Abstract
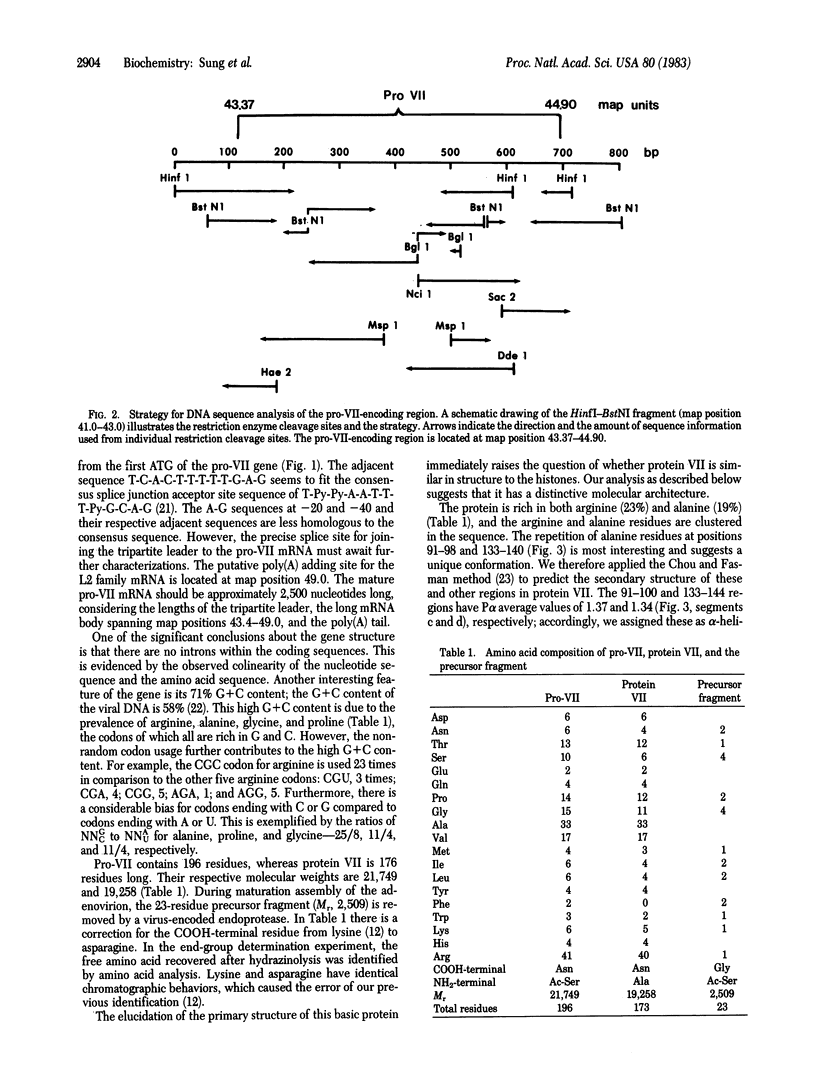
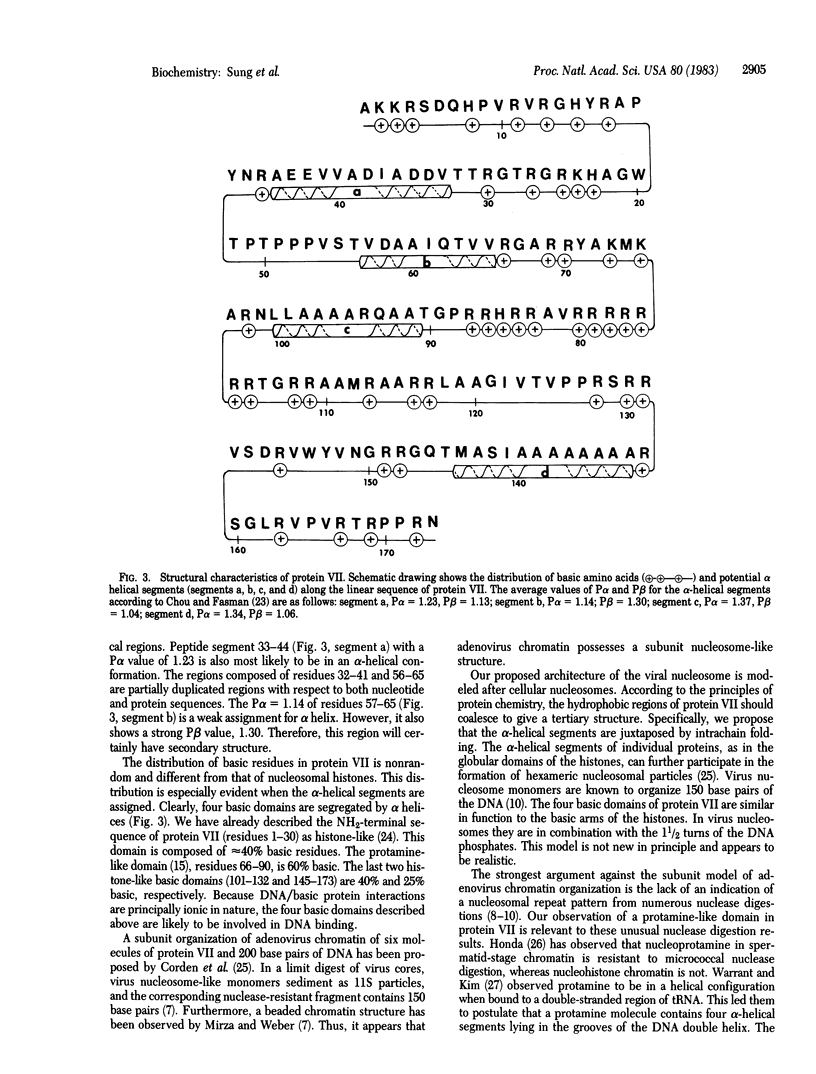
The sequences of both the gene and the corresponding protein of adenovirus major core protein VII have been determined. The precise location of this gene is between 43.37 and 44.90 map coordinates on the viral genome. Protein VII is 173 residues long and has a molecular weight of 19,258. Detailed analysis of its sequence has revealed four basic domains separated by several predicted alpha helices. It is proposed that intrachain folding of protein VII is driven by hydrophobic interactions of the alpha helices, leaving the basic domains of the protein to interact with DNA phosphates. Protein monomers may further associate with each other in the formation of hexameric nucleosome-like particles. The displacement and replacement of protein VII during the viral infectious cycle in the host cell appears to mimic the biology of nucleoprotamine during the processes of spermatogenesis and fertilization. The presence of a protamine-like domain affirms a hybrid histone/protamine molecular structure for protein VII, although it may resemble the protamine in function.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Persson H. Gene and mRNA for precursor polypeptide VI from adenovirus type 2. J Virol. 1981 May;38(2):469–482. doi: 10.1128/jvi.38.2.469-482.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brown M., Weber J. Virion core-like organization of intranuclear adenovirus chromatin late in infection. Virology. 1980 Nov;107(1):306–310. doi: 10.1016/0042-6822(80)90297-4. [DOI] [PubMed] [Google Scholar]
- Cao T. M., Sung M. T. A protamine-like domain in basic adenovirus core protein. Biochem Biophys Res Commun. 1982 Oct 15;108(3):1061–1066. doi: 10.1016/0006-291x(82)92107-6. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Roberts J. M., Lewis J. B., Broker T. R. A map of cytoplasmic RNA transcripts from lytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrids. Cell. 1977 Aug;11(4):819–836. doi: 10.1016/0092-8674(77)90294-x. [DOI] [PubMed] [Google Scholar]
- Corden J., Engelking H. M., Pearson G. D. Chromatin-like organization of the adenovirus chromosome. Proc Natl Acad Sci U S A. 1976 Feb;73(2):401–404. doi: 10.1073/pnas.73.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell E., Groff D. E., Fedor M. J. Adenovirus chromatin structure at different stages of infection. Mol Cell Biol. 1981 Dec;1(12):1094–1105. doi: 10.1128/mcb.1.12.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt E., Sundquist B., Pettersson U., Philipson L. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology. 1973 Mar;52(1):130–147. doi: 10.1016/0042-6822(73)90404-2. [DOI] [PubMed] [Google Scholar]
- Kedinger C., Brison O., Perrin F., Wilhelm J. Structural analysis of viral replicative intermediates isolated from adenovirus type 2-infected HeLa cell nuclei. J Virol. 1978 May;26(2):364–379. doi: 10.1128/jvi.26.2.364-379.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lischwe M. A., Sung M. T. A histone-like protein from adenovirus chromatin. Nature. 1977 Jun 9;267(5611):552–554. doi: 10.1038/267552a0. [DOI] [PubMed] [Google Scholar]
- Lischwe M. A., Sung M. T. Use of N-chlorosuccinimide/urea for the selective cleavage of tryptophanyl peptide bonds in proteins. Cytochrome c. J Biol Chem. 1977 Jul 25;252(14):4976–4980. [PubMed] [Google Scholar]
- Marushige K., Dixon G. H. Developmental changes in chromosomal composition and template activity during spermatogenesis in trout testis. Dev Biol. 1969 Apr;19(4):397–414. doi: 10.1016/0012-1606(69)90050-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Mirza M. A., Weber J. Structure of adenovirus chromatin. Biochim Biophys Acta. 1982 Jan 26;696(1):76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Piña M., Green M. Biochemical studies on adenovirus multiplication. IX. Chemical and base composition analysis of 28 human adenoviruses. Proc Natl Acad Sci U S A. 1965 Aug;54(2):547–551. doi: 10.1073/pnas.54.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant A., Tigges M. A., Raskas H. J. Nucleosome-like structural subunits of intranuclear parental adenovirus type 2 DNA. J Virol. 1979 Mar;29(3):888–898. doi: 10.1128/jvi.29.3.888-898.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M. T., Lischwe M. A., Richards J. C., Hosokawa K. Adenovirus chromatin I. Isolation and characterization of the major core protein VII and precursor Pro-VII. J Biol Chem. 1977 Jul 25;252(14):4981–4987. [PubMed] [Google Scholar]
- Tate V. E., Philipson L. Parental adenovirus DNA accumulates in nucleosome-like structures in infected cells. Nucleic Acids Res. 1979 Jun 25;6(8):2769–2785. doi: 10.1093/nar/6.8.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T. E., Hartford J. B., Serra M., Vandegrift V., Sung M. T. Phosphorylation and dephosphorylation of histone (V (H5): controlled condensation of avian erythrocyte chromatin. Appendix: Phosphorylation and dephosphorylation of histone H5. II. Circular dichroic studies. Biochemistry. 1977 Jan 25;16(2):286–290. doi: 10.1021/bi00621a020. [DOI] [PubMed] [Google Scholar]
- Warrant R. W., Kim S. H. alpha-Helix-double helix interaction shown in the structure of a protamine-transfer RNA complex and a nucleoprotamine model. Nature. 1978 Jan 12;271(5641):130–135. doi: 10.1038/271130a0. [DOI] [PubMed] [Google Scholar]
- Ziff E., Fraser N. Adenovirus type 2 late mRNA's: structural evidence for 3'-coterminal species. J Virol. 1978 Mar;25(3):897–906. doi: 10.1128/jvi.25.3.897-906.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



