Abstract
Objective
Impaired amyloid clearance has been proposed to contribute to β-amyloid deposition in sporadic late-onset Alzheimer's disease (AD). Low density lipoprotein receptor-related protein 1 (LRP-1) is involved in the active outward transport of β-amyloid across the blood–brain barrier (BBB). The C667T polymorphism (rs1799986) of the LRP-1 gene has been inconsistently associated with AD in genetic studies.
We aimed to elucidate the association of this polymorphism with in-vivo brain amyloid load of AD patients using amyloid PET with [11C]PiB.
Materials and methods
72 patients with very mild to moderate AD were examined with amyloid PET and C667T polymorphism was obtained using TaqMan PCR assays. The association of C667T polymorphism with global and regional amyloid load was calculated using linear regression and voxel based analysis, respectively. The effect of the previously identified modulator of amyloid uptake, the apolipoprotein E genotype, on this association was also determined.
Results
The regression analysis between amyloid load and C667T polymorphism was statistically significant (p = 0.046, β = 0.236). In an additional analysis ApoE genotype and gender were identified to explain further variability of amyloid load. Voxel based analysis revealed a significant (p < 0.05) association between C667T polymorphism and amyloid uptake in the temporo-parietal cortex bilaterally. ApoE did not interact significantly with the LRP-1 polymorphism.
Discussion
In conclusion, C667T polymorphism of LRP-1 is moderately but significantly associated with global and regional amyloid deposition in AD. The relationship appears to be independent of the ApoE genotype. This finding is compatible with the hypothesis that impaired amyloid clearance contributes to amyloid deposition in late-onset sporadic AD.
Keywords: Alzheimer's disease (AD), Low density lipoprotein receptor related protein 1 (LRP-1), C667T polymorphism, Apolipoprotein E (ApoE), Pittsburgh compound B ([11C]PiB), Positron emission tomography (PET)
Highlights
-
•
Impaired drainage systems are discussed to be causative for AD.
-
•
LRP-1 transports Aß from the brain.
-
•
LRP-1 polymorphism is associated with amyloid deposition in-vivo in-human.
-
•
Results are compatible with the hypothesis of impaired drainage systems in AD.
1. Introduction
The characteristic histopathological features of Alzheimer's disease (AD) include senile plaques and neurofibrillary tangles in conjunction with loss of neurons and synapses (Braak and Braak, 1991, Thal et al., 2002). The major constituent of senile plaques is amyloid beta peptide (Aβ). The amount and progression of amyloid deposition during the course of AD can be monitored using positron emission tomography (PET) by means of the radiotracer [11C]PiB (Pittsburgh compound B) (Grimmer et al., 2010, Villemagne et al., 2011). It is known that the ε4 allele of the apolipoprotein E (APOE) gene modulates amyloid increase gene-dose dependently (Grimmer et al., 2010). Mutations leading to an overproduction of amyloid are recognized as a major cause of aggregation of the peptide in early onset familial AD (Hardy and Selkoe, 2002). However, the reasons for β-amyloid deposition in late onset sporadic AD are less clear (Duyckaerts et al., 2009). One hypothesis is that an impaired clearance of β-amyloid contributes to cerebral amyloid deposition (Thal, 2009). This notion is supported by the finding that AD patients show identical β-amyloid production rates but decreased β-amyloid clearance rates relative to cognitive healthy controls (Mawuenyega et al., 2010).
From animal studies it is known that molecules such as β-amyloid contained in the interstitial fluid (ISF) are cleared from the brain via different pathways including internalization by neurons or the endovascular unit (Kandimalla et al., 2009), transport across the blood–brain-barrier (BBB) (Shibata et al., 2000) and extracellular degradation by proteases including NEP (Iwata et al., 2000) or IDE (Miners et al., 2008). While ISF of white matter seems to be preferentially drained into the cerebrospinal fluid (CSF) directly, the interstitial fluid of gray matter appears to flow outward via perivascular spaces which are located alongside cerebral arteries and empty into cervical lymph nodes (Carare et al., 2008, Szentistvanyi et al., 1984, Weller et al., 1998, Zhang et al., 1992). The latter drainage pathway could be impaired in late-onset AD. As a consequence, amyloid may be less efficiently cleared from the brain and become deposited in the form of β-amyloid plaques (Grimmer et al., 2012).
Low density lipoprotein receptor-related protein 1 (LRP-1) is a 600 kDa type-I transmembrane glycoprotein, the largest of the low density lipoprotein receptor family (Herz and Strickland, 2001, Van Leuven et al., 1994). It has multiple functions: transportation of cholesterol, recognition of at least 30 structurally diverse ligands, transcytosis of ligands across the BBB, and transmembrane and nuclear signaling (Herz and Strickland, 2001, Jaeger and Pietrzik, 2008). It provides a homeostatic control mechanism of amyloid clearance including cell-surface LRP-1 at the BBB and cerebrovascular cell mediating brain-to-blood amyloid clearance (Jaeger and Pietrzik, 2008). Circulating soluble LRP-1 acts as a peripheral “sink” for plasma amyloid preventing access of free amyloid to the brain, and LRP-1 in the liver mediates systemic amyloid clearance (Deane et al., 2009, Pflanzner et al., 2011, Yamada et al., 2008, Zlokovic et al., 2010).
LRP-1 may play an important role in AD, since in mice inhibition of LRP-1 by the inhibitor RAP (receptor associated protein) or by LRP-1 antibodies leads to substantial reduction in amyloid clearance (Shibata et al., 2000, Williams et al., 1992).
The C667T polymorphism in exon 3 (rs1799986) of the LRP-1 gene on chromosome 12q13–q14, which does not alter the amino acid sequence (Kang et al., 1997), has been inconsistently associated with AD (Beffert et al., 1999, Deng et al., 2006, Forero et al., 2006, Glaser et al., 2004, Hatanaka et al., 2000, Kamboh et al., 1998, Kang et al., 1997, Kang et al., 2000, Kolsch et al., 2003, Pritchard et al., 2005).
Physiologically Aβ is processed in the cell and not deposited in the PVS (perivascular spaces — the space between glia limitans and adventitial pericytes). In AD Aβ/ApoE deposits are found in the PVS and near the basement membrane (Thal, 2009) predominantly around small arteries (Weller et al., 1998) suggesting impaired perivascular drainage. LRP-1 is involved in the amyloid clearance system from the brain into circulation (Deane and Zlokovic, 2007). LRP-1 binds to ApoE-linked Aβ as well as to Aβ alone (Fuentealba et al., 2010, Jaeger and Pietrzik, 2008) and mediates endocytosis of the LRP-1/ApoE/Aβ-complex or LRP-1/Aβ-complex (Deane et al., 2008, Gylys et al., 2003). An association study found a protective effect of the TT LRP-1 polymorphism against AD only in ApoE ε4 carriers (Kamboh et al., 1998).
Since LRP-1 is involved in multiple mechanisms which could have an influence on the pathogenesis of AD and additionally impact Aβ transcytosis, internalization, processing and degradation, we wished to elucidate on how the C667T polymorphism of the LRP-1 gene is associated with cerebral amyloid load in a cohort of 72 AD patients. Because LRP-1 is associated with ApoE ε4 we also examined the association of LRP-1 and ApoE genotype on amyloid load.
2. Materials and methods
2.1. Ethics statement
The study protocol was submitted to the ethics committee of the Faculty of Medicine of the Technische Universität München, which raised no objections, and was approved by radiation protection authorities. All patients gave written informed consent and all clinical investigations have been conducted in accordance with the principles of the Declaration of Helsinki, sixth revision.
2.2. Patient recruitment, inclusion and exclusion criteria
Recruitment and inclusion criteria of the patient sample have been described elsewhere (Grimmer et al., 2010). Briefly, outpatients with very mild to moderate dementia, as rated on the Clinical Dementia Rating scale (CDR; global CDR score of 0.5–2) (Morris et al., 1989) were included. Only patients who fulfilled the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association (NINCDS–ADRDA) diagnostic criteria for Probable Alzheimer's Disease were involved (McKhann et al., 1984). To enhance the likelihood of underlying AD pathology cranial positron emission tomography with 2-deoxy-2-[18F]Fluoro-d-glucose ([18F]FDG-PET) findings typical for AD were also required for inclusion (Hoffman et al., 2000, Jagust et al., 2007), i.e. hypometabolism in the temporo-parietal and posterior cingulate cortex with relative sparing of the primary sensorimotor cortex on visual inspection (Minoshima et al., 1995). Thus, they also met the new National Institutes on Aging and the Alzheimer's Association (NIA–AA) diagnostic criteria for probable AD dementia with evidence of the AD pathophysiological process (McKhann et al., 2011). The study's participants had been referred for the evaluation of cognitive impairment by general practitioners, neurologists, psychiatrists, or other institutions, and had undergone a standardized diagnostic procedure including the Mini-Mental State Examination (MMSE) (Folstein et al., 1975) and ApoE genotyping (Zivelin et al., 1997).
All patients underwent cranial magnetic resonance imaging (MRI) on a 1.5 T scanner to assess structural brain abnormalities. In addition, [11C]PiB-PET was used to assess brain amyloid burden.
2.3. Brain imaging
Structural MRI, [18F]FDG-PET, and [11C]PiB-PET of the brain were obtained using standard procedures (Grimmer et al., 2009a, Grimmer et al., 2009b, Grimmer et al., 2010). The [11C]PiB images were co-registered to high resolution MRI scans and normalized to the MNI space using the warping parameters of the MRI to obtain inter-individually comparable images. Using the standard reference tissue model (Ziolko et al., 2006) a relative measure of global cerebral [11C]PiB uptake was obtained by calculating a cerebral cortex to cerebellar vermis (C/cv) ratio of each patient's [11C]PiB SUV 40–70 min scan to control for between-subject differences in tracer uptake using standard methods (Grimmer et al., 2009a, Grimmer et al., 2009b, Grimmer et al., 2010, Ziolko et al., 2006).
2.4. CAA
To reduce the likelihood of a co-existent cerebral amyloid angiopathy (CAA) only the patients that did not show lacunes on fluid-attenuated inverse recovery (FLAIR) MRI-images or microbleeds on T2* images were included (Goos et al., 2010).
2.5. LRP-1 C667T polymorphism genotyping
The genotypes were determined using TaqMan® polymerase chain reaction (PCR) assays (single nucleotide polymorphism (SNP) assays-on-demand) on a StepOne analyzer with StepOne software v2.1 (all assays, machine, and software were from Applied Biosystems, Carlsbad, CA, USA) according to established methods (Guo et al., 2012). Genotyping was performed in 96 well-plates; the final reaction volume was 20 μl using 20 ng of genomic DNA, 10 μl of TaqMan® master mix and 1 μl of 20 × SNP genotyping assay mix. PCR plates were read after heating at 95 °C for 10 min, followed by forty cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. Duplicate genotypes were determined for 12.5% of the samples (9 of 72) for quality control reasons.
2.6. Statistical analyses
-
(a)
To determine whether the C667T LRP-1 genotype was associated with global amyloid load, a regression analysis was performed using the C/cv[11C]PiB uptake as dependent variable and the C667T LRP-1T allele frequency as independent variable. A significance threshold of 0.05 was applied to this analysis.
-
(b)
To identify possible modifiers of the association between C667T LRP-1T allele frequency and amyloid load, a regression analysis was performed using the C/cv[11C]PiB uptake as dependent variable, the LRP-1T allele frequency as independent variable and stepwise adding of the following independent variables to the model: age, gender, ApoE genotype, and education. Variables resulting in an increased adjusted R2 were included in the final model.
-
(c)
To determine possible regional differences of the strength of the association between C667T LRP-1T allele frequency and amyloid load, a voxel based regression analysis was performed with the [11C]PiB images as dependent variable, LRP-1T allele frequency as independent variable, controlling for all variables identified in the before mentioned regression analysis (b) as modifiers using statistical parametric mapping version 8 (SPM8) (Friston et al., 1994). As a pre-processing method for this analysis, the individual spatially normalized [11C]PiB images were quantitatively normalized to the cerebellar vermis and were smoothed (Gaussian kernel of 10 × 10 × 10 mm) (Grimmer et al., 2009a, Grimmer et al., 2009b, Grimmer et al., 2010). For the voxel based regression analysis a cluster significance threshold of 0.05 corrected for multiple comparisons (family-wise error: FWE) was applied.
-
(d)
To investigate an interaction between ApoE and LRP-1 genotypes, a correlation analysis between frequency of ApoE ε4 alleles and of LRP-1T alleles was calculated. Moreover, the interaction term ApoE ε4 × LRP-1 was included as an additional independent variable in the above mentioned regression analysis (b). Further, two regression analyses were performed accordingly to the before mentioned regression analysis (b) after stratifying patients into ApoE ε4 carrier and non-carrier sub-groups.
-
(e)
Additional analyses — To exclude that the previous associations are caused by a sample bias, i.e. patients with T alleles that are in a more advanced stage of the disease, correlation analyses between frequency of T alleles and clinical severity measured by the CDR SOB as well as age were performed.
3. Results
3.1. Patients
The characteristics of the patient sample including clinical characteristics, C667T allele frequency and C/cv[11C]PiB uptake ratios are shown in Table 1.
Table 1.
Characteristics of the patient sample.
| Demographic and clinical data | |
|---|---|
| Number | 72 |
| Male:Female | 42:30 |
| LRP-1 C667T genotype CC:CT:TT | 57:14:1 |
| ApoE ε 4 alleles: 0:1:2 | 29/28/15 |
| Age at PET: mean ± SD (range) | 69.2 ± 7.86 (50–84) |
| MMSE: mean ± SD (range) | 24.7 ± 3.77 (11–30) |
| CDR SOB: mean ± SD (range) | 3.34 ± 2.126 (0.5–10.0) |
| [11C]PiB uptake ratios (C/cv): mean ± SD (range) | 1.77 ± 0.347 (1.0–2.4) |
ApoE ε4: apolipoprotein E epsilon 4; CDR-SOB: clinical dementia rating sum of boxes; C/cv: cerebral to cerebellar vermis; LRP-1: low density lipoprotein receptor-related protein 1; MMSE: Mini-Mental State Examination; PiB: Pittsburgh compound B; ROI: region of interest; and SD: standard deviation.
3.2. Conducted analysis
-
(a)
The regression analysis using the C/cv[11C]PiB uptake as dependent variable and the LRP-1 T allele frequency as independent variable was statistically significant (p = 0.046). Adjusted R2 was 0.042, and the standardized coefficient β for LRP-1T allele frequency was 0.236. This indicates that the LRP-1T allele is moderately but significantly associated with a higher global amyloid load in AD patients.
-
(b)
In the regression analysis using the C/cv[11C]PiB uptake as dependent variable and the LRP-1T allele frequency as independent variable, the variables ApoE genotype and gender were additionally included. In this model the adjusted R2 was 0.140 with a statistical significance of p = 0.004. The standardized coefficient β for LRP-1T allele frequency was 0.215 (p = 0.057). This indicates that the model controlled for copies of ApoE ε4 and gender explains 14.0% of the variability of the progression of [11C]PiB uptake, and that the frequency of the C667T T allele is positively correlated with the increase of [11C]PiB uptake.
-
(c)
The voxel-based regression analysis with the C/cv[11C]PiB uptake as dependent variable and LRP-1T allele frequency as independent variable, controlling for copies of ApoE ε4 and gender, is depicted in Fig. 1. The associations were predominantly temporo-parietal and in regions nearer to the brain surface. A scatterplot of the association at the local maximum in the right middle temporal gyrus is provided in Fig. 2.
-
(d)
The correlation analysis between the number of ApoE ε4 and of LRP-1T alleles was not statistically significant (p = 0.470). In addition, the coefficient of the variable ApoE ε4 × LRP-1T allele frequency included in the regression analysis (b) was not significant. Both results consistently indicate that there is no substantial association between ApoE and LRP-1 genotypes in this sample. However, in the sub-group analyses in the ApoE ε4 carriers and non-carriers the statistical model was significant in the ApoE ε4 carrier sub-group (p = 0.018; adjusted R2 = 0.141, standardized coefficient β for LRP-1T allele frequency = 0.378, p = 0.013) but not in the ApoE ε4 non-carrier sub-group (p = 0.794). Scatterplots of these two analyses are shown in Fig. 3.
-
(e)
The correlation analyses of the LRP-1 genotype with CDR SOB or with age were not significant either (p = 0.145 and p = 0.318, respectively).
Fig. 1.
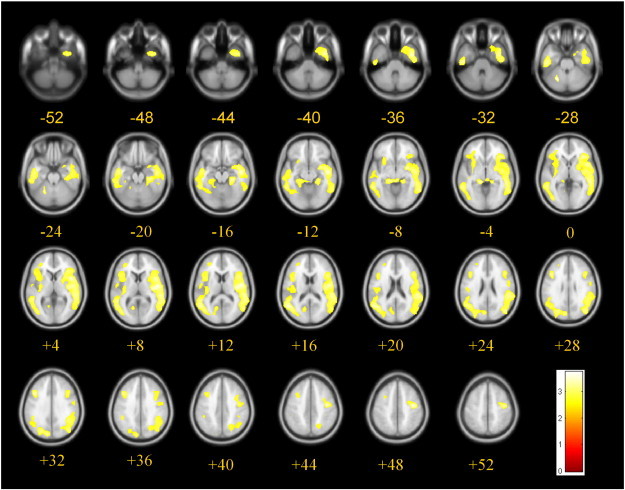
Voxel-based regression analysis between the C/cv[11C]PiB uptake ratio and the frequency of LRP-1T alleles, controlled for copies of the ApoE ε4 allele and sex. Significant (p < 0.05, corrected for multiple comparisons using family wise error) correlations are depicted in yellow and are projected on axial T1-MRI scans (average of 152 scans, implemented in SPM8), numbers indicate z-coordinates of slices in Talairach space in mm. LRP-1 T allele: Low Density Lipoprotein receptor-related protein C667T polymorphism; [11C]PiB: Pittsburgh compound B.
Fig. 2.
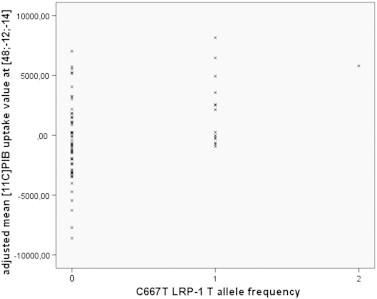
Scatter plot of the voxel-based analysis with mean [11C]PiB uptake as dependent variable and LRP-1 T allele frequency as independent variable, controlling for copies of ApoE ε4 and gender at the local maximum of x = 48, y = −12, z = −14 (coordinates in Talairach space in mm). LRP-1 T allele: Low Density Lipoprotein receptor-related protein C667T polymorphism; [11C]PiB: Pittsburgh compound B.
Fig. 3.
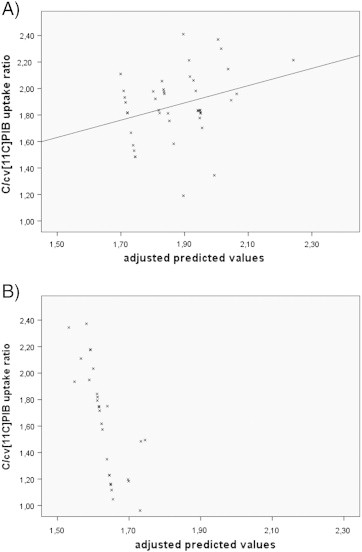
Scatterplots of sub-group regression analysis after stratifying patients into ApoE ε4 carriers and non-carriers using the C/cv[11C]PiB uptake as dependent variable, the LRP-1 T allele frequency as independent variable controlling for gender. A) in ApoE ε4 carriers: significant model (p = 0.018); adjusted R² = 0.141, standardized coefficient ß for LRP-T allele frequency = 0.378 (p = 0.013). B) in ApoE ε4 non-carriers: not significant (p = 0.794). ApoE: apolipoprotein E; LRP-1 T allele: Low Density Lipoprotein receptor-related protein C667T polymorphism; [11C]PiB: Pittsburgh compound B.
4. Discussion
In this study, we found a significant positive association between the numbers of T alleles of LRP-1 C667T polymorphism on chromosome 12 and global amyloid load in a cohort of 72 AD patients. The strength of the association showed regional variability with associations predominantly in temporo-parietal areas and in brain regions nearer to the cortex. In addition, we were able to show that this association is stronger in ApoE ε4 carriers.
In a previous in-vivo study in humans we were able to demonstrate that amyloid clearance processes are important for the development of cerebral amyloid deposits. Specifically, we showed that white matter lesions indicating impaired perivascular drainage pathways are associated with increased rates of amyloid deposition in AD patients (Grimmer et al., 2012). The current study provides further hints that amyloid clearance pathways are relevant for the development of cerebral amyloid load in AD. Thus, it may be interesting to examine pharmacological strategies increasing the transporting capacity of LRP-1 in AD patients. For example Rifampicin alters the brain efflux index of Aβ by inducing LRP-1 (Qosa et al., 2012).
A meta-analysis including 4668 AD patients and 4473 controls was not able to demonstrate a difference in the rates of T alleles between healthy controls and AD patients (Pritchard et al., 2005). This finding could potentially be explained that the control groups of these studies might have been contaminated by subjects suffering from pre-symptomatic AD. However, more likely the LRP-1 polymorphism does not protect against the disease but modifies its course.
The ApoE ε4 gene dose itself which was included as a control variable explained variability of [11C]PiB uptake (p = 0.003) which is consistent with other association studies (Drzezga et al., 2009, Grimmer et al., 2010). ApoE and LRP-1 are both involved in clearance pathways for β-amyloid (Castellano et al., 2011). The ApoE ε4 allele as well as the LRP-1T allele are potential risk factors for AD (Bahia et al., 2008).
What is interesting is that in sub-group analyses the association between LRP-1 and amyloid load was only significant in ApoE ε4 carriers while the interaction term LRP-1 × ApoE ε4 in the regression analysis of the whole sample was not significant. Differences between ApoE ε4 carriers and non-carriers with regard to LRP-1 transport would be consistent with pre-clinical findings (Castellano et al., 2011). Amyloid can be transported by LRP-1 as an ApoE–Aβ complex. The uptake of ApoE–Aβ complexes by astrocytes is mediated by LRP-1. These complexes undergo transcytosis and subsequent perivascular drainage. ApoE ε2 and ApoE ε3 as well as Aβ–ApoE ε2 and Aβ–ApoE ε3 complexes are cleared at the BBB via LRP-1 at a substantially faster rate than the Aβ–ApoE ε4 complex. (Deane et al., 2008). However, a lack of statistical power would be another possible explanation for the non-significant result in the ApoE ε4 non-carrier sub-group.
4.1. Regional variability
The voxel-based analysis of the association between LRP-1 and brain amyloid load revealed that the association occurs predominantly in temporo-parietal brain areas and appears to be somewhat stronger in cerebral regions close to the cortical surface. This observation is consistent with the assumption that deeper brain regions have other ways of drainage i.e. diffusion directly into the CSF (Szentistvanyi et al., 1984). It would also be in line with the finding that the association between amyloid measured in the CSF and cerebral amyloid is strongest in brain regions adjacent to the CSF (Grimmer et al., 2009b).
4.2. Potential limitations
One limitation is the small sample size. p values of the associations between C667T LRP-1T allele frequency and global amyloid load were near the threshold for statistical significance. After the post-hoc analysis excluding the homozygote for LRP-1T allele, β coefficients for LRP-1T allele frequency are no longer significant in analyses (a) and (b) but in the sub-group analysis in ApoE ε4 carriers (p = 0.110, p = 0.093, and p = 0.048, respectively). Therefore, it would be important to replicate our finding in larger cohorts. AD was diagnosed using the standard criteria for probable AD (McKhann et al., 1984), but the clinical diagnosis of AD was not confirmed by post-mortem examination to avoid misclassification of patients as AD. However, patients were only enrolled if they showed AD-typical findings on [18F]FDG-PET. Thus, they also fulfilled the new NIA-AA criteria of probable AD dementia with evidence of the AD pathophysiological process (McKhann et al., 2011). On the other hand, this additional inclusion criterion might have biased patient selection.
5. Conclusions
In conclusion, an LRP-1 polymorphism is significantly associated with amyloid deposition. This is compatible with the view that impaired drainage systems are causative for amyloid deposition and possibly for the development of AD.
Financial disclosures
All authors reported no biomedical financial interests or potential conflicts of interest.
Acknowledgments
This work was supported in part by the German Research Foundation (Deutsche Forschungsgemeinschaft) [HE 4560/1-2 to GH and AD, DR 445/3-1 to AD, GH and AK, DR 445/4-1 to AD], and by a Kommission Klinische Förderung grant for clinical research from the Technische Universität München [to AD and TG].
References
- Bahia V.S., Kok F., Marie S.N., Shinjo S.O., Caramelli P., Nitrini R. Polymorphisms of APOE and LRP genes in Brazilian individuals with Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2008;22:61–65. doi: 10.1097/WAD.0b013e31815a9da7. [DOI] [PubMed] [Google Scholar]
- Beffert U., Arguin C., Poirier J. The polymorphism in exon 3 of the low density lipoprotein receptor-related protein gene is weakly associated with Alzheimer's disease. Neurosci. Lett. 1999;259:29–32. doi: 10.1016/s0304-3940(98)00888-x. (S0304-3940(98)00888-X [pii]) [DOI] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Carare R.O., Bernardes-Silva M., Newman T.A., Page A.M., Nicoll J.A., Perry V.H., Weller R.O. Solutes, but not cells, drain from the brain parenchyma along basement membranes of capillaries and arteries: significance for cerebral amyloid angiopathy and neuroimmunology. Neuropathol. Appl. Neurobiol. 2008;34:131–144. doi: 10.1111/j.1365-2990.2007.00926.x. (NAN926 [pii]) [DOI] [PubMed] [Google Scholar]
- Castellano J.M., Kim J., Stewart F.R., Jiang H., DeMattos R.B., Patterson B.W., Fagan A.M., Morris J.C., Mawuenyega K.G., Cruchaga C., Goate A.M., Bales K.R., Paul S.M., Bateman R.J., Holtzman D.M. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. (3/89/89ra57 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R., Zlokovic B.V. Role of the blood–brain barrier in the pathogenesis of Alzheimer's disease. Curr. Alzheimer Res. 2007;4:191–197. doi: 10.2174/156720507780362245. [DOI] [PubMed] [Google Scholar]
- Deane R., Sagare A., Hamm K., Parisi M., Lane S., Finn M.B., Holtzman D.M., Zlokovic B.V. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J. Clin. Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R., Bell R.D., Sagare A., Zlokovic B.V. Clearance of amyloid-beta peptide across the blood–brain barrier: implication for therapies in Alzheimer's disease. CNS Neurol. Disord. Drug Targets. 2009;8:16–30. doi: 10.2174/187152709787601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Sun Y., Shi J.J., Zhang S.Z. Meta-analysis of the association of the LRP C766T polymorphism with the risk of Alzheimer's disease. Yi Chuan. 2006;28:393–398. (0253-9772(2006) 04-0393-06 [pii]) [PubMed] [Google Scholar]
- Drzezga A., Grimmer T., Henriksen G., Muhlau M., Perneczky R., Miederer I., Praus C., Sorg C., Wohlschlager A., Riemenschneider M., Wester H.J., Foerstl H., Schwaiger M., Kurz A. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology. 2009;72:1487–1494. doi: 10.1212/WNL.0b013e3181a2e8d0. (WNL.0b013e3181a2e8d0 [pii]) [DOI] [PubMed] [Google Scholar]
- Duyckaerts C., Delatour B., Potier M.C. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009;118:5–36. doi: 10.1007/s00401-009-0532-1. [DOI] [PubMed] [Google Scholar]
- Folstein M.F., Folstein S.E., McHugh P.R. Mini-mental state. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. (0022-3956(75)90026-6 [pii]) [DOI] [PubMed] [Google Scholar]
- Forero D.A., Arboleda G., Yunis J.J., Pardo R., Arboleda H. Association study of polymorphisms in LRP1, tau and 5-HTT genes and Alzheimer's disease in a sample of Colombian patients. J. Neural Transm. 2006;113:1253–1262. doi: 10.1007/s00702-005-0388-z. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Tononi G., Reeke G.N., Jr., Sporns O., Edelman G.M. Value-dependent selection in the brain: simulation in a synthetic neural model. Neuroscience. 1994;59:229–243. doi: 10.1016/0306-4522(94)90592-4. (0306-4522(94)90592-4 [pii]) [DOI] [PubMed] [Google Scholar]
- Fuentealba R.A., Liu Q., Zhang J., Kanekiyo T., Hu X., Lee J.M., LaDu M.J., Bu G. Low-density lipoprotein receptor-related protein 1 (LRP1) mediates neuronal Abeta42 uptake and lysosomal trafficking. PLoS One. 2010;5:e11884. doi: 10.1371/journal.pone.0011884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser C., Schulz S., Handschug K., Huse K., Birkenmeier G. Genetic and functional characteristics of the human in vivo LRP1/A2MR receptor suggested as a risk marker for Alzheimer's disease and other complex (degenerative) diseases. Neurosci. Res. 2004;50:85–101. doi: 10.1016/j.neures.2004.06.001. (S016801020400118X [pii]) [DOI] [PubMed] [Google Scholar]
- Goos J.D., Henneman W.J., Sluimer J.D., Vrenken H., Sluimer I.C., Barkhof F., Blankenstein M.A., Scheltens P.H., van der Flier W.M. Incidence of cerebral microbleeds: a longitudinal study in a memory clinic population. Neurology. 2010;74:1954–1960. doi: 10.1212/WNL.0b013e3181e396ea. (74/24/1954 [pii]) [DOI] [PubMed] [Google Scholar]
- Grimmer T., Henriksen G., Wester H.J., Forstl H., Klunk W.E., Mathis C.A., Kurz A., Drzezga A. Clinical severity of Alzheimer's disease is associated with PIB uptake in PET. Neurobiol. Aging. 2009;30:1902–1909. doi: 10.1016/j.neurobiolaging.2008.01.016. (S0197-4580(08)00035-3 [pii]) [DOI] [PubMed] [Google Scholar]
- Grimmer T., Riemenschneider M., Forstl H., Henriksen G., Klunk W.E., Mathis C.A., Shiga T., Wester H.J., Kurz A., Drzezga A. Beta amyloid in Alzheimer's disease: increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid. Biol. Psychiatry. 2009;65:927–934. doi: 10.1016/j.biopsych.2009.01.027. (S0006-3223(09)00110-3 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer T., Tholen S., Yousefi B.H., Alexopoulos P., Forschler A., Forstl H., Henriksen G., Klunk W.E., Mathis C.A., Perneczky R., Sorg C., Kurz A., Drzezga A. Progression of cerebral amyloid load is associated with the apolipoprotein E epsilon4 genotype in Alzheimer's disease. Biol. Psychiatry. 2010;68:879–884. doi: 10.1016/j.biopsych.2010.05.013. (S0006-3223(10)00473-7 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer T., Faust M., Auer F., Alexopoulos P., Forstl H., Henriksen G., Perneczky R., Sorg C., Yousefi B.H., Drzezga A., Kurz A. White matter hyperintensities predict amyloid increase in Alzheimer's disease. Neurobiol. Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.01.016. (S0197-4580(12)00037-1 [pii]) [DOI] [PubMed] [Google Scholar]
- Guo L.H., Westerteicher C., Wang X.H., Kratzer M., Tsolakidou A., Jiang M., Grimmer T., Laws S.M., Alexopoulos P., Bujo H., Kurz A., Perneczky R. SORL1 genetic variants and cerebrospinal fluid biomarkers of Alzheimer's disease. Eur. Arch. Psychiatry Clin. Neurosci. 2012 doi: 10.1007/s00406-012-0295-x. [DOI] [PubMed] [Google Scholar]
- Gylys K.H., Fein J.A., Tan A.M., Cole G.M. Apolipoprotein E enhances uptake of soluble but not aggregated amyloid-beta protein into synaptic terminals. J. Neurochem. 2003;84:1442–1451. doi: 10.1046/j.1471-4159.2003.01643.x. (1643 [pii]) [DOI] [PubMed] [Google Scholar]
- Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. (297/5580/353 [pii]) [DOI] [PubMed] [Google Scholar]
- Hatanaka Y., Kamino K., Fukuo K., Mitsuda N., Nishiwaki-Ueda Y., Sato N., Satoh T., Yamamoto H., Yoneda H., Imagawa M., Miki T., Ohta S., Ogihara T. Low density lipoprotein receptor-related protein gene polymorphisms and risk for late-onset Alzheimer's disease in a Japanese population. Clin. Genet. 2000;58:319–323. doi: 10.1034/j.1399-0004.2000.580410.x. [DOI] [PubMed] [Google Scholar]
- Herz J., Strickland D.K. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J.M., Welsh-Bohmer K.A., Hanson M., Crain B., Hulette C., Earl N., Coleman R.E. FDG PET imaging in patients with pathologically verified dementia. J. Nucl. Med. 2000;41:1920–1928. [PubMed] [Google Scholar]
- Iwata N., Tsubuki S., Takaki Y., Watanabe K., Sekiguchi M., Hosoki E., Kawashima-Morishima M., Lee H.J., Hama E., Sekine-Aizawa Y., Saido T.C. Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat. Med. 2000;6:143–150. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- Jaeger S., Pietrzik C.U. Functional role of lipoprotein receptors in Alzheimer's disease. Curr. Alzheimer Res. 2008;5:15–25. doi: 10.2174/156720508783884675. [DOI] [PubMed] [Google Scholar]
- Jagust W., Reed B., Mungas D., Ellis W., Decarli C. What does fluorodeoxyglucose PET imaging add to a clinical diagnosis of dementia? Neurology. 2007;69:871–877. doi: 10.1212/01.wnl.0000269790.05105.16. (69/9/871 [pii]) [DOI] [PubMed] [Google Scholar]
- Kamboh M.I., Ferrell R.E., DeKosky S.T. Genetic association studies between Alzheimer's disease and two polymorphisms in the low density lipoprotein receptor-related protein gene. Neurosci. Lett. 1998;244:65–68. doi: 10.1016/s0304-3940(98)00141-4. (S0304-3940(98)00141-4 [pii]) [DOI] [PubMed] [Google Scholar]
- Kandimalla K.K., Scott O.G., Fulzele S., Davidson M.W., Poduslo J.F. Mechanism of neuronal versus endothelial cell uptake of Alzheimer's disease amyloid beta protein. PLoS One. 2009;4:e4627. doi: 10.1371/journal.pone.0004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.E., Saitoh T., Chen X., Xia Y., Masliah E., Hansen L.A., Thomas R.G., Thal L.J., Katzman R. Genetic association of the low-density lipoprotein receptor-related protein gene (LRP), an apolipoprotein E receptor, with late-onset Alzheimer's disease. Neurology. 1997;49:56–61. doi: 10.1212/wnl.49.1.56. [DOI] [PubMed] [Google Scholar]
- Kang D.E., Pietrzik C.U., Baum L., Chevallier N., Merriam D.E., Kounnas M.Z., Wagner S.L., Troncoso J.C., Kawas C.H., Katzman R., Koo E.H. Modulation of amyloid beta-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway. J. Clin. Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch H., Ptok U., Mohamed I., Schmitz S., Rao M.L., Maier W., Heun R. Association of the C766T polymorphism of the low-density lipoprotein receptor-related protein gene with Alzheimer's disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2003;121B:128–130. doi: 10.1002/ajmg.b.20043. [DOI] [PubMed] [Google Scholar]
- Mawuenyega K.G., Sigurdson W., Ovod V., Munsell L., Kasten T., Morris J.C., Yarasheski K.E., Bateman R.J. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. (science.1197623 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS–ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging–Alzheimer's Association Workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer’s Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. (S1552-5260(11)00101-4 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miners J.S., Baig S., Palmer J., Palmer L.E., Kehoe P.G., Love S. Abeta-degrading enzymes in Alzheimer's disease. Brain Pathol. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. (BPA132 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima S., Frey K.A., Koeppe R.A., Foster N.L., Kuhl D.E. A diagnostic approach in Alzheimer's disease using three-dimensional stereotactic surface projections of fluorine-18-FDG PET. J. Nucl. Med. 1995;36:1238–1248. [PubMed] [Google Scholar]
- Morris J.C., Heyman A., Mohs R.C., Hughes J.P., van Belle G., Fillenbaum G., Mellits E.D., Clark C. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer's disease. Neurology. 1989;39:1159–1165. doi: 10.1212/wnl.39.9.1159. [DOI] [PubMed] [Google Scholar]
- Pflanzner T., Janko M.C., Andre-Dohmen B., Reuss S., Weggen S., Roebroek A.J., Kuhlmann C.R., Pietrzik C.U. LRP1 mediates bidirectional transcytosis of amyloid-beta across the blood–brain barrier. Neurobiol. Aging. 2011;32(2323):e2321–e. doi: 10.1016/j.neurobiolaging.2010.05.025. (S0197-4580(10)00239-3 [pii]) [DOI] [PubMed] [Google Scholar]
- Pritchard A., Harris J., Pritchard C.W., St Clair D., Lemmon H., Lambert J.C., Chartier-Harlin M.C., Hayes A., Thaker U., Iwatsubo T., Mann D.M., Lendon C. Association study and meta-analysis of low-density lipoprotein receptor related protein in Alzheimer's disease. Neurosci. Lett. 2005;382:221–226. doi: 10.1016/j.neulet.2005.03.016. (S0304-3940(05)00308-3 [pii]) [DOI] [PubMed] [Google Scholar]
- Qosa H., Abuznait A.H., Hill R.A., Kaddoumi A. Enhanced brain amyloid-beta clearance by Rifampicin and caffeine as a possible protective mechanism against Alzheimer's disease. J. Alzheimer’s Dis. 2012 doi: 10.3233/JAD-2012-120319. (Y25020027301T215 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Yamada S., Kumar S.R., Calero M., Bading J., Frangione B., Holtzman D.M., Miller C.A., Strickland D.K., Ghiso J., Zlokovic B.V. Clearance of Alzheimer's amyloid-ss(1–40) peptide from brain by LDL receptor-related protein-1 at the blood–brain barrier. J. Clin. Invest. 2000;106:1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentistvanyi I., Patlak C.S., Ellis R.A., Cserr H.F. Drainage of interstitial fluid from different regions of rat brain. Am. J. Physiol. 1984;246:F835–F844. doi: 10.1152/ajprenal.1984.246.6.F835. [DOI] [PubMed] [Google Scholar]
- Thal D.R. The pre-capillary segment of the blood–brain barrier and its relation to perivascular drainage in Alzheimer's disease and small vessel disease. Scientific World Journal. 2009;9:557–563. doi: 10.1100/tsw.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal D.R., Rub U., Orantes M., Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58:1791–1800. doi: 10.1212/wnl.58.12.1791. [DOI] [PubMed] [Google Scholar]
- Van Leuven F., Stas L., Hilliker C., Lorent K., Umans L., Serneels L., Overbergh L., Torrekens S., Moechars D., De Strooper B. Structure of the gene (LRP1) coding for the human alpha 2-macroglobulin receptor lipoprotein receptor-related protein. Genomics. 1994;24:78–89. doi: 10.1006/geno.1994.1584. (S0888754384715849 [pii]) [DOI] [PubMed] [Google Scholar]
- Villemagne V.L., Pike K.E., Chetelat G., Ellis K.A., Mulligan R.S., Bourgeat P., Ackermann U., Jones G., Szoeke C., Salvado O., Martins R., O'Keefe G., Mathis C.A., Klunk W.E., Ames D., Masters C.L., Rowe C.C. Longitudinal assessment of Abeta and cognition in aging and Alzheimer disease. Ann. Neurol. 2011;69:181–192. doi: 10.1002/ana.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller R.O., Massey A., Newman T.A., Hutchings M., Kuo Y.M., Roher A.E. Cerebral amyloid angiopathy: amyloid beta accumulates in putative interstitial fluid drainage pathways in Alzheimer's disease. Am. J. Pathol. 1998;153:725–733. doi: 10.1016/s0002-9440(10)65616-7. (S0002-9440(10)65616-7 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Ashcom J.D., Argraves W.S., Strickland D.K. A novel mechanism for controlling the activity of alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor-associated protein. J. Biol. Chem. 1992;267:9035–9040. [PubMed] [Google Scholar]
- Yamada K., Hashimoto T., Yabuki C., Nagae Y., Tachikawa M., Strickland D.K., Liu Q., Bu G., Basak J.M., Holtzman D.M., Ohtsuki S., Terasaki T., Iwatsubo T. The low density lipoprotein receptor-related protein 1 mediates uptake of amyloid beta peptides in an in vitro model of the blood–brain barrier cells. J. Biol. Chem. 2008;283:34554–34562. doi: 10.1074/jbc.M801487200. (M801487200 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E.T., Richards H.K., Kida S., Weller R.O. Directional and compartmentalised drainage of interstitial fluid and cerebrospinal fluid from the rat brain. Acta Neuropathol. 1992;83:233–239. doi: 10.1007/BF00296784. [DOI] [PubMed] [Google Scholar]
- Ziolko S.K., Weissfeld L.A., Klunk W.E., Mathis C.A., Hoge J.A., Lopresti B.J., DeKosky S.T., Price J.C. Evaluation of voxel-based methods for the statistical analysis of PIB PET amyloid imaging studies in Alzheimer's disease. Neuroimage. 2006;33:94–102. doi: 10.1016/j.neuroimage.2006.05.063. (S1053-8119(06)00652-5 [pii]) [DOI] [PubMed] [Google Scholar]
- Zivelin A., Rosenberg N., Peretz H., Amit Y., Kornbrot N., Seligsohn U. Improved method for genotyping apolipoprotein E polymorphisms by a PCR-based assay simultaneously utilizing two distinct restriction enzymes. Clin. Chem. 1997;43:1657–1659. [PubMed] [Google Scholar]
- Zlokovic B.V., Deane R., Sagare A.P., Bell R.D., Winkler E.A. Low-density lipoprotein receptor-related protein-1: a serial clearance homeostatic mechanism controlling Alzheimer's amyloid beta-peptide elimination from the brain. J. Neurochem. 2010;115:1077–1089. doi: 10.1111/j.1471-4159.2010.07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


