upstream Rab, and sharp boundaires between Rab signaling domains are self-organized. In support of this idea, elegant work with Caenorhabditis elegans demonstrated that Rab7 is needed to position a Rab5 GAP, TBC-2 (Chotard et al. 2010). In yeast the situation is less clear. Gyp3 inhibits both endosomal and vacuolar Rabs, Vps21 and Ypt7 (Fig. 7; Albert and Gallwitz, 1999; Brett et al., 2008; C. Ungermann, personal communication). However, Gyp3 is considerably more active against Vps21 than it is toward Ypt7 (Fig. 7). Thus, it is still possible that Ypt7 positions Gyp3 to inactivate Vps21 on the vacuole. Further work will be needed to evaluate this model. Alternatively, other factors may regulate Gyp3 localization. Rho family small G proteins control Gyp3 and Gyp4 recruitment to sites of polarized exocytosis (Park and Bi, 2007). Rho1 and Cdc42 are also found on the vacuole, where they influence actin dynamics and membrane fusion (Eitzen et al., 2001; Isgandarova et al., 2007). Thus endolyosomal compartments and secretory patches at the plasma membrane might share common mechanisms for directing the localization and activity of Gyp3, and excluding Vps21, at the initial and terminal boundaries of the endocytic pathway. These models set an obvious agenda for future experimentation.
METHODS
Strain and plasmid construction
Strains and plasmids are summarized in Table I. Expression vector pDN616 was made by PCR amplification of the CEN/ARSH4 locus from pRS415, using primers incorporating flanking, parallel LoxP sequences. Gap repair of AatII-digested pRS406 vector using the resulting PCR product yielded a low-copy replicative plasmid similar to pRS416, but with the ability to readily convert to a non-replicative, integrating plasmid after excision of the CEN locus by Cre recombinase. pDN526 was made in manner similar to pDN616, except that gap repair inserted the 2μ locus PCR-amplified from pRS425 using primers that encoded a trio of flanking restriction sites (AatII-AvrII-SphI).
Table I.
Strains and plasmids used in this study
| Name | Genotype | Reference/source |
|---|---|---|
| S. cerevisiae | ||
| SEY6210 |
MATa leu2-3,112 ura3-52 his3-200 trp1-901
lys2-801 suc2-9 |
(Robinson et al., 1988) |
| BHY10 | SEY6210 CPY-Invertase::LEU2 (pBHY11) | (Horazdovsky et al., 1994) |
| GOY23 | SEY6210 prb1Δ::LEU2 pep4Δ::LEU2 | (Odorizzi et al., 1998) |
| MBY3 | SEY6210 vps4Δ::TRP1 | (Babst et al., 1997) |
| EEY6-2 | SEY6210 vps23Δ::HIS3 | (Babst et al., 2000) |
| DNY38 | BHY10 did2Δ::HIS3 | (Nickerson et al., 2006) |
| DNY40 | BHY10 vta1Δ::HIS3 | (Nickerson et al., 2006) |
| DNY464 | BHY10 crz1Δ::KAN | This work |
| DNY206 | BHY10 vps21Δ::KAN | This work |
| DNY438 | BHY10 vps21Δ0 | This work |
| DNY423 | BHY10 ypt52Δ0 | This work |
| DNY443 | BHY10 ypt53Δ::KAN | This work |
| DNY471 | BHY10 vps21Δ0 ypt52Δ0 | This work |
| DNY449 | BHY10 ypt52Δ0 ypt53Δ::KAN | This work |
| DNY511 | BHY10 vps21Δ::URA3 ypt53Δ::natR | This work |
| DNY532 | BHY10 vps21Δ0 ypt53Δ::natR | This work |
| DNY472 | BHY10 vps21Δ0 ypt52Δ0 ypt53Δ::KAN | This work |
| DNY536 | BHY10 vps21Δ0 ypt52Δ0 crz1Δ::KAN | This work |
| DNY441 | BHY10 ypt7Δ::KAN | This work |
| DNY347 | BHY10 vac1Δ::TRP1 | This work |
| DNY349 | BHY10 vps3Δ::HIS3 | This work |
| DNY350 | BHY10 vps45Δ::HIS3 | This work |
| DNY462 | BHY10 vps8Δ::KAN | This work |
| GOY223 | BHY10 vps9Δ::HIS3 | G Odorizzi (U Colorado) |
| DNY479 | BHY10 vps9Δ::HIS3 gyp1Δ::KAN | This work |
| JMY22 | BHY10 vps9Δ::HIS3 gyp2Δ:: | This work |
| DNY494 | BHY10 vps9Δ::HIS3 gyp3Δ::KAN | This work |
| DNY495 | BHY10 vps9Δ::HIS3 gyp4Δ::KAN | This work |
| DNY522 | BHY10 vps9Δ::HIS3 gyp5Δ::KAN | This work |
| DNY496 | BHY10 vps9Δ::HIS3 gyp6Δ::KAN | This work |
| DNY524 | BHY10 vps9Δ::HIS3 gyp7Δ::KAN | This work |
| DNY521 | BHY10 vps9Δ::HIS3 gyp8Δ::natR | This work |
| DNY525 | BHY10 vps9Δ::HIS3 roy1Δ::KAN | This work |
| DNY520 | BHY10 vps9Δ::HIS3 vps21Δ::URA3 | This work |
| DNY519 | BHY10 vps9Δ::HIS3 gyp3Δ::KAN vps21Δ::URA3 | This work |
| NSY1 | SEY6210 VPS8-GFP::HIS3 | G Odorizzi (U Colorado) |
| DNY523 | NSY1 gyp3Δ::KAN | This work |
| DNY239 | BHY10 VPS3-GFP::KAN | This work |
| DNY539 | BHY10 VPS3-GFP::KAN gyp3Δ::natR | This work |
| DNY364 | SEY6210 gyp1Δ::KAN | This work |
| DNY377 | SEY6210 gyp2Δ::KAN | This work |
| DNY489 | SEY6210 gyp3Δ::KAN | This work |
| DNY490 | SEY6210 gyp4Δ::KAN | This work |
| DNY491 | SEY6210 gyp5Δ::KAN | This work |
| DNY504 | SEY6210 gyp6Δ::KAN | This work |
| DNY373 | SEY6210 gyp7Δ::HIS3 | This work |
| DNY366 | SEY6210 gyp8Δ::KAN | This work |
| DNY503 | SEY6210 gyl1Δ::KAN | This work |
| JMY16 | BHY10 gyp3Δ::KAN | This work |
| DNY533 | BHY10 gyp7Δ::KAN | This work |
| DNY531 | GOY23 gyp3Δ::KAN | This work |
| DNY514 | BHY10 vps21Δ0 roy1Δ::KAN | This work |
| DNY515 | BHY10 vps21Δ0 VPS21pr::Flag-YPT53 (pDN225A::URA3) |
This work |
| DNY512 | BHY10 vps21Δ0 ypt52Δ0 VPS21pr::Flag-YPT53 (pDN225A::URA3) |
This work |
| MRY40 | SEY6210 vps21Δ::KAN | G Odorizzi (U Colorado) |
| E. coli | ||
| TOP10F’ | F’[lacIq Tn10(tetR)] mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR nupG recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(StrR) endA1 λ- |
Invitrogen |
| BL21(DE3) | F– ompT gal dcm lon hsdSB(rB- mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) |
Stratagene/Agilent |
| NS2114Sm | F– recA λ-cre rpsL | (Seifert et al., 1986) |
| Plasmids | ||
| pRS416 | URA3 CEN/ARSH4 Amp R | (Sikorski & Hieter, 1989) |
| pRS406 | URA3 Amp R | (Sikorski & Hieter, 1989) |
| pDN616 | URA3 LoxP::CEN/ARSH4::LoxP Amp R | This work |
| pDN606 | URA3 LoxP Amp R | This work |
| pDN526 | URA3 2μ Amp R | This work |
| pDN224 | VPS21pr::Flag-YPT53 (pDN616) | This work |
| pDN225A | VPS21pr::Flag-YPT53 (pDN606) | This work |
| pGO45 | URA3 2μ AmpR GFP-CPS1 | (Odorizzi et al., 1998) |
| pGO47 | LEU2 2μ AmpR GFP-CPS1 | (Odorizzi et al., 1998) |
| pSna3-GFP | URA3 CEN AmpR SNA3-GFP | (Reggiori & Pelham, 2001) |
| pRP1 | pRSF kanR His7-MBP-(tev) | This work |
| pDN190 | pRSF kanR His7-MBP-(tev) URA3 CEN | This work |
| pDN193 | His7-MBP-(tev)-GYP7 (pDN190) | This work |
| pDN248 | His7-MBP-(tev)-Flag-GYP3 (pDN190) | This work |
| pMB175 | LEU2 AmpR 2μ VPS22 VPS25 VPS36 | (Babst et al., 2002) |
| pVW31 | URA3 2μ AmpR PGK1pr::RLuc GCN4pr::FLuc | (Steffen et al., 2008) |
| pDN228 | URA3 2μ PGK1pr::RLuc 0::FLuc | This work |
| pDN247 | PGK1pr::RLuc YPT53pr::FLuc (pDN228) | This work |
| pDN251 | URA3 CEN PGK1pr::RLuc 0::FLuc | This work |
| pDN252 | PGK1pr::RLuc SNA3-FLuc (pDN251) | This work |
| pMM171 | URA3 CEN AmpR sna3Y109A-GFP | (McNatt et al., 2007) |
| pDN263 | PGK1pr::RLuc sna3Y109A-FLuc (pDN251) | This work |
| pVJS3 | AmpR GDI-intein-CBD | (Starai et al., 2007) |
| pRP2 | VPS21pr::GFP-VPS21 (pRS416) | This work |
| pGO700 | mCherry-CPS1 (pRS414) | G.Odorizzi (U Colorado) |
| pSL5916 | GFP-Atg8 (pRS416) | S. Lemmon (U Miami) |
| pCB257 | GFP-FYVE(EEA1) (pRS424) | C. Burd (Yale) |
| pCB260 | GFP-FYVE(EEA1) (pRS425) | C. Burd (Yale) |
| AMP219 | AmpR GST-VPS21 (p//GST) | (Brett et al. 2008) |
| AMP220 | AmpR GST-YPT52 (p//GST) | (Brett et al. 2008) |
| AMP217 | AmpR GST-YPT53 (p//GST) | (Brett et al. 2008) |
| AMP218 | AmpR GST-YPT7 (p//GST) | (Brett et al. 2008) |
| pDN125 | GFP-DID2 (pRS416) | (Nickerson et al., 2010) |
| pPHY2427 | URA3 2μ AmpR CUP1pr::3xHA-ATG13 (pRS426) | P. Hermann (Ohio State) |
pDN224 was constructed by using sequence overlap extension (SOE)-PCR to fuse the VPS21 promoter to the YPT53 coding sequence and terminator using primers that also inserted a six-codon N-terminal FLAG epitope, followed by gap repair of PvuII-digested pDN616. pDN224 was passaged through NS2114Sm bacteria expressing Cre recombinase, removing the CEN locus to yield the integrating plasmid pDN225A. DNY512 and DNY515 were constructed by integrating PstI-digested pDN225A at the ura3-52 locus in DNY471 and DNY438, respectively.
High-copy dual luciferase vector pDN228 was constructed by sequential gap repair of pDN526 using PCR products derived from plasmid template pVW31 (Steffen et al., 2008) PGK1pr::RLuc::GCY1term and FLuc::CYC1term (lacking promoter) inserted at XhoI and SacI cut sites, respectively. Promoter sequence (500 bp upstream plus 3 native codons) of YPT53 was PCR-amplified from genomic DNA and inserted by gap repair into SacII-digested pDN228, introducing promoters and start codons in-frame with FLuc to yield pDN247.
Low-copy dual luciferase vector pDN251 derived from removal of the 2μ locus from pDN228 by restriction digestion with AatII and subsequent gap repair at the AatII cut site using a LoxP::CEN/ARSH4::LoxP insert. For trafficking studies, SNA3 or sna3Y109A were PCR amplified from genomic DNA or pMM174 (McNatt et al., 2007) templates, respectively, then cloned in-frame into SacII-digested pDN251 to create chimeric FLuc reporters (pDN252 and pDN261).
Bacterial expression vector pDN190 was constructed by PCR amplifying a portion of the pRS416 vector containing both the URA3 and CEN/ARSH4 loci and inserting this sequence into AgeI-digested AMP451 via gap repair in yeast. The resulting pRSF-family vector contains a T7 promoter to allow regulated expression of His7-MBP (maltose binding protein) in bacteria, but also replicates stably in yeast. His7-MBP-Gyp3 and His7-MBP-Gyp7 bacterial expression plasmids were constructed by PCR-amplification of GYP3 and GYP7 coding sequences from yeast genomic DNA, and gap repair of BamHI/SalI-digested pDN190.
Culture and media
Standard methods for culture and media were used (Guthrie & Fink, 2002). Liquid media for experiments involving added CaCl2 or NaCl was adjusted to pH 5. Synthetic media was prepared using NH4Cl and buffered with 7.5 mM succinic acid in order to avoid precipitation of Ca2+ salts (Yoshimoto et al., 2002). Microbial growth in liquid cultures was assayed at 30° C with periodic shaking using a Bioscreen-C machine (Growth Curves USA, Piscataway, NJ) and 150 μL cultures inoculated at OD600 = 0.1.
Direct competition between vps9Δ and vps9Δ gyp3Δ strains was assessed by mixing independently grown cultures to achieve equal representation in a mixed starter culture, then diluting 1:1000 in fresh YPD (pH 5) with or without 200 mM CaCl2 and shaking at 30° C. This procedure was iterated 4 times. Starter and overnight cultures were serially diluted to achieve a plate inoculum of ~150 colonies and applied to YPD plates, from which isolated colonies were restruck to YPD plates containing G418 in order to determine the presence of the gyp3Δ::KAN marker.
Trafficking and dual luciferase assays
Colorimetric assays of CPY-invertase secretion were performed essentially as described (Darsow et al., 2000). Cells for invertase assays were grown in media supplemented with 2% fructose instead of glucose. Levels of the MVB cargo Sna3 were analyzed using a Promega (Madison, WI) dual-luciferase assay system. Cells were grown to early log phase (OD600 = 0.25 to 0.4) at 30° C in synthetic medium (2% glucose) lacking uracil and supplemented with casamino acids, cycloheximide (50 μg mL−1 final) was added for 20-30 min, and cultures were harvested by low-speed centrifugation. Cell pellets containing 0.4 OD600 × ml were resuspended in 500 μL lysis buffer and incubated at RT for 25 min before vortexing with glass beads for a further 5 min at RT. 5 μL aliquots of cell lysate were analyzed in 96-well plate format using a Perkin Elmer Victor Light Model 1420 luminometer. After subtracting backgrounds, luciferase activity of Sna3-FLuc was normalized vs. the activity of soluble RLuc, expressed from the same plasmid and driven by the constitutive PGK1 promoter. Luciferase assays to examine promoter activity were performed similarly, but without bead beatng and with a cell input of 0.22 OD600 × ml. FLuc activity was further normalized by subtracting the FLuc:RLuc signal ratio resulting from promoter-less Fluc. Autophagy was initiated in log phase cells by addition of 200 ng/μL rapamycin (LC Laboratories). Plasmid-borne CUP1pr::3xHA-ATG13 expressed adequately without added copper, permitting analysis without toxic effects associated with copper induction.
Light microscopy
For microscopy, overnight yeast cultures were diluted in synthetic media and grown at 30° C, 225 r.p.m. to OD600 ~0.4, then harvested by low-speed centrifugation. For FM 4-64 labeling of endolysosomal membranes (Vida and Emr, 1995), cell pellets were resuspended in 50 μL synthetic media containing FM 4-64 (2 μm) and incubated at 30° C for 15 min. Labeled cells were recovered by centrifugation and rinsed twice in media before resuspension in media and shaking at 30° C to permit growth to OD600 ~ 0.5 before concentration of cells and imaging. Where specified, cells were labeled with FM 4-64 for 30 min without chase and imaged immediately. Fluorescence microscopy was performed using an IX71 Olympus (Melville, NY) microscope equipped with 100× UPlanFLN, NA 1.30 objective, Hg arc or custom LED illuminator, and Andor (South Windsor, CT) iXon electron-multiplying charge-coupled device driven by Andor IQ v6 software. Confocal fluorescence microscopy was performed as described (Russell et al., submitted). Plate images were captured using a Sony DSC-W30 6.0 megapixel digital camera equipped with Zeiss Vario-Tessar lens. Thin section transmission electron microscopy was performed essentially as described (Russell et al., submitted). Photoshop v9.0 (Adobe) and Canvas v9.0 (ACD) software were used for image processing and figure preparation.
Protein purification
Purifications of Gyp1TBC (Lo et al., 2012) and Rab GDI (Starai et al., 2007) were as described. His7-MBP-FLAG-Gyp3 and His7-MBP-Gyp7 were expressed in E. Coli BL21(DE3)-pRIL grown in TB medium with chloramphenicol and kanamycin to OD600 = 1.0-1.5. Protein expression was induced with 0.3 mM IPTG for 3.5 hours at 30° C and cells were harvested by centrifugation. Cells containing His7-MBP-FLAG-Gyp3 or His7-MBP Gyp7 were lysed by sonication on ice in Buffer A (50 mM sodium phosphate, pH 7.4, 500 mM NaCl, and 20 mM imidazole, pH 7.4) or Buffer B (25 mM Tris ·HCl , pH 7.9, 300 mM NaCl, and 40 mM imidazole, pH 7.9), respectively, in the presence of protease inhibitors (PMSF, leupeptin, and pepstatin), lysozyme, and DNaseI. Cell lysates were clarified by centrifugation (18,500 × g, 20 min, 4° C). GAPs were affinity purified on HisTrap FF columns (GE Healthcare) and washed extensively with Buffer A or Buffer B prior to elution with Buffer A or Buffer B supplemented with 500 mM imidazole. His7-MBP-FLAG-Gyp3 fractions were subsequently diluted with 25 mM Tris ·HCl, pH 7.5 to [NaCl] <100 mM and loaded onto a Source Q ion exchange column (GE Healthcare). His7-MBP-FLAG-Gyp3 eluted at 25 mM Tris ·HCl and 220-240 mM NaCl. Eluate fractions were concentrated and then further purified by size exclusion on Superdex 200 pre-equilibrated with Buffer C (25 mM Tris ·HCl, pH 7.5, 300 mM NaCl, and 2 mM 2-mercaptoethanol). Fractions collected from the major peak were concentrated and glycerol was added to 5% (w/v) prior to storage. His7-MBP-Gyp7 containing fractions eluted from HisTrap FF resin were concentrated and buffer exchanged on Superdex 75 pre-equilibrated with Buffer C, concentrated, and glycerol was added to 10% prior to storage. For both GAPs, aliquots in thin-wall PCR tubes were flash-frozen in liquid N2, and stored at −80° C.
GST-Rab fusions were expressed in E. Coli BL21(DE3)-pRIL grown in TB with chloramphenicol and kanamycin to OD600 = 2-2.5. Expression was induced with 0.1 mM IPTG overnight at 18° C. Cells were harvested by centrifugation and resuspended into Buffer D (50 mM TrisH ·Cl, pH 7.9, 100 mM NaCl, 0.1% Triton-X-100, and 10% glycerol) supplemented with protease inhibitors (PMSF, leupeptin, and pepstatin), lysozyme, and DNaseI. Cells were lysed with a pressure lysis apparatus (EmulsiFlex-C5; Avestin, Ottawa, ON) with minimum peak pulses of 7,500 psi, then clarified by centrifugation (18,500 × g, 20 min, 4° C). GST-Rabs were adsorbed to 2 mL of Glutathione Sepharose 4B Resin (GE Heathcare) for 3 h at 4° C. Bound GST-Rabs were washed three times with Buffer D, then Buffer D plus 500 mM NaCl, and again in Buffer D, all without Trition-X-100 or glycerol. GST-Rabs were eluted by incubating the resins in 2 mL of buffer E (50 mM Tris ·HCl, 7.9, 100 mM NaCl, 5 mM MgCl2, 10 mM glutathione, and 2 mM 2-mercaptoethanol) for 10 min at 4° C. Elution was repeated at least 3 times. Eluted fusion proteins were concentrated to 4-8 mg/mL and then glycerol was added to 10% (w/v) prior to storage. All concentration steps were performed using Amicon Ultra-15 concentrators (10,000 MWCO; Millipore). Aliquots were flash-frozen in liquid N2 and stored at −80°C.
GAP assays
The GTP hydrolysis assay is modified from Pan et al. (2006) and Lo et al. (2012). GAPs were buffer exchanged into Buffer F (20 mM HEPES · NaOH, pH 7.5, and 150 mM NaCl) using Micro Bio-Spin columns (BioRad). For GTP loading, 3.0-3.5 mg of GST-Rab was suspended in 800 μL of Buffer D containing 5-10 mM reduced glutathione and supplemented with 1 mM DTT, 25× molar excess GTP, and 15 mM EDTA. After 60 min at 25-28° C, MgCl2 was added to 20 mM and GST-Rab fusions were incubated for an additional 15 min on ice to terminate the GTP loading reaction. GTP-loaded Rabs were rapidly separated from free nucleotides at on 5 mL HiTrap desalting columns (GE healthcare) pre-equilibrated with Buffer F and run at 4° C. Eluted Rabs were concentrated using an Amicon Ultra-0.5 mL concentrator (30,000 MWCO; Millipore) to ≥2.0 mg/mL. Inorganic phosphate released by single-turnover GTP hydrolysis was monitored in real time with the EnzChek Phosphate Assay (Invitrogen) in 100 μL reaction volume in 96-well half area plates (Corning). To initiate the assay 60 μL of GTP-loaded GST-Rab was added to 40 μL of assay buffer with or without GAP. Each reaction contained a final concentration of 20 mM HEPES · NaOH, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.15 mM 2-amino-6-mercapto-7-methylpurine ribonucleoside, 0.75 U/mL purine nucleoside phosphorylase, 20 μM GTP-loaded GST-Rab, and GAPs at the indicated concentrations. Each reaction also contained 1% (w/v) bovine serum albumin, which improved assay signal-to-noise ratio and reproducibility. Qualitatively similar results were obtained in the absence of BSA. Before use BSA was exchanged into Buffer F by gel filtration on Superdex 75 (GE healthcare), and stored at 4°C. Inorganic phosphate evolution was monitored by measuring A360nm in a Perkin Elmer Victor 3 plate reader.
Subcellular fractionation, antibodies and western blotting
Cells (18 OD600 × ml) grown to late log phase were harvested by low-speed centrifugation and resuspended in 10 mM DTT, 0.1 M Tris-Cl pH 9.4 for 10 min. Cells were again pelleted and resuspended in spheroplasting buffer (50 mM HEPES-KOH pH 7.2, 1× yeast nitrogen base with casamino acids, 2% glucose, 1 M sorbitol) with Zymolyase 20T ( Seikagaku, Tokyo, Japan) subjected to further purification by ion exchange chromatography. Spheroplasts were pelleted and rinsed once in spheroplasting buffer before resuspension in ice-cold lysis buffer [20 mM HEPES-KOH pH 7.2, 50 mM KOAc, 200 mM sorbitol, 0.1 mM Pefabloc SC, 1 mM PMSF, 0.01 mM chymostatin, 1 μg/mL each of aprotinin, leupeptin and pepstatin, and 1X protease inhibitor cocktail (EDTA-free; Roche)] and lysis by 20 strokes in an ice-cold Dounce homogenizer. Lysates were cleared of cell debris and nuclear membranes by centrifugation at 1000 × g for 5 min before sedimenting the supernatant again at 15,000 × g to yield endolysosomal membrane pellet (P15) and supernatant (S15) fractions. P15 fractions were resuspended in lysis buffer and proteins were harvested from total, P15 and S15 samples by precipitation with 0.15% deoxycholic acid and 10% trichloroacetic acid followed by two washes in acetone. Protein pellets were dried under vacuum and resuspended in Laemmli sample buffer (40 μL sample buffer per OD600 × ml equivalent) before boiling 10 min. For each SDS-PAGE lane, 0.2 OD600 × ml of cell suspension was analyzed.
Membrane extraction of Rabs by GDI was done by resuspending a P15 membrane pellet from 14 OD600 × ml of cell suspension in 140 μL lysis buffer dividing the total into 25 μL (2.5 OD600 × ml of cell suspension) aliquots to which were added lysis buffer with or without 3 μM recombinant Rab GAP. Samples were incubated on ice 15 min. Some tubes received 9 μM recombinant GDI immediately prior to sedimentation at 15,000 × g for 15 min., after which pellets were resuspended in lysis buffer and proteins from both membrane-associated (P15) and soluble (S15) fractions were processed as described above. 0.2 OD600 × ml of cell suspension per sample was analyzed by SDS-PAGE and western blot.
Western blotting was performed using an Odyssey fluorescence scanner (LI-COR Biosciences, Lincoln, NE). Monoclonal anti-PGK (3′-phosphoglycerate kinase), anti-ALP (alkaline phosphatase), and anti-Vph1 antibodies were obtained from Invitrogen (Carlsbad, CA). Polyclonal anti-Ypt7 and -Vam3 antibodies were gifts of W. Wickner (Dartmouth College, Hanover, NH). Polyclonal anti-Vps21 antisera was a gift of B. Horazdovsky (Mayo Clinic, Rochester, MN). Polyclonal anti-Ape1 antisera was a gift of D. Klionsky (Univ. Michigan, Ann Arbor, MI).
Quantitative analyses
Quantification of western blots was performed using Odyssey software (LI-COR Biosciences). Summary statistics for raw data from yeast growth curves were derived using the YODA program (Olsen et al., 2010). Other statistical analyses were performed using Microsoft Excel and GraphPad Prism (v. 4 and 5). To obtain Rab-GAP catalytic specificity constants (kcat/KM) and Rab intrinsic GTP hydrolysis rates (kintrinsic), data collected from each Rab replica were adjusted to subtract baseline absorbance and then the kinetic curves for each Rab-GAP combination were simultaneously fitted by a nonlinear lest-squares method with a pseudo-first order Michaelis–Menten model (Pan et al., 2006):
where kcat/KM and kintrinsic were treated as non-zero global parameters. A∞ and Ao were obtained from the Rab-Gyp1TBC combination fit and treated as global parameters within each Rab replica.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Odorizzi, M. Cyert, and members of the Merz Lab for fruitful discussions, C. Ungermann for sharing results prior to publication, R. Plemel and J. Milnes (UW) and N. Sulkko (CU-Boulder) for plasmid and strain construction, M. Sage and V. MacKay (UW) for assistance with qPCR, M. Kaeberlein for use of the Bioscreen instrument, and B. Horazdovsky, D. Klionsky and W. Wickner for sharing antisera. D.N. is an American Cancer Society Postdoctoral Fellow. S.L. was supported in part by a University of Washington Nanotechnology Integrative Graduate Education and Research Traineeship award (NSF DGE-0504573). M.R. was supported by NIH GM065505 (to G. Odorizzi). H. Chapin was supported by NIH T32 AG000057. This work was supported by NIH GM077349 (to A.M.), and Research Scholar Grant 10-026-01-CSM from the American Cancer Society (to A.M.).
Abbreviations
- TEM
transmission electron microscopy
- GAP
GTPase accelerating protein
- MVB
multivesicular body
- LUCID
luciferase reporter of intralumenal deposition
- ESCRT
endosomal sorting complex required for transport
- GDI
GDP dissociation inhibitor
- ILV
intralumenal vesicle
Footnotes
AUTHOR CONTRIBUTIONS D.N. and A.M. conceived, and D.N. carried out, most experiments. S.L. designed and performed GTP hydrolysis assays. M.R. performed and analyzed electron microscopy. H.C. performed and analyzed assays of autophagy. J.M. and M.R. performed some of the fluorescence microscopy. All authors interpreted results. D.N. and A.M. wrote the manuscript.
REFERENCES
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem. 1999;274:33186–33189. doi: 10.1074/jbc.274.47.33186. [DOI] [PubMed] [Google Scholar]
- Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Plemel RL, Lobinger BT, Vignali M, Fields S, Merz AJ. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J Cell Biol. 2008;182:1141–1151. doi: 10.1083/jcb.200801001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Molecular and cellular biology. 1996;16:2369–2377. doi: 10.1128/mcb.16.5.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot-Frei S, Rahl PB, Nussbaum M, Briggs BJ, Calero M, Janeczko S, Regan AD, Chen CZ, Barral Y, Whittaker GR, Collins RN. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Molecular and cellular biology. 2006;26:7299–7317. doi: 10.1128/MCB.02405-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Chotard L, Mishra AK, Sylvain M-A, Tuck S, Lambright DG, Rocheleau CE. TBC-2 regulates RAB-5/RAB-7 mediated endosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2285–2296. doi: 10.1091/mbc.E09-11-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Odorizzi G, Emr SD. Invertase fusion proteins for analysis of protein trafficking in yeast. Methods Enzymol. 2000;327:95–106. doi: 10.1016/s0076-6879(00)27270-4. [DOI] [PubMed] [Google Scholar]
- Eitzen G, Will E, Haas A, Wickner W. Sequential action of two GTPases to promote vacuole docking and fusion. EMBO J. 2000;19:6713–6720. doi: 10.1093/emboj/19.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter CE, Collinson LM, Backer JM, Hopkins CR. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J Cell Biol. 2001;155:1251–1264. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Albert S, Tcheperegine SE, Burd CG, Gallwitz D, Bi E. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J Cell Biol. 2003;162:635–646. doi: 10.1083/jcb.200302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard SR, Bryant NJ, Stevens TH. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 2000;11:613–626. doi: 10.1091/mbc.11.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier J-M, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. vol. 350. Academic Press; San Diego: 2002. p. 623. [Google Scholar]
- Hama H, Tall GG, Horazdovsky BF. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J Biol Chem. 1999;274:15284–91. doi: 10.1074/jbc.274.21.15284. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, Busch GR, Emr SD. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Cowles CR, Mustol P, Holmes M, Emr SD. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J Biol Chem. 1996;271:33607–33615. doi: 10.1074/jbc.271.52.33607. [DOI] [PubMed] [Google Scholar]
- Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–59. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgandarova S, Jones L, Forsberg D, Loncar A, Dawson J, Tedrick K, Eitzen G. Stimulation of actin polymerization by vacuoles via Cdc42p-dependent signaling. J Biol Chem. 2007;282:30466–30475. doi: 10.1074/jbc.M704117200. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk R, Kierzek AM, Slonimski PP, Rytka J. The Ccz1 protein interacts with Ypt7 GTPase during fusion of multiple transport intermediates with the vacuole in S. cerevisiae. J Cell Sci. 2001;114:3137–45. doi: 10.1242/jcs.114.17.3137. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nakatsukasa K, Kotera M, Kanada A, Nishimura T, Kishi T, Mimura S, Kamura T. Non-SCF-type F-box protein Roy1/Ymr258c interacts with a Rab5-like GTPase Ypt52 and inhibits Ypt52 function. Mol Biol Cell. 2011;22:1575–1584. doi: 10.1091/mbc.E10-08-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S-Y, Brett CL, Plemel RL, Vignali M, Fields S, Gonen T, Merz AJ. Intrinsic tethering activity of endosomal Rab proteins. Nat Struct Mol Biol. 2012;19:40–47. doi: 10.1038/nsmb.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Ahnert F, Arlt H, Mari M, Peplowska K, Epp N, Griffith J, Reggiori F, Ungermann C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol Biol Cell. 2009;20:5276–5289. doi: 10.1091/mbc.E09-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DC, Kim H, Mackin NA, Maldonado-Baez L, Evangelista CC, Jr., Beaudry VG, Dudgeon DD, Naiman DQ, Erdman SE, Cunningham KW. New regulators of a high affinity Ca2+ influx system revealed through a genome-wide screen in yeast. J Biol Chem. 2011;286:10744–10754. doi: 10.1074/jbc.M110.177451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, McKittrick I, West M, Odorizzi G. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol Biol Cell. 2007;18:697–706. doi: 10.1091/mbc.E06-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Cell Mol Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–9. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosome system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Murakami CJ, Kaeberlein M. YODA: software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC bioinformatics. 2010;11:141. doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiology and molecular biology reviews : MMBR. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec A, Arsic J, Kolling R. Mapping of Vps21 and HOPS binding sites in Vps8 and effect of binding site mutants on endocytic trafficking. Eukaryotic cell. 2010;9:602–610. doi: 10.1128/EC.00286-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- Plemel RL, Lobingier BT, Brett CL, Angers CG, Nickerson DP, Paulsel A, Sprague D, Merz AJ. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, Horazdovsky BF, Hurley JH. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci U S A. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Molecular and cellular biology. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert HS, Chen EY, So M, Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B, Stenmark H, Dusterhoft A, Philippsen P, Yoo JS, Gallwitz D, Zerial M. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B, Stenmark H, Zerial M. Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J Cell Sci. 1995;108(Pt 11):3509–3521. doi: 10.1242/jcs.108.11.3509. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Jun Y, Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci U S A. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall GG, Hama H, DeWald DB, Horazdovsky BF. The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol Biol Cell. 1999;10:1873–1889. doi: 10.1091/mbc.10.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcheperegine SE, Gao XD, Bi E. Regulation of cell polarity by interactions of Msb3 and Msb4 with Cdc42 and polarisome components. Molecular and cellular biology. 2005;25:8567–8580. doi: 10.1128/MCB.25.19.8567-8580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer M, Azmi I, West M, Davies B, Katzmann D, Odorizzi G. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J Cell Biol. 2011;192:295–306. doi: 10.1083/jcb.201007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann H, Hengst L, Gallwitz D. Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p) Cell. 1992;71:1131–42. doi: 10.1016/s0092-8674(05)80062-5. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.