These data suggest an altered renal transcriptome induced by CO2 pneumoperitoneum and laparoscopic surgery characterized by up-regulation of genes associated with acute inflammation, apoptosis, and immune injury.
Keywords: Laparoscopic donor nephrectomy (LDN), Interaction network, Differentially expressed genes (DEGs), Functional analysis, Small-molecule drugs
Abstract
Background and Objectives:
The objective was to compare gene expression profiles of 6 kidneys from open donor nephrectomy with 6 kidneys removed after laparoscopic donor nephrectomy and several hours of carbon dioxide pneumoperitoneum with DNA microarrays and identify small-molecule drugs.
Methods:
The gene expression profile GSE3297 was downloaded from the Gene Expression Omnibus database, and the differentially expressed genes were identified by a bioinformatics approach. First, Osprey software was used to construct a differentially expressed gene associated network. Then, DAVID (Database for Annotation, Visualization, and Integrated Discovery) and FuncAssociate were used to perform functional analyses. Finally, the Connectivity Map was used to screen for small-molecule drugs.
Results:
A total of 285 differentially expressed genes were identified, including 148 down-regulated genes and 137 up-regulated genes. In addition, the differentially expressed genes in the most significant Gene Ontology term were CASP6, KRAS, SOCS1, ESR1, TSHB, COL1A1, and MMP14. Furthermore, several differentially expressed genes, including STAT1, STAT6, SOS2, and SOCS1, participated in the most remarkable Janus kinase–signal transducer and activator of transcription signaling pathway. Finally, luteolin—with the highest score (0.887)—was identified as the small-molecule drug.
Conclusions:
Our data show an altered renal transcriptome induced by several hours of carbon dioxide pneumoperitoneum and laparoscopic surgery characterized by up-regulation of genes associated with acute inflammation, apoptosis, and immune injury, which could potentially result in renal injury and an enhanced immune response in the recipient after transplant.
INTRODUCTION
Kidney transplantation is the preeminent treatment for chronic kidney disease and end-stage renal disease. Not only does the living kidney donation help to reduce the number of patients waiting for an organ donation, but it also has—regarding the graft function—decisive advantages over cadaveric kidney grafts.1Nevertheless, living donor nephrectomy is a surgical challenge because the operation is performed in a healthy individual. Laparoscopic donor nephrectomy (LDN) offers less postoperative pain, a quicker convalescence, and a better cosmetic result than open nephrectomy, thus possibly increasing the number of renal donors.2 However, the LDN technique itself has drawbacks, such as a longer operative time and warm ischemia time, as well as possibly a higher rate of complications.3 Moreover, it has been criticized for some risk factors, including incisional hernia, bowel obstruction, and intraoperative massive hemorrhage.4,5
Carbon dioxide (CO2) pneumoperitoneum, which may alter normal physiology and may affect kidney function and post-transplant immunity, have been widely used in LDN.6,7 Recently, it has been shown in animals that pneumoperitoneum can increase the messenger RNA expression of endothelin-1, a potent vasoconstrictor that is implicated in the pathogenesis of ischemic pain and inflammation, by the kidney.8 CO2 pneumoperitoneum can induce stress responses with increased levels of tumor necrosis factor α, interleukin 6, and heat shock protein 70 in the serum,9 and oxidative stress markers (ischemia-modified albumin) were increased.10 These studies describe the differential expression of 1 gene or a few genes known to be associated with ischemia, injury, and fibrosis. However, there is little information on global gene expression in tissues associated with laparoscopic interventions. In addition, although there are several drugs for kidney diseases, such as atorvastatin,11 erythropoietin,12 and activated vitamin D,13 there is still an urgent need to discover novel drugs for the treatment of these diseases more effectively and selectively.
In this study we compared the gene expression profiles from kidneys recovered by LDN with CO2 pneumoperitoneum with those of open donor control kidneys and identified small-molecule drugs for kidney diseases by a bioinformatics approach. Our findings provide more knowledge about kidney diseases and may help researchers understand the underlying mechanism profoundly. Meanwhile, this study could contribute to the treatment of kidney diseases.
MATERIALS AND METHODS
Source of Data
The expression profile GSE329714 was downloaded from the GEO (Gene Expression Omnibus) database based on the GPL96 platform. A total of 12 healthy adult kidney donors signed consent forms and were approved for donation according to the usual evaluation and practice of the Cleveland Clinic Renal Transplant Program. To avoid potentially significant differences in gene expression due to race, only white recipients were selected for our study. The 12 patients were divided into 2 groups, and general anesthesia and diuresis were performed in each patient. The 6 control patients underwent an extraperitoneal flank incision with only minimal manipulation of the kidney, and 2 subcapsular core renal biopsy specimens were obtained with a 15-gauge spring-loaded needle. For the 6 LDN cases, 2 core renal biopsy specimens were obtained with the same needles after the kidneys underwent 2 to 3 hours of CO2 pneumoperitoneum.
Data Preprocessing and Differentially Expressed Gene Analysis
The original data in CEL files were converted into identifiable expression form, and the missing data were imputed.15 Then, the complemented data were normalized.16
The Limma package in R language was used to identify the differentially expressed genes (DEGs) between control kidneys and LDN kidneys.17 The genes with a logFC >1 and P < .01 were considered differentially expressed.
Construction of Interaction Networks of DEGs
In this study Osprey software was used to screen for all the interactions among DEGs and construct an interaction network.18 Osprey is a network visualization system that not only represents interactions in a flexible and rapidly expandable graphical format but also provides options for functional comparisons between datasets. This system integrates with BIND (Bimolecular Interaction Network Database)19 and GRID (General Repository for Interaction Database),20 which include 50,000 interactions.
Functional Enrichment Analysis for Interaction Network
Gene enrichment analysis is a promising high-throughput strategy that increases the likelihood for investigators to identify biological processes most pertinent to their study.21 DAVID (Database for Annotation, Visualization, and Integrated Discovery), which consists of an integrated biological knowledge base and analytic tools aimed at systematically extracting biological meaning from large gene or protein lists, is one of the most popular tools for enrichment analysis.22
In this study the DEGs in the network underwent functional enrichment analysis by DAVID and a hypergeometric algorithm. P < .05 was set as the threshold.
Pathway Enrichment Analysis for Interaction Network
FuncAssociate is a Web application that discovers properties enriched in lists of genes or proteins that emerge from large-scale experimentation.23 In this study the DEGs in the interaction network were mapped into FuncAssociate, and P < .05 was set as the threshold.
Screening for Small-Molecule Drugs
The Connectivity Map (CMap) is a large public database that contains data on 7056 gene expression profiles and 6100 small-molecule treatment-control pairs. The CMap can be used to find connections among small molecules, diseases, and drugs.24 The up-regulated and down-regulated DEGs were mapped into CMap, and the small molecules with the absolute value of scores >0.8 were identified.
RESULTS
DEG Analysis Between Control Kidneys and LDN Kidneys
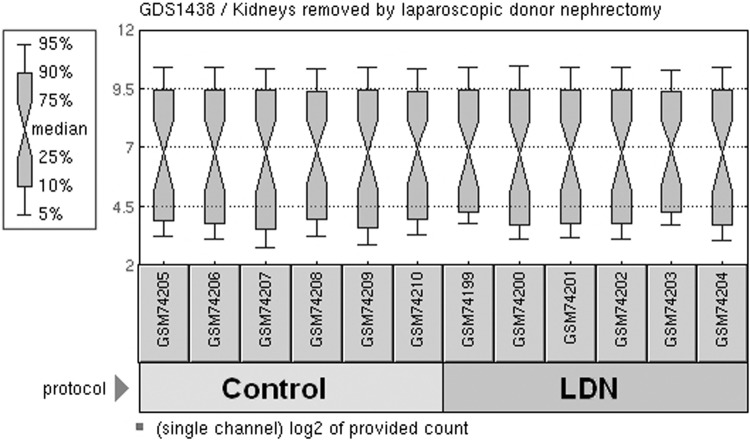
We obtained publicly available microarray dataset GSE3297 from the GEO database. After preprocessing and normalization, a total of 285 DEGs, which included 148 down-regulated genes and 137 up-regulated genes, were identified with P < .01 and logFC >1 (Figure 1).
Figure 1.
Box plots were generated using the normalized data of 6 donor kidneys from open nephrectomy and 6 kidneys removed after LDN and several hours of CO2 pneumoperitoneum. The light-green and dark-green boxes indicate the control and LDN samples, respectively. The black line in each box represents the median of each sample. All the black lines are almost in the same position, which indicates minimum variability in these datasets.
Construction of Interaction Network
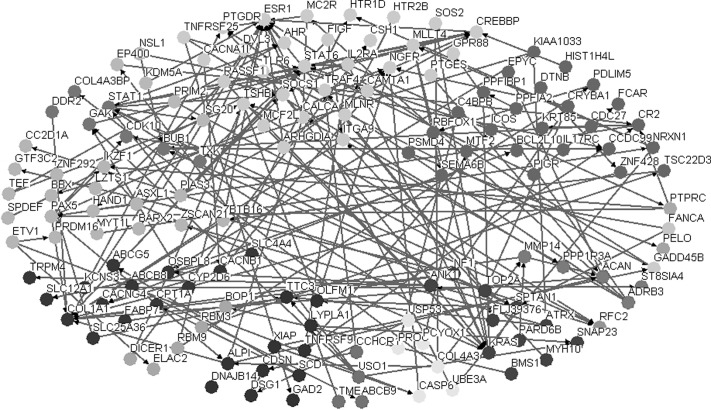
Osprey software was used to analyze the interactions of DEG protein products and establish a DEG associated network. The interaction network, which contained 89 DEGs, is shown in Figure 2.
Figure 2.
DEG associated interaction network.
Functional Enrichment Analysis
As shown in Table 1, a total of 13 significantly enriched Gene Ontology (GO) terms were identified and the most significant term was response to steroid hormone stimulus (GO 0048545, P = .001). Several DEGs, including CASP6, KRAS, SOCS1, ESR1, TSHB, COL1A1, and MMP14, were in this term.
Table 1.
Functional Enrichment Results of the Interaction Network
| GO Term | Description | Genes | P Value |
|---|---|---|---|
| 0048545 | Response to steroid hormone stimulus | CASP6, KRAS, SOCS1, ESR1, TSHB, COL1A1, MMP14 | .001 |
| 0051129 | Negative regulation of cellular component organization | BUB1, PAX5, NGFR, MMP14, TTC3, SPTAN1 | .002 |
| 0045596 | Negative regulation of cell differentiation | CAL1A1, NF1, CREBBP, ZBTB16, NGFR, PRDM16, TTC3 | .002 |
| 0042981 | Regulation of apoptosis | PTPRC, ESR1, ZBTB16, STAT1, BCL2L10, PROC, TNFRSF9, CASP6, KRAS, SOS2, NGFR, TRAF4, NF1 | .003 |
| 0006915 | Apoptosis | SATA1, CASP6, TSC22D3, KRAS, SOS2, ZBTB16, NGFR, GADD45B, AHR, TRAF4, BCL2L10 | .003 |
| 0043067 | Regulation of programmed cell death | PTPRC, ESR1, ZBTB16, STAT1, BCL2L10, PROC, TNFRSF9, CASP6, KRAS, SOS2, NGFR, TRAF4, NF1 | .003 |
| 0010941 | Regulation of cell death | PTPRC, ESR1, ZBTB16, STAT1, BCL2L10, PROC, TNFRSF9, CASP6, KRAS, SOS2, NGFR, TRAF4, NF1 | .003 |
| 0012501 | Programmed cell death | SATA1, CASP6, TSC22D3, KRAS, SOS2, ZBTB16, NGFR, GADD45B, AHR, TRAF4, BCL2L10 | .003 |
| 0043627 | Response to estrogen stimulus | CASP6, SOCS1, ESR1, TSHB, MMP14 | .004 |
| 0008283 | Cell proliferation | CALCA, PTPRC, COL4A3BP, CREBBP, BUB1, PELO, BOP1, MMP14, ISG20 | .004 |
| 0009725 | Response to hormone stimulus | SOCS1, ESR1, MMP14, STAT1, COL1A1, TSHB, KRAS, CASP6 | .006 |
| 0019221 | Cytokine-mediated signaling pathway | STAT6, KRAS, SOCS1, STAT1 | .008 |
| 0045637 | Regulation of myeloid cell differentiation | CALCA, IKZF1, ZBTB16, PRDM16 | .008 |
Pathway Enrichment Analysis
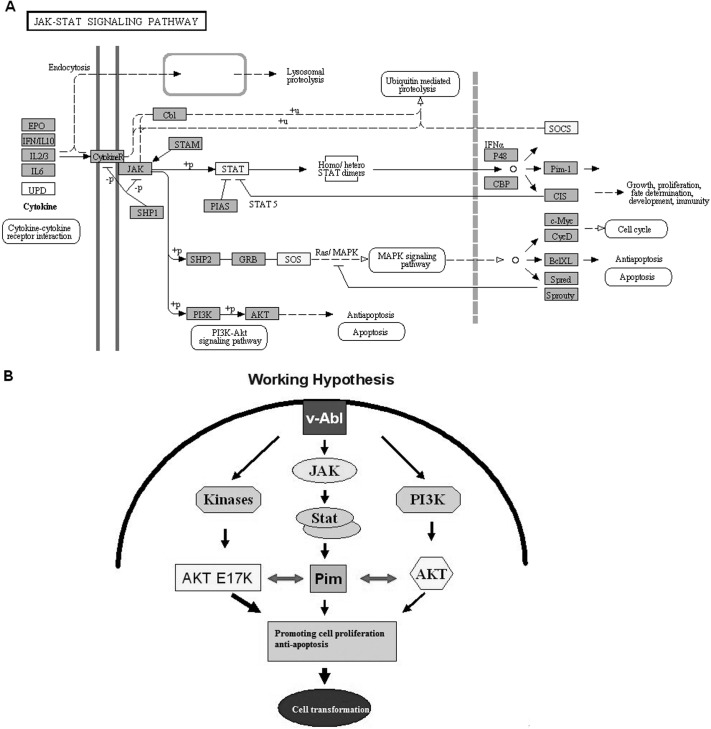
Only 1 pathway were screened out in our study, namely the Janus kinase (JAK)–signal transducer and activator of transcription (STAT) (JAK-STAT) pathway (P = .009). The DEGs that participated in this pathway were STAT1, STAT6, SOS2, and SOCS1 (Figure 3A). The signal transduction process in this JAK-STAT signaling pathway is shown in Figure 3B.
Figure 3.
A, JAK-STAT signaling pathway in interaction network. B, Working hypothesis, showing signal transduction process in pathway. The yellow boxes in A represent the DEGs.
Small-Molecule Drugs
A total of 14 small molecules with a highly significant correlation are shown in Table 2, including 6 negatively related molecules and 8 positively related molecules. Among these molecules, luteolin—with the highest correlation (score of 0.887)—had the potential to be the small-molecule drug for kidney diseases.
Table 2.
Enriched Significant Small-Molecule Drugs
| CMap Name | Score | P Value |
|---|---|---|
| Prestwick-691 | –0.877 | .004 |
| Cromoglycic acid | –0.876 | .031 |
| Isometheptene | –0.852 | .001 |
| Trihexyphenidyl | –0.852 | .006 |
| Disopyramide | –0.826 | .012 |
| Atracurium besylate | –0.824 | .011 |
| 8-Azaguanine | 0.818 | .002 |
| Fenoterol | 0.824 | .011 |
| Phenoxybenzamine | 0.829 | .001 |
| Propylthiouracil | 0.831 | .001 |
| Chrysin | 0.846 | .007 |
| 0297417–0002B | 0.861 | .005 |
| Medrysone | 0.871 | .000 |
| Luteolin | 0.887 | .000 |
DISCUSSION
In this study we compared the gene expression profiles of kidneys recovered by LDN with CO2 pneumoperitoneum and those of open donor control kidneys. A total of 285 DEGs, including 148 down-regulated genes and 137 up-regulated genes, were identified. In addition, about 13 GO terms were screened out, and the most significant term was response to steroid hormone stimulus. A total of 7 DEGs were in this term, including CASP6, KRAS, SOCS1, ESR1, TSHB, COL1A1, and MMP14.
KRAS is a central player in intracellular signaling. It may be activated by the epidermal growth factor receptor or possibly other receptor tyrosine kinases. Through interaction with phosphatidylinositol-3 kinase and activation of the downstream effectors such as mammalian target of rapamycin, KRAS indirectly modulates cell survival.25 Through the Braf/mitogen-activated protein kinase pathway, it also influences cell proliferation.26
As part of a negative-feedback regulation, SOCS1 down-regulates cytokine signaling through direct inhibition of the JAK tyrosine kinase and the signaling cascade of activated cytokine receptors, thereby attenuating cytokine-initiated signal transduction.27 Moreover, other studies have shown that SOCS1 also down-regulates toll receptor signaling through direct and indirect mechanisms. Both cytokine receptor and toll receptor signaling pathways mediate important functions in survival, maturation, and differentiation of various types of cells and in the regulation of immune function.28
The protein product of ESR1, estrogen receptor α, is a ligand-activated transcription factor that plays an essential role in directing tissue-specific gene expression.29 COL1A1 is a pro-fibrogenic extracellular matrix gene. Its protein product collagen-associated proteoglycan, which belongs to an extracellular matrix family, was up-regulated at the active/chronic inflammatory processes that accompany tissue damage.30 Matrix metalloproteinases that cleave interstitial collagens also play a crucial role in regulating matrix remodeling.31 MMP14 (MT1-MMP) is a pericellular type I collagenase that is critical for the postnatal development of the mesenchyme.32 In our study COL1A1 and MMP14 were both up-regulated. This indicated that proteolytic collagen turnover is active in the LDN kidney tissues.
In addition, the JAK-STAT signaling pathway was identified as the most significant pathway and the DEGs that participated in this pathway were STAT6, SOCS1, SOS2, and STAT1. Members of the signal transducers and STATs are major substrates of JAKs.33 STAT3, STAT1/4, and STAT6 are essential for differentiation of Th17, Th1, and Th2, respectively.34 STAT1 has been previously associated with certain kidney diseases. The SOS2 (salt overly sensitive 2) gene encodes a serine/threonine-type protein kinase, which has the potential to activate SOS1.35 The suppressors of cytokine signaling family members, including SOCS1 to SOCS7 in mammals, are important regulators of cytokine signaling pathways.36 Nakajima et al37 have shown that STAT1 is activated by proteinuria in proximal tubular cells. Our results are consistent with the previous study, which suggests that these DEGs may induce kidney diseases by affecting the JAK-STAT signaling pathway. In a word, this activation results in the production of cytokines and growth factors, as well as the induction of immune responses, subsequently leading to kidney diseases.
Furthermore, luteolin—which had the highest connectivity—was screened out as a small-molecule drug in our research. Luteolin is a natural antioxidant with less pro-oxidant potential than the flavonol quercetin, the best-studied flavonoid, but apparently with a better safety profile. It displays excellent radical scavenging and cytoprotective properties, especially when tested in complex biological systems in which it can interact with other antioxidants like vitamins. Luteolin displays specific anti-inflammatory effects at micromolar concentrations, which are only partly explained by its antioxidant capacities.38 The anti-inflammatory activity includes activation of antioxidative enzymes, suppression of the Nuclear Factor-kappa B pathway, and inhibition of proinflammatory substances. In vivo, luteolin reduced increased vascular permeability and was effective in animal models of inflammation after parenteral and oral application.38 In our study we showed that luteolin may have the potential to treat kidney diseases.
In conclusion, the expression profile was compared between LDN and open control kidneys, and the significant DEGs and small-molecule drug luteolin were identified. Laparoscopy and CO2 pneumoperitoneum cause an up-regulation of genes associated with acute inflammation and immune injury, which could potentially result in renal injury and an enhanced immune response in the recipient after transplant. Our findings may help researchers to understand kidney diseases more profoundly and develop novel therapeutic drugs for these diseases. However, more investigations should be conducted to explore the pathogenesis of kidney diseases and develop more drugs to combat them.
Contributor Information
Ya-hong Xu, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Jian-Li, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Xiao-ping Ma, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Yi Lu, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Ping Chen, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Shun-wen Luo, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Zhi-gang Jia, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Yang Liu, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
Yu Guo, Department of Urology, 452nd Hospital of Chinese People's Liberation Army, Chengdu, China..
References:
- 1. Hoda M, Hamza A, Greco F, Wagner S, Fischer K, Fornara P. Early and late graft function after laparoscopic hand-assisted donor nephrectomy for living kidney transplantation: comparison with open donor nephrectomy. Urol Int. 2010;84:61–66 [DOI] [PubMed] [Google Scholar]
- 2. Stamatakis L, Mercado MA, Choi JM, et al. Comparison of laparoendoscopic single site (LESS) and conventional laparoscopic donor nephrectomy at a single institution. BJU Int. 2013;112:198–206 [DOI] [PubMed] [Google Scholar]
- 3. Yuan H, Liu L, Zheng S, et al. The safety and efficacy of laparoscopic donor nephrectomy for renal transplantation: an updated meta-analysis. Transplant Proc. 2013;45:65–76 [DOI] [PubMed] [Google Scholar]
- 4. Chien C-H, Wang H-H, Chiang Y-J, Chu S-H, Liu H-E, Liu K-L. Change in renal function after laparoscopic donor nephrectomy for kidney transplantation. Transplant Proc. 2010;42:692–695 [DOI] [PubMed] [Google Scholar]
- 5. Sundaram CP, Bargman V, Bernie JE. Methods of vascular control during laparoscopic donor nephrectomy. J Endourol. 2006;20:467–470 [DOI] [PubMed] [Google Scholar]
- 6. Chiu AW, Chang LS, Birkett DH, Babayan RK. The impact of pneumoperitoneum, pneumoretroperitoneum, and gasless laparoscopy on the systemic and renal hemodynamics. J Am Coll Surg. 1995;181:397–406 [PubMed] [Google Scholar]
- 7. McDougall EM, Monk TG, Wolf JS, Jr, et al. The effect of prolonged pneumoperitoneum on renal function in an animal model. J Am Coll Surg. 1996;182:317–328 [PubMed] [Google Scholar]
- 8. Ambrose JA, Onders RP, Stowe NT, et al. Pneumoperitoneum upregulates preproendothelin-1 messenger RNA. Surg Endosc. 2001;15:183–188 [DOI] [PubMed] [Google Scholar]
- 9. Han C, Ding Z, Fan J, Sun J, Qian Y. Comparison of the stress response in patients undergoing gynecological laparoscopic surgery using carbon dioxide pneumoperitoneum or abdominal wall-lifting methods. J Laparoendosc Adv Surg Tech A. 2012;22:330–335 [DOI] [PubMed] [Google Scholar]
- 10. Aran T, Unsal MA, Guven S, Kart C, Cetin EC, Alver A. Carbon dioxide pneumoperitoneum induces systemic oxidative stress: a clinical study. Eur J Obstet Gynecol Reprod Biol. 2012;161:80–83 [DOI] [PubMed] [Google Scholar]
- 11. Bianchi S, Bigazzi R, Caiazza A, Campese VM. A controlled, prospective study of the effects of atorvastatin on proteinuria and progression of kidney disease. Am J Kidney Dis. 2003;41:565–570 [DOI] [PubMed] [Google Scholar]
- 12. Phrommintikul A, Haas SJ, Elsik M, Krum H. Mortality and target haemoglobin concentrations in anaemic patients with chronic kidney disease treated with erythropoietin: a meta-analysis. Lancet. 2007;369:381–388 [DOI] [PubMed] [Google Scholar]
- 13. Kovesdy CP, Ahmadzadeh S, Anderson JE, Kalantar-Zadeh K. Association of activated vitamin D treatment and mortality in chronic kidney disease. Arch Intern Med. 2008;168:397–403 [DOI] [PubMed] [Google Scholar]
- 14. Kurian SM, Flechner SM, Kaouk J, et al. Laparoscopic donor nephrectomy gene expression profiling reveals upregulation of stress and ischemia associated genes compared to control kidneys. Transplantation. 2005;80:1067–1071 [DOI] [PubMed] [Google Scholar]
- 15. Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525 [DOI] [PubMed] [Google Scholar]
- 16. Fujita A, Sato JR, Rodrigues Lde O, Ferreira CE, Sogayar MC. Evaluating different methods of microarray data normalization. BMC Bioinformatics. 2006;7:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2005. p. 397–420 [Google Scholar]
- 18. Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Willis RC, Hogue CW. Searching, Viewing, and Visualizing Data in the Biomolecular Interaction Network Database (BIND). Curr Protoc Bioinformatics. 2006;Chapter 8:Unit 8.9 [DOI] [PubMed] [Google Scholar]
- 20. Breitkreutz BJ, Stark C, Tyers M. The GRID: the general repository for interaction datasets. Genome Biol. 2003;4:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 23. Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935 [DOI] [PubMed] [Google Scholar]
- 25. Lo Ré AE, Fernández-Barrena MG, Almada LL, et al. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J Biol Chem. 2012;287:25325–25334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wickia A, Herrmannb R, Christoforic G. Kras in metastatic colorectal cancer. Swiss Med Wkly. 2010;140:w13112. [DOI] [PubMed] [Google Scholar]
- 27. Tamiya T, Kashiwagi I, Takahashi R, Yasukawa H, Yoshimura A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler Thromb Vasc Biol. 2011;31:980–985 [DOI] [PubMed] [Google Scholar]
- 28. Zhang J, Li H, Yu JP, Wang SE, Ren XB. Role of SOCS1 in tumor progression and therapeutic application. Int J Cancer. 2012;130:1971–1980 [DOI] [PubMed] [Google Scholar]
- 29. Schoen J, Sharbati S, Ritter J, Jewgenow K. Feline gonads exhibit tissue specific alternative splicing of oestrogen receptor alpha (ESR1). Reprod Domest Anim. 2012;47:30–34 [DOI] [PubMed] [Google Scholar]
- 30. Chen S-H, Li D, Xu C. Downregulation of Col1a1 induces differentiation in mouse spermatogonia. Asian J Androl. 2012;14:842–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sounni NE, Dehne K, van Kempen L, et al. Stromal regulation of vessel stability by MMP14 and TGFβ. Dis Model Mech. 2010;3:317–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chun T-H, Inoue M, Morisaki H, et al. Genetic link between obesity and MMP14-dependent adipogenic collagen turnover. Diabetes. 2010;59:2484–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fisher KH, Brown S, Zeidler MP. Designing RNAi Screens to Identify JAK/STAT Pathway Components. In: JAK-STAT Signalling. New York, NY: Springer; 2013:81–97 [DOI] [PubMed] [Google Scholar]
- 34. Adamson AS, Collins K, Laurence A, O'Shea JJ. The Current STATus of lymphocyte signaling: new roles for old players. Curr Opin Immunol. 2009;21:161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chakraborty K, Sairam RK, Bhattacharya R. Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol Biochem. 2012;51:90–101 [DOI] [PubMed] [Google Scholar]
- 36. Wang T, Gorgoglione B, Maehr T, et al. Fish suppressors of cytokine signaling (SOCS): gene discovery, modulation of expression and function. J Signal Transduct. 2011;2011:905813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakajima H, Takenaka M, Kaimori JY, et al. Activation of the signal transducer and activator of transcription signaling pathway in renal proximal tubular cells by albumin. J Am Soc Nephrol. 2004;15:276–85 [DOI] [PubMed] [Google Scholar]
- 38. Seelinger G, Merfort I, Schempp CM. Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. 2008;74:1667–1677 [DOI] [PubMed] [Google Scholar]