SUMMARY
During interphase centrosomes are held together by an extended proteinaceous linker that connects the proximal ends of the mother and daughter centriole. This linker is disassembled at the onset of mitosis in a process known as centrosome disjunction, thereby facilitating centrosome separation and bipolar spindle formation. The NIMA-related kinase Nek2A is implicated in disconnecting the centrosomes through disjoining the linker proteins C-Nap1 and rootletin. However, the mechanisms controlling centrosome disjunction remain poorly understood. Here, we report that two Hippo pathway components, the mammalian sterile 20-like kinase 2 (Mst2) and the scaffold protein Salvador (hSav1), directly interact with Nek2A regulating its ability to localize to centrosomes and phosphorylate C-Nap1 and rootletin. Furthermore, we show that the hSav1-Mst2-Nek2A centrosome disjunction pathway becomes essential for bipolar spindle formation upon partial inhibition of the kinesin-5 Eg5. We propose that hSav1-Mst2-Nek2A and Eg5 have distinct but complementary functions in centrosome disjunction.
The centrosome is the main microtubule organizer of mammalian cells and as such is crucial for the formation of the mitotic spindle and accurate chromosome segregation. The single centrosome of G1 cells, comprising two centrioles surrounded by pericentriolar material, is duplicated once per cell cycle during S phase leading to the generation of two centrosomes, each with two centrioles 1, 2. These two centrosomes are then held together by a proteinaceous linker that extends between the proximal ends of the older two centrioles 3-5. To facilitate centrosome separation at mitotic entry, this linker is disassembled in a process known as centrosome disjunction 6.
Centrosome disjunction is regulated by phosphorylation. The serine/threonine kinase that is implicated in this process is the NIMA-related kinase Nek2 that upon overexpression induces premature centrosome splitting 7. C-Nap1 and rootletin are the two components of the centrosomal linker that are phosphorylated and displaced from centrosomes by Nek2 kinase at the onset of mitosis 3, 6-10. C-Nap1 is localized to the proximal ends of the older (mother) centrioles and serves as a docking site for rootletin, which bridges the space between the two centrosomes. Depletion of either C-Nap1 or rootletin causes premature centrosome splitting consistent with their proposed function as essential linker proteins 4, 9.
Nek2 exists primarily as two splice variants, Nek2A and Nek2B. Nek2A, the longest splice variant, contains an N-terminal kinase domain and a C-terminal regulatory domain. The non-catalytic domain contains two coiled coil regions. The first coiled coil immediately downstream of the catalytic domain serves as a homodimerization domain that allows Nek2A kinase to undergo autophosphorylation and activation 11. The function of the second C-terminal coiled coil region remains unknown. Interestingly, the shorter splice variant Nek2B that lacks the second C-terminal coiled coil region is less potent in initiating centrosome splitting upon overexpression. This suggests that the C-terminus of Nek2A has an important but yet unknown role in centrosome separation 12.
The Hippo pathway is a conserved signal transduction cascade that in mammals is best known for its function in organ size control 13-16. However, Hippo pathway components have also been implicated in mitotic processes, such as alignment of mitotic chromosomes, formation of stable kinetochore-microtubule attachments and cytokinesis 17-20. Strikingly, certain Hippo pathway components, including the Lats and Mst1/2 kinases and the scaffolding protein hSav1, localize to centrosomes 21, 22. However, with the exception of Mst1, reported to control centrosome duplication 23, the centrosomal functions of Hippo pathway components remain unknown.
In this study we unravel a novel aspect of the regulation of centrosome disjunction. We show that the hSav1 and Mst2, two Hippo pathway components, directly interact with Nek2A. Our data suggest that Mst2 phosphorylates Nek2A thereby recruiting Nek2A to centrosomes and promoting phosphorylation and displacement of centrosomal linker proteins. Additionally, we demonstrate that the hSav1-Mst1/2-Nek2A pathway cooperates with forces provided by the kinesin motor Eg5 in centrosome disjunction.
RESULTS
Nek2A interacts with Hippo pathways components via a SARAH domain
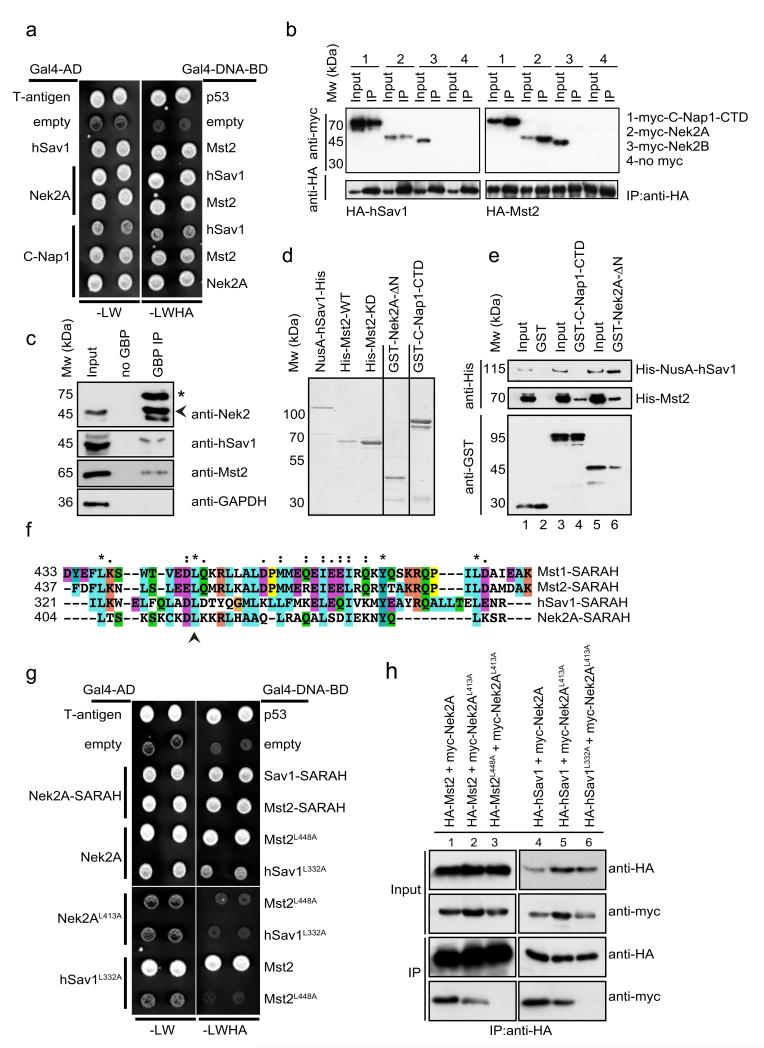
hSav1 is a scaffold protein that interacts with multiple Hippo pathway components via its protein-protein interaction domains 24, 25. To gain insight into the function of hSav1 at the centrosome, we sought to identify new interaction partners of hSav1 using the yeast two-hybrid system. In addition to Mst1 and Mst2, which are well-characterized interactors of hSav1 26, 27, we identified the NIMA-related kinase Nek2A as a binding partner of hSav1 (Fig. 1a). Further analysis revealed that Nek2A and its substrate CNap1 interact with hSav1, Mst1 and Mst2 but not with Lats1, another Hippo pathway component, in the yeast-two-hybrid system (Fig. 1a and Fig. S1).
Figure 1. Interactions between Nek2A, C-Nap1, hSav1 and Mst2.
a- Yeast two-hybrid analyses of the interactions between hSav1, Mst2 and Nek2A. Yeast cells with either Gal4-DNA-BD or Gal4-AD plasmid derivatives were mated on YPD plates and selected for –LW and –LWHA plates. Growth on –LW plates indicates mating and on –LWHA plates interaction of bait and prey encoded proteins. Colonies from non-interactors (e.g. empty plasmids) appear darker on –LW plates because cells did not express ADE2 and therefore accumulate a red pigment.
b- Co-IP of hSav1, Mst2, C-Nap1 and Nek2. HEK293 cells were cotransfected with the indicated constructs followed by anti-HA immunoprecipitation. Immunoprecipitated proteins were analysed by SDS-PAGE and immunoblotting.
c- Co-IP of endogenous hSav1, Nek2A and Mst2. Extracts of LAP-Nek2A cell line which have near-endogenous levels of Nek2A were prepared and Nek2A was immunoprecipitated with GFP-binder protein (GBP) coupled (GBP IP) or uncoupled (no GBP) to NHS-activated sepharose beads. The asterisk indicates LAP-Nek2A and the arrowhead Nek2. Of note is that LAP-Nek2A is difficult to detect in cell extracts with anti-Nek2 antibodies without enrichment by immunoprecipitation because of its low expression. LAP-Nek2A also immunoprecipitates endogenous Nek2 due to dimerization of Nek2 45.
d- NusA- and 6His-tagged hSav, GST-tagged C-Nap1-CTD and Nek2-ΔN were purified from E. coli, whereas wild-type and kinase-dead versions of Mst2 (Mst2-KD: K56R) were purified from Sf21 insect cells. Recombinant proteins were analysed by SDS-PAGE and visualized by Simply Blue Safe staining (Invitrogen).
e- Direct interactions between hSav1, Mst2, Nek2A and C-Nap1. Purified, recombinant GST-tagged truncations of C-Nap1 and Nek2A were incubated with recombinant His-tagged hSav1 or Mst2. C-Nap1 and Nek2A proteins were precipitated with glutathione sepharose beads and bound proteins were analysed by immunoblotting.
f- Presence of a putative SARAH domain in Nek2A. Sequence alignment of the C-terminal coiled-coil of Nek2A with the SARAH domains of hSav1 and Mst1/2 kinases.
g- Mutations in the SARAH domains of Nek2A, Mst2 and hSav1 impair interactions. Yeast two-hybrid analyses of indicated Leu-Ala mutants.
h- Myc-tagged Nek2A constructs together with HA-tagged hSav1 or Mst2 constructs were coexpressed in HEK293 cells. HA-tagged proteins were immunoprecipitated with anti-HA antibodies. Co-immunoprecipitation of myc-tagged proteins was determined by immunoblotting.
Next we used co-immunoprecipitation (co-IP) experiments to validate the yeast two-hybrid results. Nek2A and C-Nap1-CTD (the C-terminal domain of CNap1) but not Nek2B were found in complexes with hSav1 and Mst2 in HEK293 cells (Fig. 1b). Furthermore, Nek2A interactions were confirmed by co-IP of nonoverexpressed LAP-Nek2A protein from HeLa cell extracts (Fig. 1c).
We then asked whether Nek2A and C-Nap1 interact with hSav1 and Mst2 directly. Purified recombinant GST, GST-C-Nap1-CTD and GST-Nek2A-ΔN (lacking the N-terminal kinase domain) were incubated with His-tagged Mst2 or His-NusA-hSav1 (Fig. 1d). Mst2 and hSav1 but not His-NusA alone bound specifically to GST-Nek2A-ΔN (Fig. 1e, lane 6, data not shown). In addition, Mst2 but not hSav1 was found to associate with GST-C-Nap1-CTD (Fig. 1e, lane 4). Taken together, our data suggest that Nek2A and C-Nap1 interact directly with Mst2.
Mst1, Mst2 and hSav1 share a common homo- and hetero-dimerization motif known as the SARAH domain 25. Interestingly, the extreme C-terminal coiled-coil of Nek2A (residues 403-439) is similar to the SARAH domains of Mst1, Mst2 and hSav1 (Fig. 1f) and a conserved leucine residue, essential for homo-dimerization of Mst1 25, is also present in Nek2A (L413). We therefore hypothesized that Nek2A interacts with Mst2 and hSav1 via this putative SARAH domain. Accordingly, no interactions between SARAH domain containing proteins were observed in the yeast two-hybrid system when the critical leucine residues were mutated to alanine (Nek2AL413A, Mst2L448A and hSav1L332A) simultaneously in both interaction partners (Fig. 1g). Similar results were observed in co-IP experiments. Whereas Mst2 or hSav1 showed decreased binding to Nek2AL413A (Fig. 1h, lanes 2 and 5), Mst2L448A or hSav1L332A completely failed to interact with Nek2AL413A (Fig. 1h, lanes 3 and 6). In conclusion, Nek2A, Mst2 and hSav1 interact directly with each other via C-terminal SARAH domains.
Centrosomal recruitment of Nek2 is mediated by Hippo pathway components
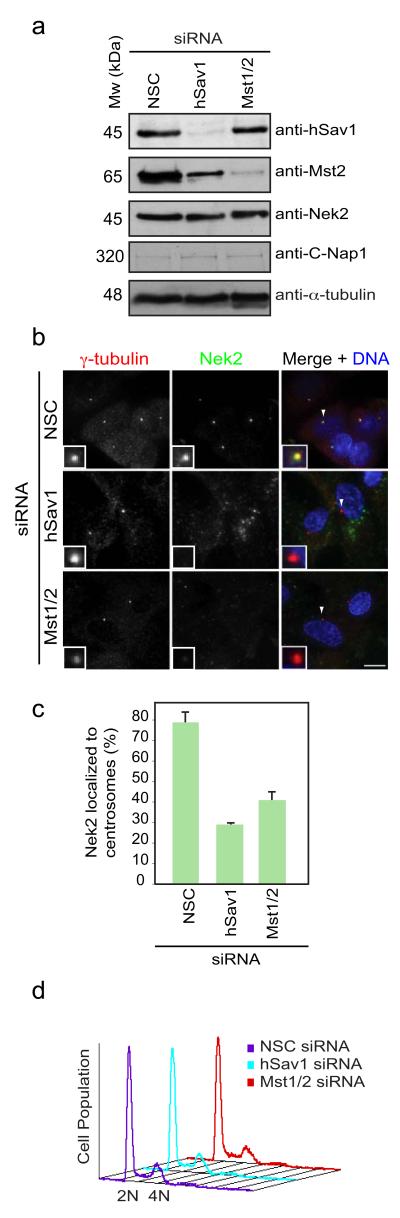
To establish a possible functional cooperation between Mst1/2, hSav1 and, Nek2 we investigated whether centrosomal localizations of proteins were interdependent. To exclude possible complementation of the highly related Mst1 and Mst2 kinases, we used siRNA oligos that targeted both kinases (Mst1/2). The specificity of the antibodies used in immunofluorescence microscopy was confirmed by siRNA depletion and subsequent decrease of centrosomal signal of the corresponding proteins (Fig. S2a-e). Centrosomal localizations of Mst1/2 and hSav1 were not altered in cells depleted of Nek2 or C-Nap1 (Fig. S2f-i). In contrast, Nek2 binding to centrosomes was strongly reduced in RPE-1 (Fig. 2a-c) and MCF7 cells (Fig. S3a-c) depleted of Mst1/2 or hSav1. In ~60% of hSav1 or Mst1/2 depleted cells Nek2 was no longer detected at centrosomes. In the remaining cells the centrosomal Nek2 signal was strongly reduced (data not shown). In addition, we noticed that depletion of hSav1 resulted in decreased levels of Mst1/2 (Fig. 2a), which is consistent with the idea that hSav1 acts as an Mst1/2 activator 27. Since Nek2 protein levels fluctuate throughout the cell cycle 28, it is formally possible that the observed changes in the Nek2 signal at centrosomes are an indirect outcome of changes in the cell cycle profile after siRNA treatment. However, measurements of the DNA content by flow cytometry indicated no change in cell cycle distribution upon hSav1 or Mst1/2 depletion (Fig. 2d). Thus, the Hippo pathway components hSav1 and Mst1/2 are important for the recruitment of Nek2 to centrosomes.
Figure 2. hSav1 and Mst1/2 are responsible for the centrosomal localization of Nek2.
a- Extracts of non-specific control (NSC), hSav1 or Mst1/2 siRNA-treated RPE-1 cells were analysed by immunoblotting using anti-tubulin, -hSav1, -Mst1/2, -C-Nap1 and -Nek2 antibodies.
b- RPE-1 cells were treated with NSC, hSav1 or Mst1/2 siRNA for 72 h. Cells were fixed and co-stained with anti-Nek2 and anti-γ-tubulin antibodies. Arrows indicate the centrosomes in insets. Scale bar, 10 μm.
c- Quantification of the analysis shown in Fig. 2b. Results are from three independent experiments. n>50 cells counted for each condition. Data are mean ± SEM.
d- Cells of Fig. 2b were analysed by flow cytometry. Note that there is no significant difference in cell cycle distribution between cells treated with NSC, hSav1 or Mst1/2 siRNAs.
We then analysed whether other components of the Hippo pathway are required for the centrosomal binding of Nek2. siRNA depletion of Lats1/2 kinases, Rassf1A or the transcriptional co-activator YAP did not alter Nek2 localization at centrosomes (Fig. S3d,e). Depletion of Mob1 had a modest effect on Nek2 binding to centrosomes, which may be explained by its interaction with Mst1/2 29. Taken together our data suggest that Mst1/2 and hSav1 are the only Hippo pathway components that have a clear function in targeting Nek2A to centrosomes.
Mst1/2 regulates the function of Nek2 at the centrosome
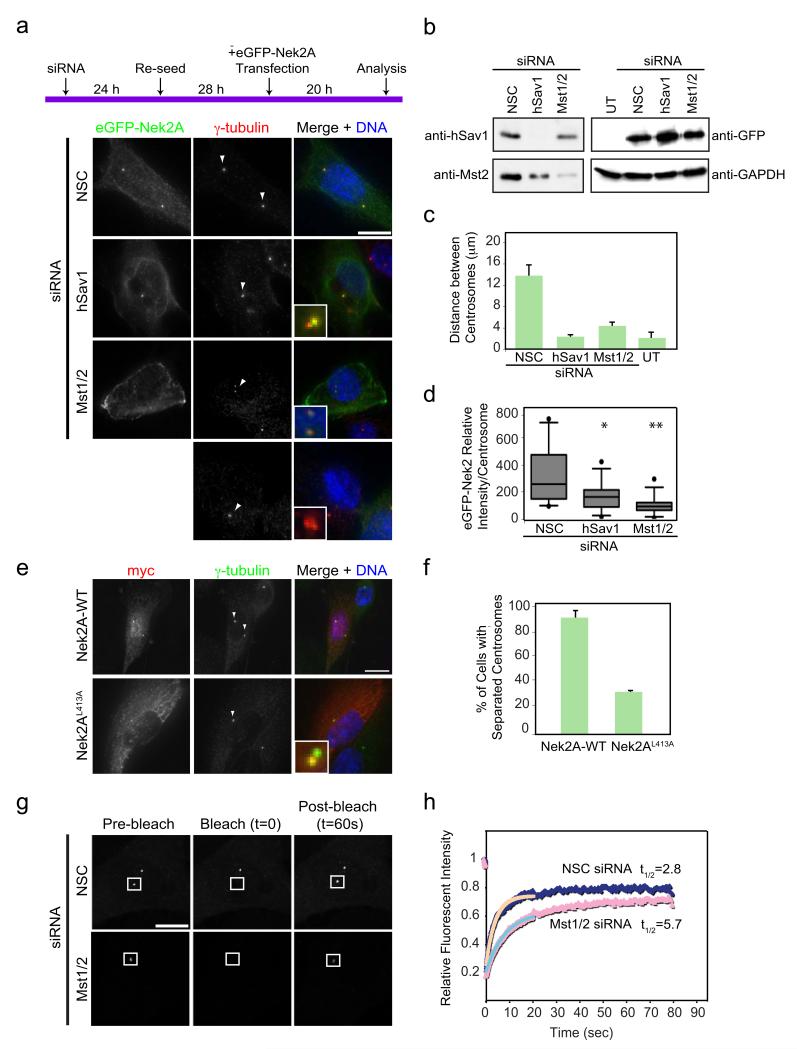
As depletion of Mst1/2 resulted in a significant decrease in the centrosomal signal of Nek2 (Fig. 2b), we hypothesized that lack of Mst1/2 may also reduce the ability of overexpressed Nek2A to induce premature centrosome splitting. To test this notion, siRNA treated asynchronous RPE-1 cells were transfected with eGFP-Nek2A expressed from the CMV promoter and then analysed for induction of centrosome splitting as outlined in Fig. 3a. In non-specific control (NSC) siRNA treated cells, the centrosomes were readily split upon overexpression of eGFP-Nek2A (Fig. 3a,c). Importantly, siRNA depletion of hSav1 or Mst1/2 dramatically reduced the efficiency of Nek2A to induce premature centrosome splitting although eGFP-Nek2A was expressed to similar levels in all cases (Fig. 3b). In addition, binding of eGFP-Nek2A to centrosomes was reduced in cells depleted of hSav1 or Mst1/2 (Fig. 3d). Together, this data suggest that hSav1 and Mst1/2 are essential for the Nek2A induced premature centrosome splitting in interphase cells. In support of this view, the SARAH domain defective Nek2AL413A mutant that shows only reduced interaction with hSav1 and Mst2 (Fig. 1h) failed to promote efficient centrosome splitting upon overexpression (Fig. 3e,f).
Figure 3. Mst1/2 and hSav1 regulate the centrosomal function and dynamics of Nek2.
a- RPE-1 cells were treated with the indicated siRNA oligos for 52 h and subsequently transfected or not transfected (UT = untransfected) with eGFP-Nek2A to induce centrosome splitting. Cells were fixed and stained with antibodies against γ-tubulin. Inlets are magnifications of the marked (arrow) centrosomal signals. Scale bar, 10 μm.
b- Extracts of cells from Fig. 3a were analysed by immunoblotting with the indicated antibodies.
c- Distances between the two centrosomes of cells in Fig. 3a. Results are from three independent experiments; 20 cells were analysed for each condition. Data are mean ± SEM.
d- Intensity of centrosomal association of Nek2-GFP was quantified from cells shown in Fig. 3a. Results are from two independent experiments. n=20 cells analysed for each condition. Box-and-whiskers plots: boxes show the upper and lower quartiles (25-75 %) with a line at the median, whiskers extend from the 10 to the 90 percentile and dots correspond to the outliers (*NSC/hSav1 p=0.032, **NSC/Mst2 p=0.0022).
e- U2OS cells were transfected with myc-tagged WT Nek2A or the Nek2AL413A mutant and stained with anti-myc and anti-γ-tubulin antibodies. Scale bar, 10 μm.
f- Quantification of cells shown in Fig. 3e. Results are from three independent experiments. n=20 cells for each condition. Data are represented as mean ± SEM.
g- Centrosomal dynamics of U2OS-Nek2-GFP cells were measured by FRAP. Images were taken at 65 ms intervals for 1 s before bleaching (indicated as pre-bleach) and after laser bleaching of the indicated areas, images were acquired every 65 ms for 20 s followed by an acquisition of half a second for 60 s. Scale bar, 10 μm.
h- FRAP data were analysed by ImageJ, the relative GFP intensity within the bleached area was recorded as a measure of time. Background intensities were subtracted and corrected for acquisition bleaching. The data were fitted using a single exponential equation and each data point represents the average intensity from 20 cells (p = 0.0002).
Next, we asked whether Mst1/2 regulates dynamic behavior of Nek2A at the centrosome. Previous fluorescence recovery after photobleaching experiments (FRAP) revealed that eGFP-Nek2A is a rapid-exchanging protein at the centrosome with a turn over rate (t1/2) of 3 s 30. Using the same U2OS cells with tetracycline inducible eGFP-Nek2A we confirmed this result (t1/2 2.8 s, 75% recovery) and, in addition, showed that t1/2 of centrosomal eGFP-Nek2A increased two-fold upon depletion of Mst1/2 (t1/2 5.7 s, 70% recovery; Fig. 3g,h). Importantly, U2OS tetracycline inducible eGFP-Nek2A cells showed the same Mst1/2 dependent centrosome splitting phenotype (Fig. S4a-d) as cells in Fig. 3a. Taken together, these data are consistent with the notion that Mst1/2 regulates not only the steady state level but also the dynamic behavior of Nek2A at centrosomes. The simplest way to interpret this data is that hSav1-Mst1/2 increases the affinity of Nek2A for centrosomal binding sites meaning that in the absence of Mst1/2 there is both reduced maintenance and slower recruitment of Nek2A.
Mst2 regulates centrosome separation by phosphorylating Nek2A
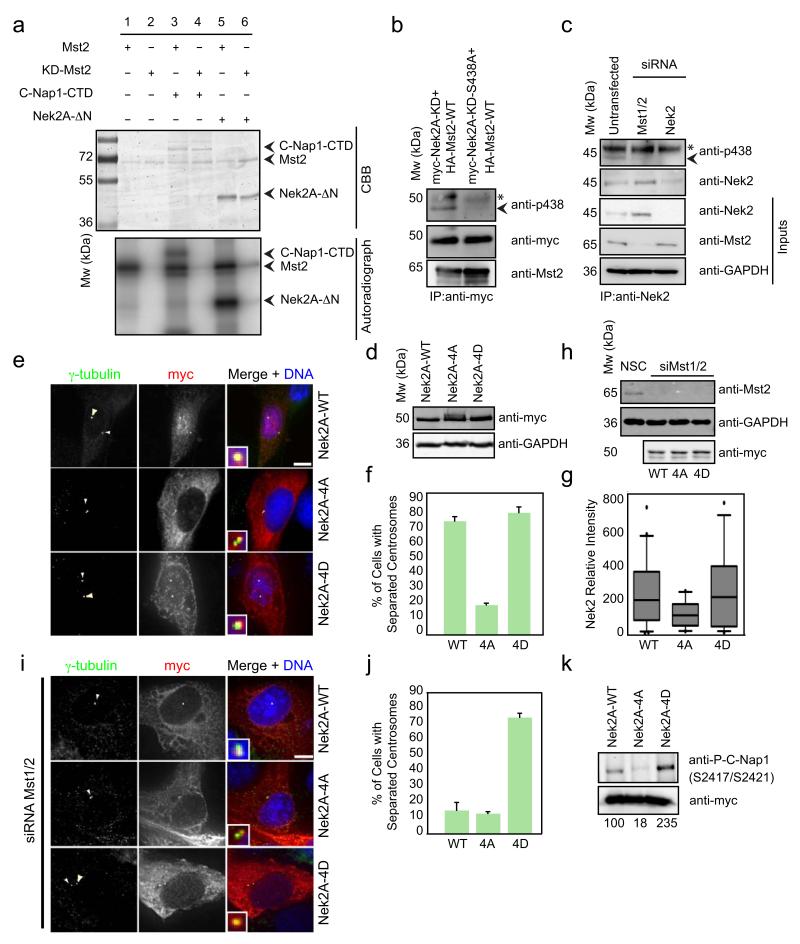
Our in vivo analyses clearly demonstrate that hSav1 and Mst1/2 cooperate with Nek2A in centrosome separation. To gain a molecular understanding of this regulation, we examined the phosphorylation of C-Nap1 and Nek2A by Mst2 in vitro. Mst2 but not Mst2-KD (K56R kinase dead mutant) was able to phosphorylate CNap1-CTD (Fig. 4a, lane 3) and Nek2A-ΔN (lane 5). MS/MS mass spectrometry analysis of Nek2A-ΔN phosphorylated by Mst2 in vitro identified four phosphorylation sites (S356, S365, T406 and S438) (Fig. S5a). The first two serine residues are located in the MT-binding region of Nek2A 30, whereas the two latter residues are in the SARAH domain (Fig. S5b).
Figure 4. Mst2 phosphorylates Nek2A and regulates the ability of Nek2A to induce centrosome disjunction.
a- Recombinant C-Nap1-CTD and Nek2-ΔN were incubated with wild-type or kinase-dead versions (KD) of Mst2 in the presence of γ-32P-ATP. Proteins were separated by SDS-PAGE and analysed by Coomassie Brilliant Blue (CBB) staining and autoradiography.
b- Phosphorylation of Nek2A by Mst2. Myc-tagged Nek2A-WT or nonphosphorylatable Nek2A-S438A were transfected together with HA-Mst2-WT into HEK293 cells. Myc-Nek2A constructs were immunoprecipitated from HEK293 cells and analysed by immunoblotting with anti-myc, -pS438 and –Mst2 antibodies. The asterisk indicates the IgG heavy chain and the arrowhead P438-Mst2.
c- Phosphorylation of endogenous Nek2A by Mst2. HeLa cells left untreated or treated with Mst1/2 or Nek2 siRNA oligos, were collected and Nek2 was immunoprecipitated with anti-Nek2 antobodies. Samples were analysed with the indicated antibodies. The asterisk indicates the IgG heavy chain and the arrowhead P438-Mst2.
d,e- Myc-tagged Nek2A (WT, -4A, or -4D) constructs were transfected into U2OS cells. (d) The extracts were analysed by immunoblotting with anti-myc and -GAPDH antibodies. (e) Cells were stained with anti-myc and anti-centrin antibodies. Arrows indicate the centrosomes in insets. Scale bar, 10 μm.
f- Quantification of Fig. 4e. The distance between two centrosomes was measured. Results are from three independent experiments. n = 20 cells for each condition. Data are mean ± SEM.
g- Intensity of centrosomal association of myc tagged Nek2 constructs was quantified from cells shown in Fig. 4e. Results are from three independent experiments. n = 20 cells analysed for each condition. Box-and-whiskers plots: boxes show the upper and lower quartiles (25-75 %) with a line at the median, whiskers extend from the 10 to the 90 percentile and dots correspond to the outliers.
h- U2OS cells were treated with siRNA oligos against Mst1/2 kinases and transfected with myc tagged Nek2A constructs 48 h later. Extracts were analysed with anti-Mst1/2, anti-myc and anti-GAPDH antibodies.
i- Cells of Fig. 4h were stained with anti-myc and anti-γ-tubulin antibodies. Arrows indicate the centrosomes in insets. Scale bar, 10 μm.
j- Quantification of Fig. 4i. The distance between two centrosomes was measured. Results are from three independent experiments. n = 20 cells for each condition. Data are mean ± SEM.
k- Extracts from RPE-1 cells transfected with myc-tagged Nek2A constructs. Whole cell extracts were analysed by immunoblotting using anti-myc and phospho-specific C-Nap1 antibodies (S2417/S2421). Phospho-C-Nap1 was normalized to anti-myc signal. The result is a representative of 2 independent experiments.
To confirm that Mst2 phosphorylates Nek2A in vivo, we raised phospho-specific antibodies against one of the phosphorylation sites in the SARAH domain of Nek2A (pS438). The specificity of the antibodies was confirmed using recombinant, kinase dead Nek2A-K37R incubated with either kinase-dead (Mst2-KD; K56R) or wild type Mst2 (Mst2-WT) (Fig. S5c). Next, we coexpressed Mst2 with wild-type or the serine-to-alanine S438A mutant of Nek2A in HEK293 cells and immunoprecipitated Nek2A. The pS438 antibody recognized Nek2A, but not Nek2AS438A (Fig. 4b, arrowhead) indicating that Mst2 is able to phosphorylate Nek2A at S438 in vivo.
In an additional experiment we show that endogenous levels of Mst2 phosphorylate Nek2A in vivo (Fig. 4c). Immunoprecipitations of Nek2A of untransfected or siRNA depleted cells (Mst1/2, Nek2A) were analyzed with anti-pS438 antibodies. Nek2A-pS438 was detected in wild type cells but not in Mst2 depleted cells (Fig. 4c, arrowhead). Thus, Mst2 phosphorylates Nek2A in vivo.
Next, we analyzed the function of Nek2A phosphorylation by Mst2. For this we examined the ability of non-phosphorylatable Nek2AS356A,S365A,S406A,S438A (Nek2A-4A) and the corresponding phospho-mimicking serine-to-aspartate Nek2AS356D,S365D,S406D,S438D (Nek2A-4D) mutant to induce centrosome splitting in interphase. When expressed at identical levels wild type Nek2A and Nek2A-4D but not Nek2A-4A induced centrosome splitting (Fig. 4d-f). Furthermore, Nek2A-4D but not wild type Nek2A or Nek2A-4A bypassed the requirement of Mst1/2 for centrosome separation (Fig. 4h-j). Together, this shows that phosphorylation of Nek2A by Mst2 is necessary for centrosome splitting.
We further investigated how phosphorylation of Nek2A by Mst2 promotes centrosome splitting. Phosphorylation by Mst2 did not affect specific kinase activity of Nek2A towards C-Nap1 in vitro (Fig. S5d). However, Nek2A-4A accumulated less on centrosomes than Nek2A-4D (Fig. 4g), consistent with reduced binding of Nek2A to centrosomes in Mst1/2- or hSav1-depleted cells (Fig. 3d). Mst2 may therefore affect localized Nek2A kinase activity at centrosomes. To test this notion, we raised phospho-specific antibodies against two phosphoserines of C-Nap1 (pS2417/pS2421), which are phosphorylated by Nek2A (A.M.F., unpublished data). The specificity of the affinity-purified antibodies was confirmed with recombinant C-Nap1 (Fig. S5e). In vivo Nek2A-4A was much less efficient in phosphorylating C-Nap1 than Nek2A or Nek2A-4D (Fig. 4k). This data suggest that phosphorylation of Nek2A by Mst2 promotes recruitment of Nek2A to centrosomes which then allows efficient phosphorylation of centrosomal linker proteins resulting in centrosome disjunction.
hSav1-Mst2-Nek2A and the kinesin Eg5 cooperate in centrosome disjunction
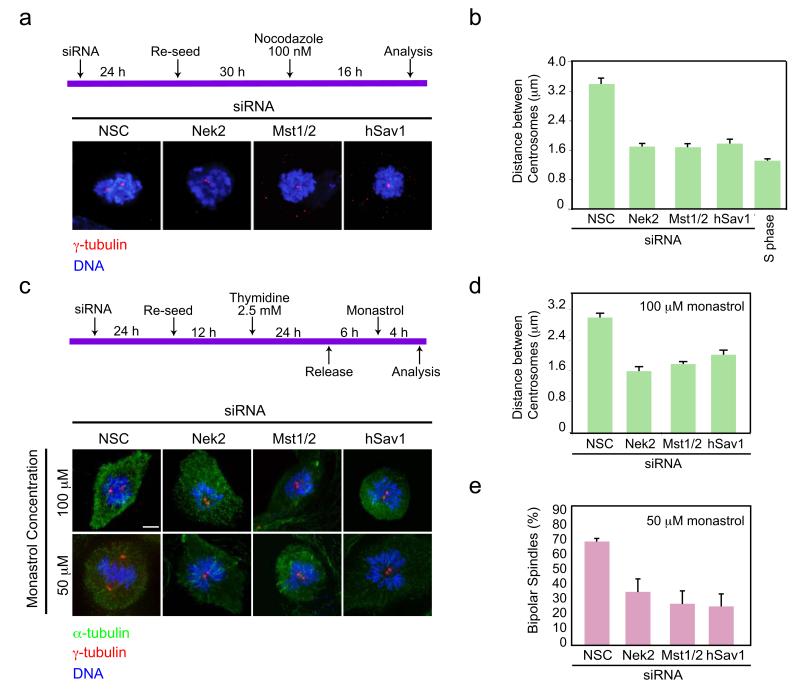
Based on the above results, we expected that centrosome separation would occur with reduced efficiency in the absence of Nek2A, Mst1/2 or hSav1 because the centrosome linkage is not readily resolved. However, major defects in mitotic progression with reduced Nek2A levels have not been observed (Fig. S6a,b) 31. We therefore reasoned that MT-dependent pushing forces provided by the kinesin motor Eg5 32 might overcome the need for Nek2A in centrosome disjunction. On this basis, impairing these forces by MT depolymerization or Eg5 inhibition 33 should unmask the role of Nek2A, Mst1/2 and hSav1 in centrosome disjunction. To test this hypothesis, cells depleted of Nek2, Mst1/2 or hSav1 were incubated either with nocodazole or monastrol as outlined in the schemes in Fig. 5a,c. Whilst control prometaphase-arrested cells treated with NSC siRNA in the presence of nocodazole showed an average centrosome separation distance of 3.3 μm, this was reduced to 1.6 μm in Nek2, Mst1/2 or hSav1 depleted cells. The latter is comparable to the distance between the two linked centrosomes of S phase cells (Fig. 5b). Therefore, we conclude that in the absence of MTs, hSav1, Mst1/2 and Nek2A become essential for centrosome disjunction.
Figure 5. hSav1, Mst1/2 and Nek2A regulate centrosome disjunction together with Eg5.
a- RPE-1 cells were treated with the indicated siRNA oligos. Cells were then incubated with nocodazole for 16 h to block MT-dependent centrosome splitting. Prometaphase arrested cells were fixed and stained with γ-tubulin antibodies. Scale bar, 5 μm.
b- Distances between the two centrosomes were analysed from the data in Fig. 5a. Results are from three independent experiments. The S phase distance was measured from cells treated with aphidicolin. n>30 cells counted for each condition. Data are mean ± SEM.
c- RPE-1 cells were treated with the indicated siRNA oligos, enriched in G2 by a single thymidine block/release and subsequently treated with monastrol for 4 h to inhibit Eg5-dependent centrosome splitting. Cells were fixed and stained with α- and γ-tubulin antibodies. Scale bar, 5 μm.
d- Distances between the two centrosomes were analysed from the data in Fig. 5c of cells treated with 100 μM monastrol. Results are from three independent experiments. n >30 cells counted for each condition. Data are mean ± SEM.
e- Cells treated with 50 μM monastrol were analysed for their ability to form bipolar spindles. Results are from three independent experiments. n >30 cells counted for each condition. Data are mean ± SEM.
Testing the effect of Eg5 inhibition on centrosome separation further strengthened this conclusion. At high concentrations of monastrol (100 μM), although cells were unable to form a bipolar spindle 33, the two centrosomes were noticeably separated presumably because they had undergone disjunction and so were free to start to diffuse away from each other. In contrast, the intercentrosomal distance was dramatically reduced in the absence of Nek2, Mst1/2 or hSav1 (Fig. 5d) suggesting that the linker between the two centrosomes was still intact. Strikingly, after partial inactivation of Eg5 (50 μM monastrol) cells were able to form bipolar spindles in the presence (Fig. 5c,e; NSC) but not absence of Nek2, Mst1/2 or hSav1 34 (Fig. 5c,e), while with fully active Eg5 Nek2, Mst1/2 or hSav1 depleted cells were capable of forming normal bipolar spindles (Fig. S6a,b). In other words, under conditions of reduced Eg5 activity, centrosome separation and bipolar spindle formation became dependent on hSav1-Mst1/2-Nek2A pathway. Similar results were obtained with another Eg5 inhibitor, VS83 35 which targets Eg5 by a different mechanism than monastrol (Fig. S6c-g) and by combination of Eg5 and Nek2 siRNA depletion (data not shown). This strongly suggests that Eg5-dependent MT pushing forces cooperate jointly with the hSav1-Mst1/2-Nek2A pathway to enable centrosome disjunction and spindle formation.
The C-Nap1/rootletin linker can be disjoined either by hSav1-Mst1/2-Nek2A pathway or by kinesin Eg5
Overexpressed Nek2A causes centrosome splitting by phosphorylation of the centrosomal linker proteins C-Nap1 and rootletin which then become dispersed into the cytoplasm 8, 9. Whether this is also the function of endogenous Nek2A is less clear. Our data suggest that in cells with reduced Eg5 activity, the hSav1-Mst1/2-Nek2A pathway acts as the driving force behind centrosome disjunction (Fig. 5). This enabled us to study the function of, hSav1, Mst1/2 and Nek2A in centrosome disjunction without overexpression of Nek2A and to assess the relative contributions of the hSav1-Mst1/2-Nek2A pathway and the Eg5 motor.
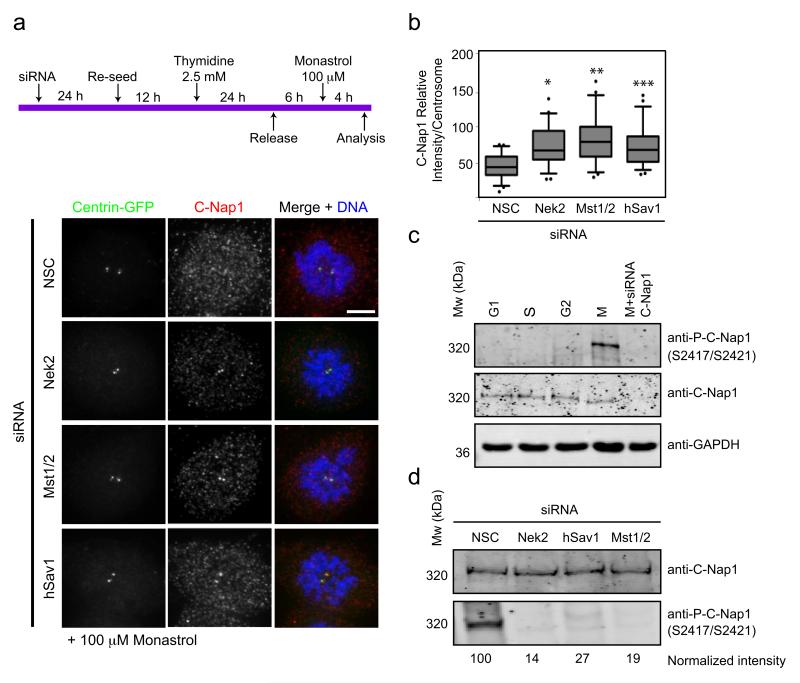
In cells without Eg5 activity the hSav1-Mst1/2-Nek2A pathway disjoins the centrosomes (Fig. 5). In these cells the centrosomal linker C-Nap1 became dispersed from centrosomes in the presence of Nek2, Mst1/2 and hSav1 but not in their absence (Fig. 6a,b). Thus, in the absence of Eg5 activity endogenous Nek2A, assisted by hSav1 and Mst1/2, displaces C-Nap1 from centrosomes.
Figure 6. C-Nap1 phosphorylation and displacement is regulated by hSav1, Mst1/2 and Nek2A.
a- RPE-1 centrin-GFP cells were treated with the indicated siRNA oligos, enriched in G2 by a single thymidine block/release and subsequently treated with 100 μM monastrol for 4 h to inhibit Eg5-dependent centrosome splitting. Cells were fixed and stained with anti-C-Nap1 antibodies. Scale bar, 5 μm.
b- Intensity of centrosomal C-Nap1 of cells from Fig. 6a was measured. The average background intensity was subtracted and the intensities were normalized to corresponding centrin signal. Results are from three independent experiments. n >30 cells counted for each condition. Box-and-whiskers plots: boxes show the upper and lower quartiles (25-75 %) with a line at the median, whiskers extend from the 10 to the 90 percentile and dots correspond to the outliers. (*NSC/Nek2 p=0.0002, **NSC/Mst2 p<0.0001, ***NSC/hSav1 p= 0.0003).
c- HeLa Kyoto cells were synchronized by a double thymidine block and released into nocodazole. Samples were collected after 0, 5, 9 and 14 h of release (indicated as G1, S, G2 and M fractions, respectively; according to DNA distribution in flow cytometry analysis). Whole cell extracts were analysed by immunoblotting using C-Nap1 and PC-Nap1 (S2417/S2421) antibodies. GAPDH was used as a loading control.
d- Extracts from RPE-1 cells treated with the indicated siRNA oligos were arrested by nocodazole. Whole cell extracts were analysed by immunoblotting using anti-C-Nap1 and phospho-specific anti-C-Nap1 antibodies (S2417/S2421). Phospho-C-Nap1 was normalized to the anti-C-Nap1 signal. The result is a representative of 3 independent experiments.
We next asked whether Nek2A-dependent phosphorylation is responsible for the dissociation of C-Nap1 from centrosomes. To address this, we used phospho-specific anti-P-C-Nap1 antibodies that recognize two Nek2 phosphorylation sites (Fig. S5e). In vivo, this antibody only detected C-Nap1 in mitotic extracts, despite the presence of C-Nap1 throughout the cell cycle (Fig. 6c). We depleted Nek2, hSav1 or Mst1/2 by siRNA and arrested cells with nocodazole in mitosis to see whether these proteins are required for C-Nap1 phosphorylation. Depletion of either one of the three proteins decreased the amount of phosphorylated C-Nap1 by ~70-85% in comparison to control nocodazole-arrested cells (Fig. 6d). We conclude that the hSav1-Mst1/2-Nek2A pathway promotes phosphorylation of C-Nap1 at the beginning of mitosis.
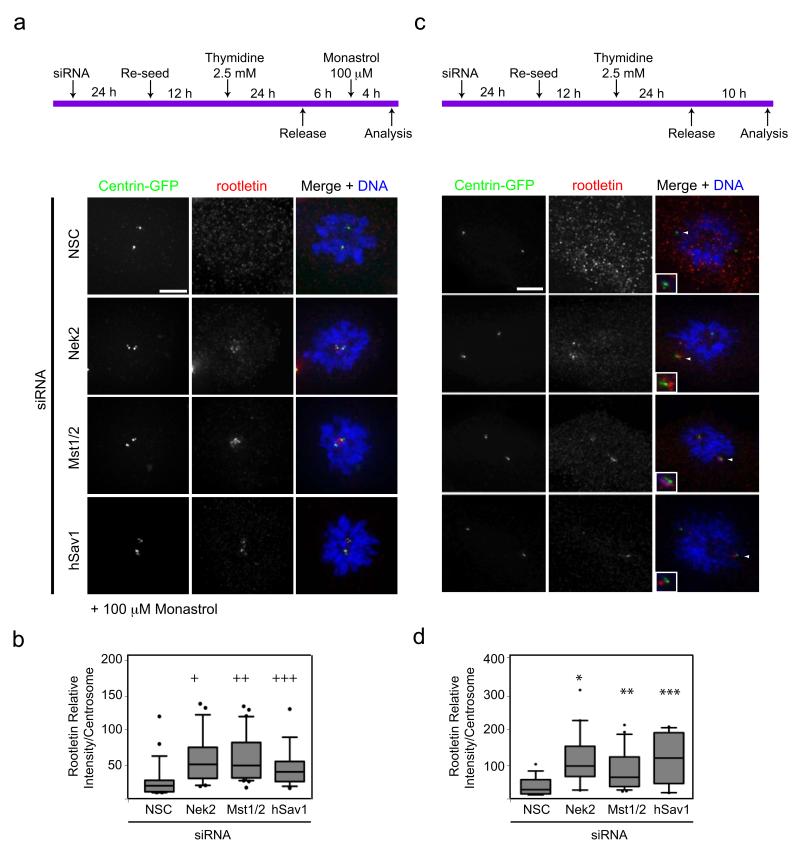
To get further insights into the individual functions of hSav1-Mst1/2-Nek2A module and the motor protein Eg5, we investigated the centrosomal localization of rootletin, a filamentous linker protein which physically connects interphase centrosomes 5, 9. In the presence of monastrol spindle formation failed and the dispersal of the linker protein rootletin became dependent on Nek2, Mst1/2 and hSav1 (Fig. 7a,b), as was observed for C-Nap1 (Fig. 6a,b). Without monastrol centrosomes became separated and spindles formed even when Nek2, Mst1/2 or hSav1 were depleted (Fig. S6a,b). However, in the absence of Nek2, Mst1/2 or hSav1 rootletin remained associated with the separated centrosomes (Fig. 7c,d). This suggests that hSav1, Mst1/2 and Nek2A are required to displace the linker proteins C-Nap1 and rootletin from centrosomes irrespective of whether centrosomes are separated by Eg5 motor forces or not.
Figure 7. Centrosomal localization of the linker protein rootletin is regulated by hSav1, Mst1/2 and Nek2A.
a- The experimental outline using RPE-1 centrin-GFP cells is described in Fig. 6a. Cells were fixed and stained with anti-rootletin antibodies. Scale bar, 5 μm.
b- Intensity of the centrosomal rootletin signal of Fig. 7a was measured and analysed as described in Fig. 6b (+NSC/Nek2 p=0.001, ++NSC/Mst2 p=0.0001, +++NSC/hSav1 p=0.03).
c- The experimental outline using RPE-1 centrin-GFP cells is described on top. Cells were fixed and stained with rootletin antibodies. Scale bar, 5 μm.
d- Intensity of centrosomal rootletin of cells in Fig. 7c was measured and analysed as described in Fig. 6b (*NSC/Nek2 p<0.0001, **NSC/Mst2 p=0.0005, ***NSC/hSav1 p<0.0001).
DISCUSSION
After duplication, the two centrosomes are connected together by a proteinaceous linker that needs to be resolved before mitosis to allow spindle formation and segregation of sister chromatids. The NIMA-related kinase, Nek2, is implicated in centrosome separation and reported to displace linker proteins from centrosomes through phosphorylation at the beginning of mitosis 4, 7. In this study we demonstrate that the ability of endogenous levels of Nek2A to phosphorylate centrosomal linker proteins is regulated by the hSav1-Mst1/2 kinase module and that the Eg5-MT motor pathway functions in parallel to hSav1, Mst1/2 and Nek2A in centrosome separation. This provides the first direct evidence that endogenous Nek2A plays a role in centrosome separation and defines a new function for hSav1 and Mst2, two components of the Hippo pathway that control organ size in flies and mammals 13. Our results indicate that other Hippo pathway components (Rassf1, Lats1/2, YAP) are not involved in the regulation of Nek2A suggesting that hSav1 and Mst1/2 are specifically used for the regulation of centrosome disjunction. In this respect, it is worth noting that the Mob1-Mst1-NDR1 module was recently found to be involved in centrosome duplication 23 suggesting that different modular cassettes of the Hippo pathway might be used to regulate distinct cellular processes.
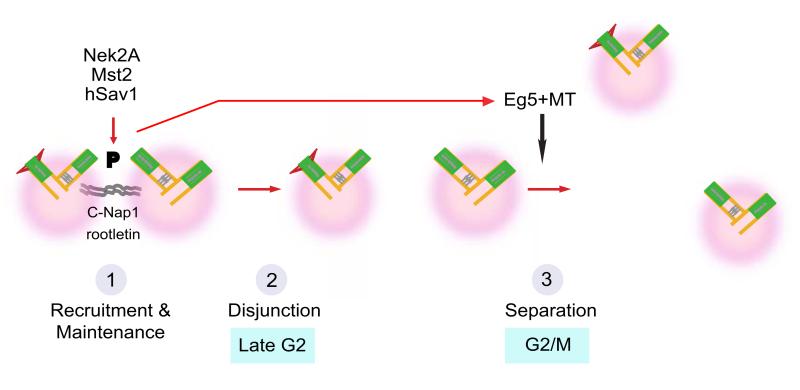
Our data are consistent with a model in which hSav1 and Mst1/2 interact via their SARAH domains with the C-terminus of Nek2A. Phosphorylation by Mst2 results in recruitment of Nek2A to centrosomes, increasing its localized activity (Fig. 8, step 1) without affecting the specific activity of Nek2A kinase. Whereas protein levels of Mst2 are maintained throughout the cell cycle, Mst2 kinase activity is regulated and peaks in G2/M phase (Fig. S7) 36. In addition, Nek2A levels increase in G2 in a similar manner 37. We propose that this cell-cycle dependent regulation of Mst2 and Nek2A controls the timing of centrosome disjunction. In this model, accumulation of Nek2A and activation of Mst2 in G2/M would allow to attain a threshold level of Nek2A activity at centrosomes necessary to displace the linker proteins C-Nap1 and rootletin (Fig. 8, step 2).
Figure 8. Model for centrosome separation in mitotic entry.
Centrosome disjunction in cells with hSav1-Mst1/2-Nek2A and Eg5-MTs. Step 1: Mst1/2 aided by hSav1 phosphorylates Nek2A. This regulates the affinity and dynamics of Nek2A for centrosomes. Step 2: Nek2A phosphorylates the centrosomal linker proteins C-Nap1 and rootletin resulting in linker dissociation. Step 3: The Eg5-MT pathway cooperates with hSav1-Mst1/2-Nek2A in centrosome disjunction and separation. The hSav1-Mst1/2-Nek2A pathway becomes essential for spindle formation when Eg5 activity is reduced.
In this study we present the first evidence that the hSav1-Mst1/2-Nek2A module has overlapping functions in centrosome disjunction/separation with the Eg5-MT motor pathway (Fig. 8, step 3) 32, 33. Similarly, in budding yeast dissolution of the linker connecting the two centrosomes (named spindle pole bodies) is driven by kinesin 5 motor proteins and regulation of the linker protein Sfi1 38, 39. Our data suggest that forces provided by the Eg5 motor protein can break or help to dissolve the linkage between the two centrosomes when Nek2A activity is reduced. Impairment of the Eg5 pathway makes the hSav1-Mst2-Nek2A module essential for centrosome separation and spindle formation. We thus provide an explanation for why knockdown of hSav1 or Nek2A alone do not block centrosome disjunction or cause a significant delay in mitotic progression 31.
Our knowledge of how the centrosome cycle is regulated at the molecular level has profound implications for the understanding of the development of aneuploidy in cancer cells. It is known that defects in the centrosome cycle lead to chromosome instability and the development of cancer 1, 40, 41. The finding that Hippo pathway components regulate centrosome disjunction in parallel to the Eg5-MT pathway means that impairment of this tumour suppressor pathway 42 sets the stage for a catastrophic mitosis as soon as additional, probably only marginal, defects arise.
Materials and methods
Plasmid constructions
Mammalian expression vectors for Nek2A and C-Nap1-CTD were as described 3, 6, 7, 37. Nek2A mutants were generated by site directed mutagenesis using pRcCMV-myc-Nek2A as template 7. cDNAs of hSav1 and Mst2 and their corresponding mutants were subcloned into the mammalian expression vector pCMV-HA (Clontech). For yeast two-hybrid analysis, cDNAs were cloned into pGADT7 and pGBKT7 DNA (Clontech). For bacterial expression cDNA of hSav1 was subcloned into a derivative of pQE vector (Qiagen) carrying a NusA tag. GST-tagged Nek2A-ΔN was cloned into pGEX-5X-1 vector (GE Healthcare). GST-tagged C-Nap1-CTD was described previously 3. cDNAs of wild-type and kinase dead versions of Mst2 were cloned into pFastBac-HT-A vector (Invitrogen) for expression in insect cells. Sequences and details of the plasmids are available upon request.
Antibodies
Following antibodies were used: anti-centrin (this study), anti-hSav1 (Abnova), anti-Mst2 (specific to Mst2) (Novus Biologicals), anti-Mst1/2 (recognizes both Mst kinases) (Santa Cruz), anti-P-Mst1/2-T183/T180 (Cell Signalling), anti-Nek2 (recognizes both isoforms) (Abcam), anti-P-Nek2 (this study) anti-C-Nap1 (BD Transduction Laboratories), anti-P-C-Nap1 (this study), anti-Eg5 (kind gift from T. Mayer), anti-γ-tubulin (Sigma-Aldrich), anti-α-tubulin (clone WA3), anti-myc (9E10), anti-HA (Covance), anti-His (Qiagen), anti-GST (Clone 21, A2), anti-GAPDH (Cell Signaling Technology). Secondary antibodies were goat HRP coupled anti-rabbit or anti-mouse IgGs and rabbit HRP coupled anti-goat IgGs (Jackson Laboratories). Mouse true blot antibodies (eBiosciences) were used for elimination of IgG heavy and light chains. Secondary antibodies for indirect immunofluorescence were donkey anti-rabbit IgGs coupled to Alexa Fluor 488, Alexa Fluor 594 or Alexa Fluor 647, donkey anti-mouse IgGs coupled to Alexa Fluor 555 or Alexa Fluor 488, and donkey anti-goat IgG coupled to Alexa Fluor 555 (Invitrogen).
Centrin antibodies were raised against total protein of human centrin 2 in rabbits and affinity purified. Phospho-specific C-Nap1 and Nek2 antibodies were raised in rabbits and guinea pigs, respectively, against the phosphorylated peptides by Peptide Specialty Laboratories (Heidelberg, Germany). Antibodies were first incubated with the non-phosphorylated peptide followed by purification with the phosphorylated peptide coupled to sepharose beads.
Cell lines and treatments
The RPE-1 centrin-GFP cell line was provided by A. Khodjakov and the HeLa LAP-tagged Nek2A cell line was provided by T. Hyman. The U2OS-eGFP-Nek2A cell line 6 was induced with 1 μg/ml doxycycline for 24 h for eGFP-Nek2A expression. HeLa, MCF-7 and U2OS cells were maintained in DMEM medium, HEK293 and hTERTRPE-1 cells were grown in DMEM F-12 medium supplemented with heat inactivated 10% fetal bovine serum (FBS), 2 mM glutamine at 37°C in a humidified atmosphere with 5% CO2.
For transient transfection of cDNAs and siRNAs, Lipofectamine 2000 reagent was used as described by the manufacturer (Invitrogen). For RNA interference experiments cells were transfected with 100 μM siRNA duplexes (see Table S1).
For double-thymidine block and release experiments, cells were incubated in 2 mM thymidine (Sigma) containing medium for 16–19 h, released in fresh medium without thymidine for 6-8 h. Cells were again treated with 2 mM thymidine and incubated for another 16–19 h. To arrest cells in S phase aphidicolin was used in final concentration of 1.6 μg/ml. For nocodazole block, cells were treated in 100 nM nocodazole for 15-16 h. For monastrol/VS83 experiments, cells were synchronized by a single thymidine block for 24 h. After 6 h of release, 100/50 μM of monastrol or 25/12.5 μM VS83 were added and incubated for 4 h.
Flow cytometry
Cell cycle profiles were determined by measuring cellular DNA content via flow cytometry. Briefly, cells were washed twice with ice-cold PBS and resuspended in 1.5 ml of 70% ice cold-ethanol. Fixed cells were collected by centrifugation, washed twice with PBS, treated with 0.1 mg/ml of RNaseA for 30 min, 37°C and stained with 0.01 mg/ml propidium iodide solution. Profiles were determined with a FACScan II instrument (BD Biosciences) and analyzed using CellQuest.
Immunofluorescence microscopy
For indirect immunofluorescence, cells were fixed with ice-cold methanol for 5 min, permeabilized with 0.1% Triton X-100 for 10 min, blocked with 10% FCS for 30 min and stained with antibodies in 3% BSA-PBS. DNA was stained with Hoechst 33342 (0.2 g/ml, Calbiochem).
Imaging was performed at 25°C on a DeltaVision RT system (Applied Precision) with an Olympus IX71 microscope equipped with FITC, TRITC and Cy5 filters (Chroma Technology), a plan-Apo 100× NA 1.4 and 60× NA 1.4 oil immersion objective (Olympus), a CoolSNAP HQ camera (Photometrics), a temperature controller (Precision Control) and Softworx software (Applied Precision).
Fluorescence intensity and centrosomal distance measurements
ImageJ (http://rsb.info.nih.gov/ij) was used to define an area around the centrosome and two areas near the centrosome (background), and to measure the mean fluorescence intensity. Unsplit centrosomes were measured together, whereas split centrosomes were measured separately. Average of background intensities were subtracted from each measurement in each channel. Intensities of unsplit pairs were divided by 2 to get the average intensity at each centrosome. Signal intensities were normalized against a core centrosomal protein (centrin or γ-tubulin). Images from one data set were acquired at the same day and exposure times were set equal between different samples. To measure distances between centrosomes in prometaphase cells, two poles were identified (according to centrin or γ-tubulin signals) from raw data and distance was determined by ImageJ.
FRAP analysis
For fluorescence recovery after photobleaching (FRAP) analysis, U2OS:eGFP-Nek2A cells were cultured in glass bottom dishes (MatTek) and expression of eGFP-Nek2 was induced with 1 μg/ml doxycycline for 24 h. Imaging was performed in Perkin Elmer spinning disc confocal ERS-FRET on Nikon TE2000 inverted microscope using a 60X oil objective (NA 1.4). Bleaching was done with 10 iterations and 100% laser power (488-nm argon laser). Images were captured before bleaching with a 65 ms interval for 1 second. After bleaching, images were acquired every 65 ms for 20 s, followed by an acquisition of 2 frames per s for 60 s. For each time point, the fluorescence intensity of the photobleached region of interest was determined using ImageJ. Background intensities were subtracted from the signal intensities and the fluorescence intensities were normalized for acquisition bleaching. For recovery and half-life calculations following single exponential formula was used to fit the curves: I(t) = y0+Ae−kt where the y0 represents the plateau of the curve, A indicates the mobile fraction and k is the rate constant.
Yeast-two-hybrid analysis
A yeast two hybrid screen with hSav1 as bait was performed with a Matchmaker pre-transformed HeLa cDNA library (Clontech). In addition, a yeast two hybrid screen with a defined set of mitotic regulators was performed. In both cases, yeast colonies were selected on double or quadruple selective media (-Leu, Trp, or -Leu, Trp, His, Ade).
Protein purification
NusA-hSav1-His, His-C-Nap1-CTD, GST-C-Nap1-CTD and GST-Nek2-ΔN were expressed in BL21 (DE3) Codon Plus and purified by affinity chromatography. His-Mst2-WT and His-Mst2-KD were expressed in Sf21 insect cells and purified by nickel affinity chromatography followed by an ion-exchange chromatography. Nek2A-WT was purchased from New England Biolabs.
In vitro binding assays
Equimolar amounts of recombinant GST or GST-fused proteins were incubated with recombinant His-hSav1-NusA or His-Mst2 (1.0 μg per reaction) in pull-down buffer (50 mM Tris/Cl pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1% NP-40) in a total reaction volume of 200 μl on a rocking platform for 1.5 h at 4°C. Fifty μl of glutathione sepharose 4B bead slurry (GE Healthcare) were then added to each reaction, followed by rocking for an additional 1.5 h at 4°C. Beads were washed four times using pull-down buffer followed by heating in sample buffer. Input and bound proteins were analysed by immunoblotting.
Immunoprecipitation
HEK 293 cells cotransfected with myc-Nek2 or myc-C-Nap1-CTD constructs together with either HA-hSav1 or HA-Mst2 were scraped off the plates and resuspended into BIPA buffer (20 mM Tris/HCl pH 7.5, 120 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5% NP-40, plus Complete EDTA-free protease inhibitor cocktail [Roche]). Cells were lysed by repeated freeze-thaw cycles. Cellular debris were sedimented, the supernatant was incubated with primary antibodies followed by addition of protein A/G sepharose 4B (GE Healthcare). Beads were washed four times with BIPA buffer followed by heating in Laemmli sample buffer. Immunoprecipitations of endogenous proteins were carried out as described above, using asynchronous HeLa LAP-Nek2A cells with near endogenous levels of Nek2AGFP generated using BAC constructs 43. Nek2A-GFP was then immunoprecipitated using GFP-binder-protein 44 coupled NHS-activated sepharose beads. For quantifications of the immunoprecipitated materials, anti-myc signals were measured and corrected for corresponding immunoprecipitated anti-HA signals. Signals were then normalized to wild-type.
Kinase assays
Myc-Nek2A wild-type, Nek2AS356A,S365A,S406A,S438A (Nek2A-4A) and Nek2AS356D,S365D,S406D,S438D (Nek2A-4D) constructs were transiently expressed for 24 h in HEK293 cells. Cells were scrapped off in NEB lysis buffer (50 mM HEPES at pH 7.5, 100 mM NaCl, 10 mM MgCl2, 5 mM MnCl2, 5 mM KCl, 2 mM EDTA, 5 mM EGTA, 1 mM DTT, 0.1% NP-40, protease inhibitor cocktail) and lyzed as described above. Myc-Nek2A constructs were immunoprecipitated using monoclonal anti-myc antibody (clone 9E10) coupled to protein A/G sepharose beads (GE Healthcare). Affinity purified kinases were washed three times in lysis buffer with ATP and twice in kinase buffer without ATP or substrate (50 mM Tris-Cl at pH 7.7, 10 mM MgCl2, 1 mM DTT). Kinases bound to sepharose beads were incubated with 500 ng recombinant C-Nap1-CTD, 1 μCi/nmol γ-32P-ATP with a total ATP concentration of 4 μM for 30 min at 30°C and the reaction was stopped by adding sample buffer and boiling at 95°C. Samples were analysed by SDS-PAGE, Simply Safe Blue staining (Invitrogen) and autoradiography. Kinase assays with recombinant proteins were performed with kinase buffer using 100 ng of Mst2 (either WT or KD), together with 250 ng of substrate (C-Nap1-CTD or Nek2-ΔN). Reactions were incubated for 30 min at 30°C. In order to identify phosphorylation sites, 200 ng Mst2 was incubated with 1 μg of Nek2-ΔN in the presence of 5 mM ATP for 90 min at 30°C. Phospho-sites were identified by LC-MS/MS analysis.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by DFG grant Schi295/3. A.M.F. acknowledges support from the Wellcome Trust, Cancer Research UK and the Associate for International Cancer Research (AICR). We acknowledge Dr. A. Khodjakov for the RPE-1 centrin-GFP cell line, Dr. T.Hyman for LAP-Nek2A cell line and Dr. T. Mayer for Eg5 antibodies and VS83 inhibitor. We are grateful to Drs. O. Gruss, G. Pereira for helpful comments and Dr. A. Khmelinskii for critical reading of the manuscript. We thank Dr. T. Ruppert for the MS/MS analysis and Nikon Imaging Center (Heidelberg, Germany) for FRAP experiments. We also thank Dr. S.R. Scholz for initial help with experiments, E. Eroglu and G. Bozkurt for their help with protein purification, and E. Ercan for help in tissue culture.
REFERENCES
- 1.Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nature Reviews Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 3.Fry AM, et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell. 2006;17:1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell Sci. 2002;115:3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 9.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahmanyar S, et al. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry AM. The Nek2 protein kinase: a novel regulator of centrosome structure. Oncogene. 2002;21:6184–6194. doi: 10.1038/sj.onc.1205711. [DOI] [PubMed] [Google Scholar]
- 12.Hames RS, Fry AM. Alternative splice variants of the human centrosome kinase Nek2 exhibit distinct patterns of expression in mitosis. Biochem J. 2002;361:77–85. doi: 10.1042/0264-6021:3610077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 14.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, et al. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 2008;27:1231–1242. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, et al. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat. Cell Biol. 2004;6:609–617. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- 18.Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr. Biol. 2009;19:675–681. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- 19.Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yabuta N, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J. Biol. Chem. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 21.Guo C, et al. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 22.Toji S, et al. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- 23.Hergovich A, et al. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 24.Valverde P. Cloning, expression, and mapping of hWW45, a novel human WW domain-containing gene. Biochem. Biophys. Res. Commun. 2000;276:990–998. doi: 10.1006/bbrc.2000.3582. [DOI] [PubMed] [Google Scholar]
- 25.Hwang E, et al. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9236–9241. doi: 10.1073/pnas.0610716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 27.Callus BA, Verhagen AM, Vaux DL. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 2006;273:4264–4276. doi: 10.1111/j.1742-4658.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- 28.Fry AM, Schultz SJ, Bartek J, Nigg EA. Substrate-specificity and cell-cycle regulation of the Nek2 protein-kinase, a potential human homolog of the mitotic regulator Nima of Aspergillus nidulans. J. Biol. Chem. 1995;270:12899–12905. doi: 10.1074/jbc.270.21.12899. [DOI] [PubMed] [Google Scholar]
- 29.Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hames RS, et al. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fletcher L, Cerniglia GJ, Nigg EA, Yend TJ, Muschel RJ. Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat. Res. 2004;162:128–135. doi: 10.1667/rr3211. [DOI] [PubMed] [Google Scholar]
- 32.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 33.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 34.Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Sarli V, Huemmer S, Sunder-Plassmann N, Mayer TU, Giannis A. Synthesis and biological evaluation of novel EG5 inhibitors. Chembiochem. 2005;6:2005–2013. doi: 10.1002/cbic.200500168. [DOI] [PubMed] [Google Scholar]
- 36.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes MJ, et al. Early mitotic degradation of Nek2A depends on Cdc20- independent interaction with the APC/C. Nat. Cell Biol. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- 38.Crasta K, Lim HH, Giddings TH, Jr., Winey M, Surana U. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat. Cell Biol. 2008;10:665–675. doi: 10.1038/ncb1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson VE, Prudden J, Prochnik S, Giddings TH, Jr., Hardwick KG. Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function. Mol. Biol. Cell. 2007;18:2047–2056. doi: 10.1091/mbc.E06-10-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 41.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Fry AM, Arnaud L, Nigg EA. Activity of the human centrosomal kinase, Nek2, depends on an unusual leucine zipper dimerization motif. J. Biol. Chem. 1999;274:16304–16310. doi: 10.1074/jbc.274.23.16304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.