Abstract
We report a novel chemical sensing array based on metal oxide nanoparticles as a portable and inexpensive paper-based colorimetric method for polyphenol detection and field characterization of antioxidant containing samples. Multiple metal oxide nanoparticles with various polyphenol binding properties were used as active sensing materials to develop the sensor array and establish a database of polyphenol standards that include epigallocatechin gallate, gallic acid, resveratrol, and Trolox among others. Unique charge-transfer complexes are formed between each polyphenol and each metal oxide on the surface of individual sensors in the array, creating distinct optically detectable signals which have been quantified and logged into a reference database for polyphenol identification. The field-portable Pantone/X-Rite© CapSure® color reader was used to create this database and to facilitate rapid colorimetric analysis. The use of multiple metal-oxide sensors allows for cross-validation of results and increases accuracy of analysis. The database has enabled successful identification and quantification of antioxidant constituents within real botanical extractions including green tea. Formation of charge-transfer complexes is also correlated with antioxidant activity exhibiting electron transfer capabilities of each polyphenol. The antioxidant activity of each sample was calculated and validated against the oxygen radical absorbance capacity (ORAC) assay showing good comparability. The results indicate that this method can be successfully used for a more comprehensive analysis of antioxidant containing samples as compared to conventional methods. This technology can greatly simplify investigations into plant phenolics and make possible the on-site determination of antioxidant composition and activity in remote locations.
Keywords: antioxidant, database, portable sensors, metal oxide, field analysis, combinatorial analysis, charge transfer complexes, colorimetric readout
Introduction
Many phenolic antioxidants (AOXs) have been recently found to play unique and specific bioactive roles in health and disease [1-4]. Structural characteristics dictate the physiological role each molecule plays, and thus, each polyphenolic compound has the potential to function uniquely in vivo [5]. As overall health-related applications of individual polyphenols are determined, public and food manufacturer interest in how to assess and increase intake of these potentially beneficial compounds, continues to grow. In order to assess the polyphenolic content of foods, beverages and botanicals, analytical tools capable of identification and quantification of polyphenols must be identified and applied. Additionally, as debate continues, regarding whether the primary mechanism of action for many “antioxidants” is related to their broad anti-oxidative properties, or to their ability to participate in specific metabolic pathways causing vasodilating, anti-inflammatory and antimicrobial effects [6],[7, 8] it is still valuable to determine in vitro antioxidant capacity of antioxidant-containing food and drink. Determination of this value may predict possible physiological activities and also appears to correlate well with the total concentration of phenolics. Thus, a method capable of (1) polyphenol identification and quantification as well as (2) analysis of antioxidant activity is in high demand.
Many botanicals high in antioxidant content have been found in remote locations, e.g., the Amazon rainforest or the jungles of the Niger, where advanced laboratory equipment is often not easily accessible [9]. Thus, for convenience and sample freshness, use of a portable on-site antioxidant analysis device is preferable to methods that require sample preservation and transportation. Herein, we report a fully field-portable nanoparticle-based sensing array for determination of (1) polyphenolic content as well as (2) antioxidant activity. It functions as a reagentless, easy-to-use, inexpensive sensing system that resembles a series of small sensing spots that form characteristic colors when in contact with antioxidants. Color change results from redox and surface chemistry reactions involving immobilized metal-oxide sensing nanoparticles which are capable of accepting electrons donated by antioxidants [10]. The resulting polyphenol radical intermediate binds to the nanoparticle surface forming a charge transfer complex. This complex has unique spectral properties, visible to the naked eye, and capable of branding each sample as uniquely identifiable from all others. The electron donating capacity of each polyphenol toward surface cations on a nanoparticle is used to reveal antioxidant activity of each sample, while the unique color of charge transfer complexes of each compound is used to distinguish and quantify varietal polyphenols. The resulting ability of this sensing array to identify and quantify polyphenolic constituents as well as to determine the antioxidant activity of field samples sets it apart from all other available antioxidant assays, which cannot perform such broad capabilities. Currently, there are no assays which combine antioxidant activity analysis with identification of primary antioxidant components in a sample. Presently used antioxidant activity assays include: the oxygen radical absorbance capacity (ORAC) [11], the ferric reducing antioxidant power (FRAP) [12], and the Copper Reducing Antioxidant Capacity (CUPRAC) [13] assays, among others [14]. Presently used methods for identification and quantification of polyphenolic compounds include: high-performance liquid chromatography (HPLC), coupled with ultraviolet-visible spectroscopy (UV-Vis), mass spectrometry (MS) or nuclear magnetic resonance (NMR) [6] as well as thin layer chromatography (TLC) as a valid but lesser used method [15]. All of these methods are costly, time consuming, non-portable, and require advanced training before use.
The main objectives of this work were to develop a mutiarray sensing system for field analysis of polyphenol-containing samples, and to establish a database of antioxidant standards that could be referred to for rapid sample characterization and screening of antioxidants. We discuss first, the development of this portable combinatorial metal oxide sensing array with respect to sensor fabrication; choice of a field-portable, highly-reproducible color analysis and documentation tool; creation of a database; and the use of each metal-oxide sensing nanoparticle for cross-validation of results. We then present the method by which this database and combinatorial analysis system can be used to characterize and classify polyphenol-containing field samples, on-site, with respect to antioxidant composition and activity. To demonstrate practicality in the field, our method was applied for the successful analysis of one mixture, and one field sample. Antioxidant activity values were validated against the conventional oxygen radical absorbance capacity assay (ORAC) and compositional analysis was validated against previously published results. Our findings, presented here, demonstrate a novel approach for analysis of food antioxidants using a multiarray combinatorial sensor system. Field-detection of antioxidants using this sensing array and portable database could find numerous applications in food industry, botanical and naturopathic medicine.
Experimental
Reagents and Equipment
Cerium (IV) oxide nanoparticles, or ceria (CeO2), 20 wt. % colloidal dispersion in 2.5% acetic acid (289744), was purchased from Sigma Aldrich. The average particle size of the 10-20 nm ceria nanoparticles was verified by scanning electron microscopy (SEM) and particle size distribution (PSD). Ammonium titanyl oxalate monohydrate (229989), titanium dioxide, rutile (<100nm), silicon dioxide (10-20 nm), sodium acetate and acetic acid were purchased from Sigma Aldrich. Zinc oxide nanoparticles (10-30 nm) were purchased from SkySpring Nanomaterials Inc. Iron oxide (20-40 nm) and zirconium dioxide (40 nm) was purchased from US Research Nanomaterials, Inc. Filter paper (P5; medium porosity; slow flow rate) was purchased from Fisher Scientific and used as received. Fluorescein sodium salt, and (2,2’-azobis(2-amidino-propane) dihydrochloride (AAPH) were from Fisher Scientific. The antioxidants 6-Hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox), capsaicin (CP) and epigallocatechin gallate (EGCG) were from Sigma Aldrich; L-ascorbic acid (AA), gallic acid (GA), genistein (G), rutin (R), and curcumin (CC) were from Acros; vanillic acid (VA) and quercetin (Q) were from Alfa Aesar; caffeic acid (CA) was from Spectrum Chemical; resveratrol (RV) and ellagic acid (EA) were from TCI America; keracyanin chloride (KC) was from Fluka; and rosmarinic acid (RA)was from Enzo. Antioxidants used at Bastyr University for ORAC analysis include Trolox, EGCG, GA, AA, and CC, and were all purchased from Sigma Aldrich. Gunpowder green tea, Camellia Sinensis was attained from the Potsdam, NY Food Co-op.
The CapSure® handheld color analysis device manufactured by Pantone/X-Rite © was used to analyze color response from all metal oxide sensor types. A fluorescence 96-well plate reader (Gemini EM fluorescence plate reader by Molecular Devices) was used to perform the ORAC assay for validation and inter-assay comparison purposes. Graphpad Prism 5 software was used to normalize data and analyze area under the curve (AUC) values for the ORAC assay. All ORAC assays were performed in the Tierney Research Laboratory at Bastyr University; metal oxide sensor calibrations and real sample analysis took place at Clarkson University.
Fabrication of the Metal Oxide Sensors
Paper sensors were prepared following a procedure similar to that previously described for immobilization of CeO2 nanoparticles onto cellulose[10, 16]. In brief, 11 cm diameter filter paper rounds were dipped into baths of the various metal oxide dispersions (4% TiO2; 2% SiO2; 2% ZnO in 2.5% HAc; 2% ZrO2 in 2.5% HAc, 0.1% Fe2O3 in 2.5% HAc, and 4% CeO2 in 2.5% HAc) and dried in the oven at 100C for 5 minutes, then allowed to dry completely at room temperature. Individual sensors were then cut in circular paper disks of 9/32” diameter. Titanyl oxalate (Ti(IV)oxo) sensors were prepared following procedures similar to those used by Xu[17]. 20 uL of 1M titanyl oxalate salt solution was deposited onto the surface of individual circular paper sensors (9/32” diameter) then allowed to dry completely. The solution was applied directly to pre-cut sensor disks rather than applied to the large surface area paper before cutting to avoid disturbance and removal of resulting titanyl oxalate crystals. Before sample analysis, all sensor types (except ceria) were pre-treated with 10 uL of 1 M NaOH, to activate −OH functionalities and increase sensitivity of polyphenol detection as described in Figure S1 (supplemental). Sensors were allowed to dry completely before use. Gallic acid was used as the standard for assessing reactivity of these metal oxides towards polyphenolic compounds.
Colorimetric Measurements
All color responses were read with the Pantone© CapSure® handheld color analysis device. The color reading window was placed directly over the sample to block external light interferences. Upon sample reading, an immediate output of red, green and blue (RGB) color intensities is displayed on screen, along with a Pantone© ID code. These antioxidant- and concentration- specific color intensities and Pantone© IDs were documented for future reference and identification of samples. The RGB and Pantone© ID values are also automatically stored in the CapSure® device, which can hold up to 100 readings at a time. A standard calibration curve was created for each polyphenol, displaying the linear relationship of blue color intensity (BCI) to concentration of analyte. The blue color channel is used as opposed to green or red, because it provides the most sensitive readings allowing detection of low concentrations of analyte, also responding with the greatest slope. Calibration curves created from the CapSure® system are intended for use in quantitative analysis of antioxidant samples. Pantone© IDs are intended for use in matching unknown samples to a polyphenolic standard within the database, allowing simultaneous determination of both sample identity and concentration.
Sample Analysis and Quantitation
Analysis of all aqueous samples was performed by adding 20 uL of sample to each paper sensor disk and allowing 90 minutes for complete drying. Analysis of samples suspended in organic solvents, such as ethanol or acetone, was performed by applying 10 uL of solution to the paper and allowing 2 minutes for complete drying. After drying, sensors were placed onto the back of a black sheet of contact paper, with pre-cut holes (¼” diameter) for sample framing and easy documentation within a notebook. Color responses of each sensor were then read using the CapSure®. All polyphenolic compounds were tested with four metal oxide sensor types (cerium oxide, titanyl oxalate, iron oxide, and zinc oxide). Gallic acid, the antioxidant activity standard, was analyzed using all seven metal oxide sensor types for comparison. 10-20 concentrations ranging from 0.01 – 250 mM were used to create standard calibration curves. Samples were analyzed in triplicate to determine the reliability of estimates.
Standard reference calibrations with matching Pantone© IDs were created for all samples in the database. Two methods have been used to determine concentration: Pantone© ID matching and interpolation of BCI into a calibration curve. The method of use depends on whether the sample is of unknown or known identity. The first method of ID matching can characterize samples of unknown polyphenol identity by assigning a Pantone© ID code to a sample's color response on a variety of sensor types, and matching those codes within their respective databases to determine the possible polyphenolic identity and concentration. The second method is used for quantification of known samples only. This method involves interpolation of the BCI of sensor response into the BCI vs. log (mM) calibration curve for a given polyphenol. The equation y=mx + b where y=BCI and x=log (mM sample) is used for this calculation.
To demonstrate the capacities of the metal oxide sensor array to determine the primary polyphenolic component of real samples and mixtures, one mixture of known composition (7mM GA, 3mM EA), as well as a real sample (gunpowder green tea) were analyzed using four sensor types (ceria, titanyl oxalate, zinc oxide and iron oxide) and their respective databases. Analysis of these mixtures was performed using each database to determine color ID matches between polyphenol standards and various concentrations of the mixture or real sample. A calibration curve of each mixture was created and Pantone© IDs were assigned in triplicate to each concentration. IDs were matched within each respective polyphenol database to determine the composition of the mixture. Polyphenol identities and concentrations assigned to the original solution by each sensor type were calculated and compared by relating the concentrations at which the mixture and a polyphenol standard produced identical color IDs. The equation used is: (mM polyphenol) / (% v/v mixture) X 100 = mM of responding polyphenol in the original mixture. Through comparison of results determined by the four metal oxide databases, select signals were eliminated. Only polyphenol identities which appeared as a match using all sensor types were considered further to identify the active polyphenolic component in the sample. This systematic matching using four metal oxide assays allows for semi-quantitative determination of the active polyphenol in the real sample matrix.
Antioxidant activity of each sample was also analyzed in terms of gallic acid equivalents (mM GAE) using established methods.1 In brief, for antioxidant activity calculation, 10-20 serial dilutions of the sample in water (3:1) were prepared and applied to metal oxide sensors. Color intensities were read using the CapSure®. BCIs were recorded and graphed vs. the log (concentration, %). The slope of the resulting linear regression was compared to that of GA as a standard. Gallic acid equivalence was determined using the equation: (slopesample/slopeGA) = mM GAE.
Validation & Inter-assay Comparison
For validation purposes, the ORAC assay was carried out on a set of known polyphenol standards (CC, GA, Trolox, EGCG, and AA) as well as one complex botanical sample, gunpowder green tea. These results, in terms of GAE, were compared to results from the CeO2, ZnO, Fe2O3 and Ti(IV)oxo sensors. The assay was carried out following the procedures from ZenBio Laboratories and Henning.[18] Briefly, experiments were performed in a 96 well plate: 75 μL of 1.9 μM fluorescein in a 75 mM sodium acetate buffer (pH 5.44) was added to all wells, followed by 50 μL of samples: antioxidants (0.078-50 μM), tea (0.08 g/L diluted from 10 g/L (2 g tea in brewed for 5 minutes in 200 mL of 80°C water), Trolox control (50 uM) or buffer as a blank. The plate was incubated at 37°C for 10 minutes before addition of 75 μL of 240 mM AAPH oxidant. Fluorescence readings (excitation 485 nm, emission 538 nm) were taken every minute for 1.5 hours in order to monitor the rate of oxidation of fluorescein by AAPH-generated peroxyl radicals. Antioxidant activity is thus proportional to a decrease in fluorescein oxidation (or fluorescence quenching) rate due to the peroxyl radical scavenging ability of the sample. Decrease in oxidation rate is monitored by an increase in area under the fluorescence (RFU vs. time) curve.
ORAC data analysis was performed by first normalizing the data relative to the starting point, and then utilizing Graphpad for automated area under the curve (AUC: fluorescence vs. time) calculations. Net AUC was calculated for each sample using the equation: AUCsample - AUCblank = AUCnet. For analysis of antioxidant standards (GA, AA, Trolox, CC, EGCG), a calibration curve of AUCnet vs. concentration (mM) was created for the dilution series tested. The slope of this curve (AUCnet /mM) was compared to that of Trolox using the equation (slopesample/ slopeTrolox) to determine ORAC value in terms of mM Trolox with equivalent activity to 1 mM sample (or Trolox equivalents: mM TE). For assays which utilized internal standards (gunpowder green tea), ORAC value is calculated by comparison of net AUC of sample to that of Trolox, using the equation: (net AUCsample/ net AUCTrolox)*(0.01mM Trolox/ 0.02g/L sample) = (mM TE/g tea). This was used to determine Trolox antioxidant activity equivalence with consideration of in-well concentrations of both the standard and sample.
High-throughput Analysis using the Metal Oxide Sensor Array
The ability to discern between Pantone© ID matches and discriminate overlapping signals is demonstrated through the use of multiple metal oxide sensors. These sensors were selected from a variety of metal oxide indicators found to be colorimetrically responsive to polyphenols. CeO2, Ti(IV)oxo, ZnO and Fe2O3 sensors were selected as examples from this group of nanoparticle indicators to use in tandem for accurate and cross-validated results. Multiple antioxidant identity matches found in each database were pared down to one final match for an unknown sample, by comparing results found using four sensor types. The recurrence of one ID match in four databases confirms sample identity and eliminates false positive readings. In this way, cross-validation of results through systematic use of multiple sensor types leads to confident sample identification. The use of an array of complementary sensors for combinatorial sample analysis therefore increases accuracy and representativeness of the results.
Results and Discussion
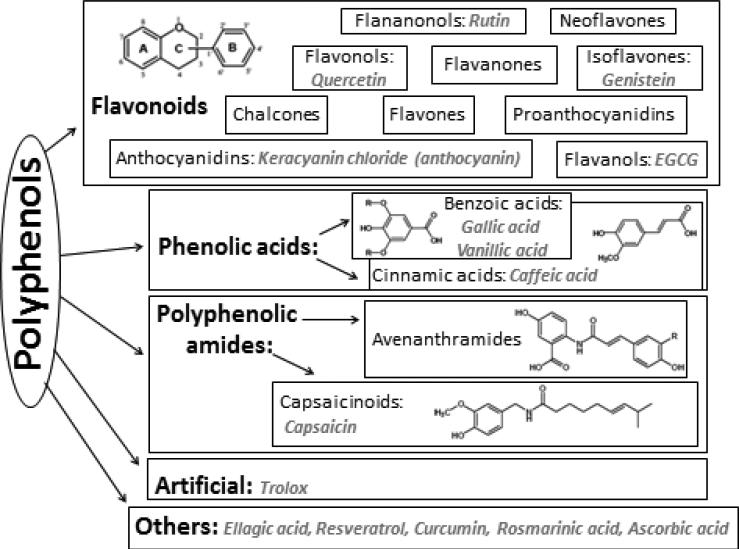
Figure 1 provides an overview of the various classes of polyphenols selected for this study. The antioxidants investigated here represent each of the main families of polyphenolic compounds, as described by Tsao et al., 2010.[19] KC was chosen to represent the anthocyanin family; and EGCG, the flavanols. Both KC and EGCG are monomeric units of procyanidins. Q, represents the flavonols; R, the flavanonols; and G, the isoflavones; all of which are flavonoids. RV, RA, CC, and EA are non-flavanoid polyphenols. GA and VA are used to represent the benzoic acids; and CA, the cinnamic acids. These three are all classified as phenolic acids. CP represents the capsaicinoids, also classified as polyphenolic amides. AA, a non-phenolic antioxidant used to protect polyphenols and the artificial antioxidant analogue of vitamin E (α-tocopherol), Trolox, were also included as reference materials in the database.
Figure 1.
Classification of polyphenolic compounds used to establish the antioxidant database as determined by structural properties. The 15 compounds studied herein are in italics, under their respective categories. At least one compound from each broad category of PPs was studied to provide standards to represent a wide variety of botanical samples which could be analyzed in the future using this sensing array.
Adaptation of the Combinatorial Sensing Array for Field Use using a Portable Color Analysis Tool
A portable color analysis tool, the CapSure®, manufactured by Pantone LLC of X-Rite Inc.© was chosen to adapt the metal oxide sensing array for field use and to facilitate rapid color quantification. The technology uses internal LED lights while blocking external light to illuminate the sample, allowing for sensitive detection of color change on the paper surface with enhanced accuracy. The device gives an immediate digital output of red, green, and blue (RGB) color intensities of the sensor using an 8-bit per channel scale of 0-255.The CapSure® matches each color reading to an internally catalogued color ID from the Pantone/X-Rite© color catalogue. Pantone© can print accurate reproductions of each color code for visual sample matching, adding a tangible dimension to color analysis. Pantone© color IDs can be used to directly match sample colors within a previously recorded database of colors for given polyphenols. In this way, identity and concentration of unknown samples can be immediately determined by matching color ID codes within the database. The CapSure® can store up to 100 results at a time, allowing storage and analysis of large data sets. The device calibrates internally after each hour of use, allowing for excellent inter-user reproducibility. Therefore, it is possible for all standard reference calibrations and color values to be cumulatively shared amongst users. The ease-of-use, speed of analysis, light-blocking and automated features are clear advantages, making the CapSure® highly suitable for field use, as compared to non-portable office scanners or cameras which allow for light interferences. The unique colors produced by the sensors can be logged as reference standards to which sample responses can be compared for immediate identification and quantification. The database facilitates access and upload of reference materials from any location with possible online file sharing and interactive capabilities[20, 21].
Operational Principle of the Metal Oxide Sensor Array
The antioxidant detection mechanism using immobilized metal oxides on paper is based on the ability of polyphenol antioxidants to form surface complexes of characteristic colors with metal oxides. Previously, we have demonstrated the ability of CeO2 nanoparticles to form charge transfer complexes with polyphenols with enhanced spectral and optical properties, in which CeO2 nanoparticles act as color indicators [10]. We establish herein, that the rich OH functionalities of other types of metal oxides also promote formation of metal oxide-AOX complexes with unique spectral properties and characteristic colors. Such color responses are used in this work for analytical quantification and cross-validation. The binding and the corresponding color change is dependent on the concentration of the antioxidant and it is specific to the type of antioxidant, as well as the type of metal oxide. To fabricate the array of complementary sensors, metal oxides of Ti(IV)oxo and ZnO, SiO2, ZrO2 , TiO2, Fe2O3 and CeO2 nanoparticles were stabilized on paper to create individual sensors that can be assembled in an array-type design for cross-validation of sample identity. The resulting metal oxide array provides a sensing platform with multiple colorimetric readouts, as each sensor in the array generates a unique representation of the antioxidant identity and composition. Moreover, the responses of the individual sensors are used to cross-validate sensor data and distinguish overlapping signals. The resulting metal oxide array is reagentless, inexpensive and easy-to-use.
Fabrication and characterization of the High-Throughput Metal Oxide Sensor Array using Multiple Metal Oxide Indicators
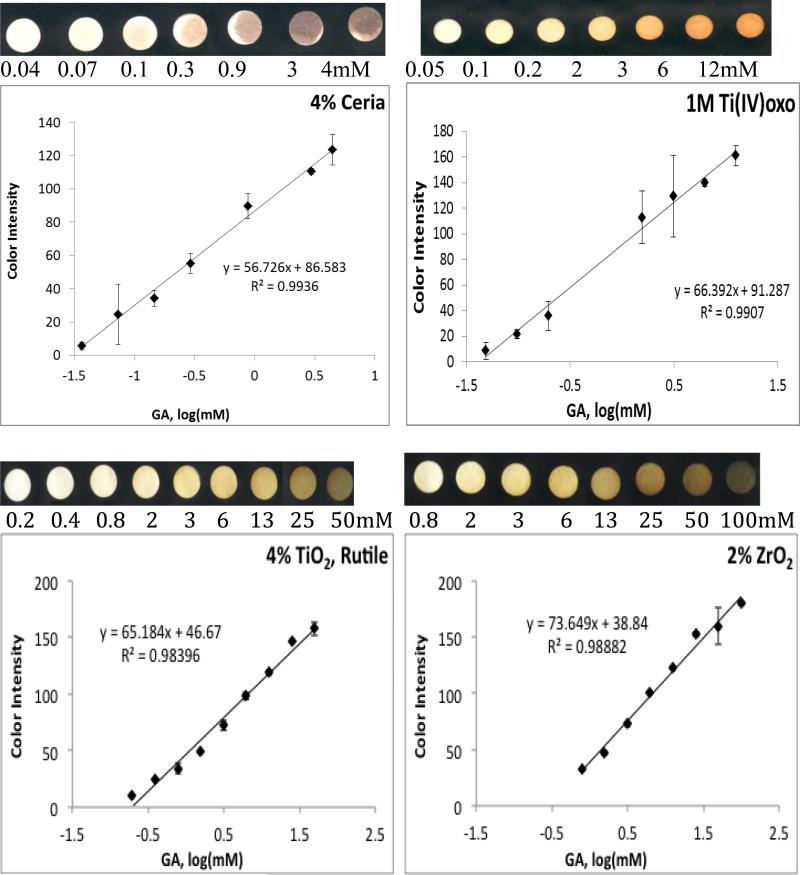
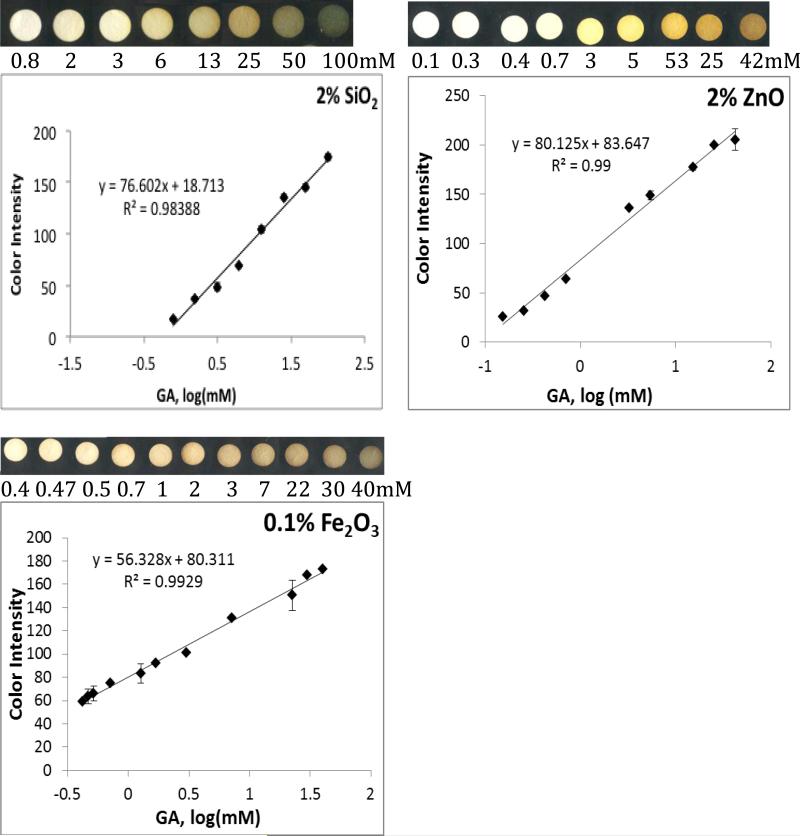
The metal oxides used to fabricate the array have activated OH surface functionalities and can attach OH rich polyphenols by forming surface stabilized complexes with enhanced charge transfer properties. Interestingly, each of the metal oxides provides a distinct color when functionalized on paper and introduced to antioxidants; therefore each antioxidant has a unique signature and characteristic ID depending on the metal oxide indicator used. All metal oxide sensors demonstrate linear calibration curves with good reproducibility for the antioxidants tested. Figure 2 shows colorimetric responses and calibration curves of each of the metal oxide sensor to GA. While CeO2 and Fe2O3 form a brown color on the paper in the presence of antioxidants, Ti(IV)oxo forms a bright orange complex, TiO2, ZrO2 and SiO2 a yellow-green and ZnO a bright yellow color. These results demonstrate that each of these metal oxides can provide an optical signature of the antioxidant activity, identity and concentration. The unique color scheme produced by each sensor type makes it possible to screen, using each metal oxide sensor, a complete database of polyphenolic compounds at various concentrations and subsequently document and reference discernibly unique Pantone color IDs for each sample. This screening can produce multiple databases, one for each metal oxide, multiplying the number of reference points which further enhances the confidence interval, increasing precision and reliability of the method.
Figure 2.
Calibration curves of blue color intensity (BCI) of various metal oxide sensors as a function of antioxidant (GA) concentration. Metal oxide sensors were made from rutile TiO2, SiO2, ZnO, and ZrO2, ceria, Fe2O3 and Ti(IV)oxo.
Essentially, each metal oxide nanoparticle indicator generates a new antioxidant assay. The side-by-side use of various metal oxide assays for sample quantitation allows validation of results. Such a practice is analogous to other analytical assays used to validate quantification of other analytes (e.g. total phenolic content as determined by the Folin-Ciocalteu (FC)[22] vs. Prussian Blue assays[23]; or total protein content as determined by the Biuret[24] vs. Bradford methods[25]). The slopes of color intensity vs. concentration calibration curves for GA detection on each metal oxide sensor type are compared in Figure 2. Based on the magnitude of slope, the most sensitive metal oxide indicator is ZnO followed by SiO2, ZrO2, Ti(IV)oxo, TiO2, CeO2, and Fe2O3 in order of decreasing slope. Greater slope values allow for greater distinction between small variations in the concentration of samples, and thus are described as having greater sensitivity. While slope sensitivity of ZnO may be the greatest, the lowest quantifiable concentration (LOQ) is provided by the ceria sensor, with an LOQ of 0.04 mM GA. The CeO2 sensor is followed by Ti(IV)oxo, TiO2, ZnO, Fe2O3, SiO2, and ZrO2 sensors in order of increasing LOQ (0.05, 0.2, 0.4, 0.4, 0.8, 0.8mM GA, respectively). Depending on the goal of each investigator the analytical parameters in Figure 2 can be used to select proper sensor types.
To achieve sensitive colorimetric responses to polyphenol antioxidants on ZnO, SiO2, ZrO2, Ti(IV)oxo, Fe2O3, and TiO2, these oxides were first pre-treated with a strong base, NaOH, which activates the OH functionalities by forming Me(OH) 2- 4and high surface area clusters of MexOy(OH)zz-2y-2x with enhanced OH functionalities after dehydration. The high density of OH functional groups enhances binding of the polyphenols increasing sensitivity. The CeO2 sensors, however, were found to function optimally between pH 2-12, while showing decreased activity at extreme pHs, such as those created on the other sensor types following pretreatment with 1M NaOH. We note that among the metal oxides tested herein, CeO2 is the only metal oxide possessing redox active properties[26] and this redox reactivity is implicated in reaction with polyphenols by first forming a quinoid-type compound before binding to the particle surface. Due to its redox reactivity and dual oxidation state, extreme changes in the pH, and therefore the ratio between Ce(III) and Ce(IV) was found to affect the ability of the CeO2 nanoparticles to form complexes with polyphenols at extreme pH values (See Figure S1, Supplemental Information).
Polyphenol Database: CapSure® Calibrations as Reference Standards
The fifteen compounds shown in Figure 1 were used as reference standards to develop the antioxidant database. These compounds are representative of different classes of polyphenol antioxidants, determined based on structural characteristics and largely defined by ring structures and type as well as number of functional groups. Representation of each polyphenol class within the database provides breadth of scope for antioxidant standards and creates the possibility that the majority of samples analyzed could match a reference standard. The response of each metal oxide sensor to reference compounds was first determined and used to construct calibration curves. The antioxidant activity in terms of GAE for each polyphenol was obtained by comparing the slopes of their calibration curves to that of a GA standard. Pantone © IDs corresponding to each polyphenol response on four sensor types were established and stored within a database of Pantone © IDs for each polyphenol on each sensor type. The corresponding ID codes can be found in the Supplementary Information. The IDs can be stored into the electronic reader for immediate automated Pantone © ID matching to facilitate rapid sample characterization. Analytical parameters of the polyphenols studied using the four metal oxide sensor types (i.e. CeO2, Ti(IV)oxo, ZnO and Fe2O3) are displayed in Table S1-S4 (supplemental) and include: antioxidant activity, equation of the calibration curve, linear range, and correlation coefficients (R2).
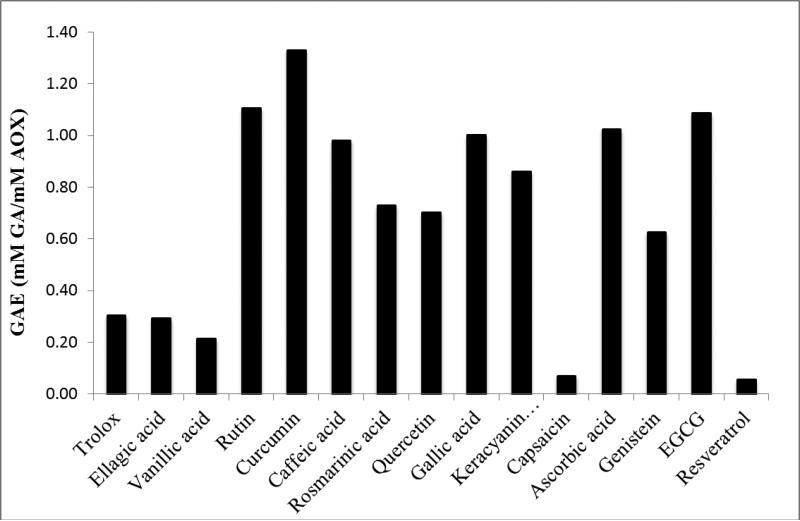
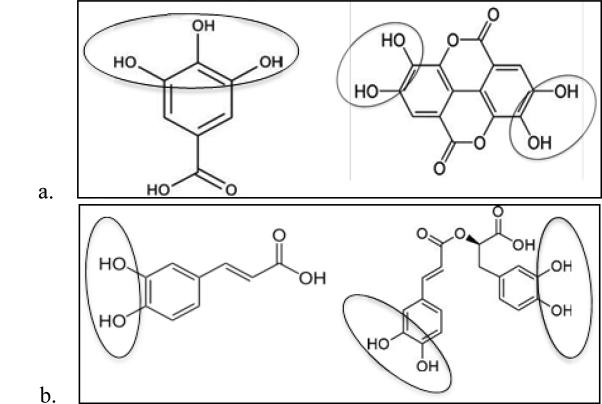
Figure 3 shows a comparison of antioxidant activity in terms of GAE (or mM GA with equivalent electron donating capacity to 1 mM sample) of the fifteen compounds studied here, using Ti(IV)oxo paper sensors as an example. Structurally, antioxidant activity appears to correlate with the number and location of hydroxyl groups, rather than the class of polyphenol; with ortho positioning often being correlated with increased antioxidant activity[5]. AOX activity as determined by the Ti(IV)oxo sensors ranges from 0.05-1.30 mM GAE. Ten polyphenols have GAEs above the median, 0.60 mM GAE. Nine of these contain at least one ortho positioned set of OH groups and between 2-9 hydroxyl functionalities. The other five polyphenols have much lower activities ranging from 0.05-0.30 mM GAE. These have only 1-3 OH groups none of which are in an ortho position (with the exception of EA). Hydroxyl groups are responsible for electron donation, which can take place through three main mechanisms (hydrogen atom transfer (HAT), single proton loss followed by electron transfer (SPLET), and electron transfer followed by proton loss (ETPT))[27]. The increased number of hydroxyl groups can increase antioxidant activity by increasing the likelihood of electron donation to a surface cation on a metal oxide nanoparticle. The ortho positioning of hydroxyl groups structurally facilitates bi-dentate binding, creating a claw-like structure which can bind easily to a redox active species. Notably, it can be observed that formation of dimers from polyphenolic monomers can have an inhibitory effect on antioxidant activity depending on whether or not hydroxyl groups are involved in binding. EA and GA have extremely different antioxidant capacities (0.29 and 1mM GAE respectively on Ti(IV)oxo; 0.43 and 1.00 on ceria; and 0.81 and 1.00 on zinc oxide), which can be attributed to the 3 OH groups in ortho position on the phenolic ring of GA as compared to its dimer form (EA), which has 4 OH groups and is constituted from two GA monomers. Formation of the dimer causes the loss of one OH group from each GA monomer, and thus offers less OH groups in favorable orientation for binding and formation of charge transfer complexes with metal oxides (Fig. 4a). In contrast, RA and CA have similar antioxidant activities on titanyl oxalate sensors (0.73 and 0.98, respectively) and nearly the same GAE on CeO2 and Fe2O3 sensors (0.80 and 0.81, respectively for CeO2; and 0.95 and 1.01 for Fe2O3). RA is a dimer of CA, and joining two CAs together appears to have a negligible effect on its antioxidant power. The ability of this new complex to maintain antioxidant activity is due to the fact that no functional groups involved in electron donation are modified during the binding process (Fig. 4b). Preservation of these functional groups allows for activity to be maintained. Additionally, the molecular structure of RA as compared to EA is much more flexible, allowing for the molecule to rotate about molecular bonds in order to bind in a bi-dentate fashion using both ortho OH groups; whereas the rigid phenolic structure of EA does not allow mobility about bonds and keeps the two sets of ortho OH groups far from one another forcing bi-dentate binding to occur through only one set of hydroxyl groups, not both simultaneously.
Figure 3.
Comparison of antioxidant activity (GAE, or mM gallic acid with equivalent electron donating capacity to 1 mM sample) assigned by titanyl oxalate sensors to demonstrate variability in electron donating capacity of each polyphenol.
Figure 4.
Visual depiction of changes in hydroxyl functionalities resulting from formation of dimers from monomers caffeic acid (a) and gallic acid (b). The dimers rosmarinic acid (a) and ellagic acid (b) exhibit unique changes in antioxidants activities as compared to their monomers; a characteristic likely caused by the effect of complex formation on the number and orientation of their hydroxyl groups.
Comparison of antioxidant activity values for fifteen polyphenolic compounds analyzed using four sensor types (CeO2, Ti(IV)oxo, ZnO and Fe2O3) can be found in Figure S5 (supplemental). It was observed that some polyphenols have an affinity or an aversion to one or more metal oxide, creating varied antioxidant activity values depending on the sensor used; a phenomenon observed when comparing most other antioxidant activity assays as well[28]. The entire range of antioxidant activity values, however, only spans from 0.05 - 1.90 mM GAE on all sensor types; indicating that the maximum distance between 15 samples would be merely 0.12 mM GAE and that as a result, slight variations in activity can significantly affect the ranking of a polyphenol within this group. Thus, only extreme differences in polyphenol ranking order between sensor types are discussed here. As a general trend, it was observed that Trolox has very low antioxidant activity (i.e. electron donating capacity toward metal oxides), exhibited by its ranking within the bottom five antioxidants (ranked from high to low with respect to GAE) using all assay types. AA, on the other hand, consistently ranked within the top seven antioxidants using all assay types; and GA hovered within the middle four to seven. Some extreme differences between sensor types can be observed, however, as in the case of capsaicin, vanillic acid and rutin for example. CP ranked as having the second lowest GAE on three of four sensor types, but it had the fifth highest activity on CeO2 sensors, showing a preferential electron transfer and subsequent binding to ceria as compared to other metal oxides. Similarly, VA appeared within the bottom three GAE values on three of four assay types, but had the fourth highest value on ZnO sensors. R ranked within the top three GAE values on all sensors butCeO2, which ranked it as eleventh out of fifteen. The differential responses can be due to differences in the sensor surface including surface morphology, surface area and orientation of the polyphenols upon binding to the oxide surfaces. These results suggest that the sensitivity of the assays varies with the metal oxide used in the sensor design. The differential responses are used to discriminate polyphenol antioxidants and constitute the basis for establishing the database for the identification and quantification of polyphenols in mixtures.
The full version of the database of Pantone ID codes for each sensor type is provided in the Supplementary Information, along with linear calibration equations for each polyphenol. These equations were used to determine the antioxidant activity (mM GAE) of each polyphenol studied herein and can be used for quantification as well. For sample identification in the field, the Pantone ID catalogue of antioxidant standards can be either printed and visually referenced, or stored into an electronic device to provide immediate and automated color matching for rapid sample characterization.
Analytical Characterization and Validation
The Polyphenol Database was further tested and validated through confirmation of the polyphenolic identity and concentration through color ID matching.
We demonstrate below the use of this database to (a) determine sample identity and concentration of an isolated polyphenolic compound, and (b) to identify the primary polyphenolic compound existing within mixtures and real sample matrices. The use of this approach for assessing the antioxidant activity of samples is also demonstrated.
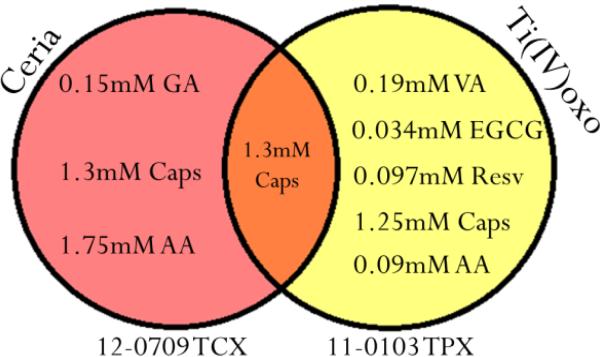
When analyzing isolated polyphenolic samples of known identity, concentration can be interpolated quantitatively using the calibration curve corresponding to the given polyphenol, and the BCI of the sample as the y-value in its linear equation. The accuracy of this method was found to have an average recovery error of ± 0.60 mM when confirming concentrations of VA, RA and EGCG using a BCI vs. calibration curve. For unknown samples, sequential readings with different types of metal oxide sensors are used to confirm identity and concentration of the sample. The accuracy of this method is displayed in Figure 5 wherein a sample of capsaicin (1mM) was placed onto two sensor types: CeO2 and Ti(IV)oxo. Using the Pantone© ID matching for each color response, only one possible identity was revealed corresponding to 1.3 mM capsaicin. This process is represented as a Venn diagram in Figure 5, which describes the sample identification process using two or more sensor types. The advantage of this technology is that an otherwise completely unknown sample can be both identified and quantified in a timely manner without the use of any advanced instrumentation (e.g. UV or fluorescence detectors). As an example of application, this process has also been carried out on real samples in order to determine the primary acting polyphenolic constituent.
Figure 5.
Demonstration of the technique used to narrow down matching ID codes between a sample of unknown composition and a polyphenolic identity. An ID code for a sample applied to each sensor type is matched within each respective database to find matches. Those matches which do not appear in all databases used are eliminated.
Demonstration of Applicability of the Antioxidant Database for Real Sample and Mixture Analysis using the Multiarray Sensor
Four sensor types (Ti(IV)oxo, CeO2, ZnO and Fe2O3) were used to analyze one mixture of known composition (7mM GA, 1mM EA) and a real sample (gunpowder green tea). Color IDs of the sample response on each sensor type were matched within the respective databases, and systematically analyzed as described above.
Results of mixture analysis are shown in Table 1, which displays the identities of each mixture as determined by the four sensor types. These results reveal the active antioxidant in the mixture. Visual examples of color matching between the sample and an individual polyphenol standard are also shown. These exact color matches constitute the basis for the hypothesis that within a mixture, one polyphenol can be preferentially active.
While the activity of each compound in a mixture influences its ability to compete for binding sites on the sensor, the concentration of each can also determine the probability of reaction with the sensing nanoparticles. The variety of antioxidant activities and concentrations of each polyphenol in a mixture makes every field sample unique. Thus, the use of multiple sensor types and their respective database is necessary to determine whether the mixture's identity, as determined by the metal oxide sensor array, truly reveals one primary active component, or if it could be a false match coming from signal overlap. If a mixture is identified as just one compound using all sensor types, it can be reasonably assumed that only one polyphenol is responding due to mixture conditions (such as activity and concentration of the polyphenols contained). This would reveal which antioxidant in the mixture preferentially performs electron transfer to the sensing surface.
Analysis of the laboratory-prepared mixture containing 7mM GA and 3mM EA showed only one possible polyphenolic identity that agreed on all four sensor types: GA. The concentration determined by averaging results from four databases was determined to be 9 ± 4 mM GA. This matched the true identity of the solution which contained 7 mM GA that likely outcompeted the 3mM EA for binding sites on the sensing surface.
For the analysis of gunpowder green tea, all sensor types determined the identity of the responding polyphenol to be: 0.9 ± 0.2 mM EGCG. This concentration matches published EGCG concentrations commonly found in green tea (1.2 ± 0.6 mM, as calculated from Henning's data that presented concentration of EGCG and other catechins in eight commercial green tea varieties[18]). EGCG is highly active (1.27, 1.08, 1.08, and 0.72 mM GAE as determined by ceria, titanyl oxalate, iron oxide, and zinc oxide sensors, respectively) and constitutes the majority of the catechin content in green tea (46 ± 5% of the catechin content in green tea[18]). Therefore, the concentration and activity of EGCG are likely the cause for its ability to out-compete other catechins in tea for binding sites on the sensor surface. Out-competition by EGCG allows for the creation of many EGCG-MeO charge-transfer complexes creating spectral changes that yield a colorimetric response which matches the same Pantone® ID code as a documented EGCG standard.
These results successfully demonstrate the ability to utilize this metal-oxide sensing array for systematic identification of the primary active polyphenolic constituent in a mixed matrix such as a botanical extract or a food sample. This technique allows for quick analysis of antioxidant activity and composition using just one assay. With current technology, this type of analysis is very time consuming and complicated as it requires the use of various antioxidant activity assays (i.e. FRAP [29, 30], ORAC [29, 31, 32], DPPH [33, 34], ABTS [29, 34] alongside HPLC [29, 31, 35, 36] or GC/MS [34] for content analysis.
Validation of Antioxidant Activities vs. the ORAC assay
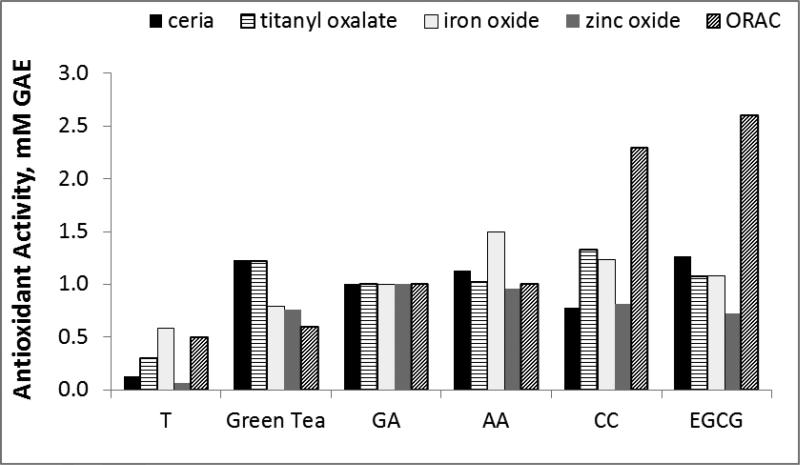
The performance of four metal oxide paper sensors was compared to a conventional method for antioxidant activity analysis, the ORAC assay. The ORAC assay is the USDA adopted method for antioxidant activity analysis, and represents antioxidant activity in terms of Trolox equivalence (mM Trolox with equal activity to 1 mM or 1 g of sample). ORAC monitors peroxyl radical scavenging capacity, which indicates hydrogen atom transfer (HAT). The metal oxide sensing array monitors single electron transfer to a active metal. Metal oxide sensor data are quantified in terms of GAE, and thus, for this section, in order to obtain comparative data, the ORAC assay was also quantified in terms of GAE, instead of the usual Trolox equivalents for the ORAC assay. Figure 5 graphically depicts the comparison of antioxidant activities as determined by the ORAC and the four metal oxide assays. It should be noted that every antioxidant assays ranks samples differently in terms of antioxidant activity. This is primarily because of the unique radical sources / redox-active reagents and the individualized assay conditions required for each assay. Within the range of paper sensors used, each will respond differently to various polyphenols based on structural and chemical reactivity differences between nanoparticle indicators; and all paper sensors will respond differently than the ORAC assay due to different electron acceptors (metal oxide NPs vs. peroxyl radicals) The non-comparability of antioxidant activity values between assays is a major challenge in the field, and has been cited as a reason for the retraction of the USDA ORAC database from public view in 2012[37]. Nonetheless, GAE values determined by the five assay types follow a very similar trend for all antioxidant samples tested, and reveal GAE values that are close to one another. Average activity values range from 0.3 – 1.4 mM GAE with standard deviations of GAE assigned by five assays averaging 0.3 mM GAE, depending on the sample. EGCG and CC, for example, respond more strongly in the ORAC assay than in the metal oxide sensing array, indicating that they may have a preference to peroxyl radical scavenging as opposed to electron donation to inorganic particles. Through averaging activity values determined using the five assay types and considering standard deviation, a standard T-test has determined Trolox to be statistically lower in activity than the other five samples (p<.006), indicating that the metal oxide sensing array is proficient in identifying major differences in antioxidant capacity with comparable sensitivity to the ORAC assay. This validates the metal oxide sensor array as an excellent assay for the determination of antioxidant activity.
Conclusion
In summary, we met our stated objectives, which were to develop a metal oxide based sensing array for field application in the characterization of antioxidant-containing samples. We established a novel method for analysis of such samples using our multiarray sensing system, based on formation of unique colored charge transfer complexes between polyphenols and metal oxide nanoparticles. Using the sensing array and an automated CapSure® color reader we have developed a database of calibration curves and color ID codes for a variety of polyphenolic compounds. We have demonstrated applicability of the sensing concept on a variety of metal oxide type materials, which we found to provide unique characteristic responses to polyphenols. The differential response of each sensor is significantly important as it provides a basis for distinguishing individual components based on their distinct interaction with the metal oxide. Colorimetric CapSure® readings can be immediately converted into information characterizing a polyphenolic sample, by matching color IDs obtained with the reference standards included in the accompanying database. This technique allows for simple, inexpensive, immediate and extremely portable identification and quantification of polyphenolic constituents; as well as determination of any sample's antioxidant activity. This database is particularly useful for field investigations of plants in remote locations.
This is the first portable database of its kind that facilitates remote analysis of any antioxidant-containing sample without the use of sophisticated laboratory instrumentation. The only equipment needed for analysis aside from the sample, are the sensing papers and the handheld color reader with the electronically stored database of standards. The assay has been used to characterize real samples in terms of the primary active polyphenolic constituent, but its application could also be extended for use as a detector alongside instrumentation for separation of mixtures (e.g. HPLC or other separation techniques) for complete characterization of polyphenol containing samples. In addition to the easy-to-use, rapid and field detection capabilities, the assay is inexpensive, reproducible and compares well with conventional antioxidant activity assays. The results of this study indicate the suitability and potential of this technology to replace conventional techniques for complete characterization of polyphenol-containing samples. Numerous possibilities are envisioned for use of this assay in the field, in a laboratory, or perhaps even at home; by trained and untrained professionals alike.
Supplementary Material
Figure 6.
Antioxidant activity of five polyphenol standards and one real sample, as determined by the ORAC assay and four metal oxide paper sensors. All assays were quantified in terms of GAE equivalents (mM GAE with equivalent activity to 1mM or 1g sample).
Acknowledgements
ES acknowledges Kaleb Lund, Leanna Standish, the Bastyr University Tierney Research Laboratory and Clarkson University for hosting ORAC research. This material is based upon work supported by the National Science Foundation under Grant No. 0954919 as well as work supported by the National Center for Complementary and Alternative Medicine at the National Institutes of Health under pre-doctoral training Grant No. T32AT00815. TF, DJ and AM acknowledge financial support from the Clarkson University Honors Program and the Clarkson University Research Experiences for Undergraduates Program. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, the National Institutes of Health, or Clarkson University.
Biography
Erica Sharpe received her Bachelor of Arts in Chemistry and Music from the State University of New York at Potsdam in 2008. She is currently a PhD candidate in Chemistry & Biomolecular Science at Clarkson University. She has completed two competitive research fellowships in botanical medicine at Bastyr University that were funded by the National Center for Complementary Medicine (NCCAM) of the National Institutes of Health (NIH) in 2011 and 2012. She has also completed research in Italy's National Institute Nutritional Research, (INRAN) in 2013. Her research focus is on the development of portable assays for the detection of various analytes (e.g. glucose, hydrogen peroxide, antioxidants) as well as for determination of antioxidant activity. The assays are intended for rapid analysis of dietary sources, botanical medicines, biological fluids, and more.
Dr. Ryan Bradley received his Bachelor of Science in Chemical Engineering in 1997, his Doctor of Naturopathy (ND) from Bastyr University in 2003, and his Master of Public Health (MPH) in Epidemiology from the University of Washington in 2009 during his training in the KL2 Multi-disciplinary Clinical Research Program at the University of Washington Medical Center. He is currently an Associate Research Scientist at the Bastyr University Research Institute at Bastyr University California in San Diego, CA and an adjunct professor of Nutrition at the National College of Natural Medicine in Portland, OR. Dr. Bradley also practices as a naturopathic doctor at the Pacific Pearl Institute for Health and Healing in La Jolla, CA. Prior to joining the Pacific Pearl, Dr. Bradley served as the Director for the Center for Diabetes & Cardiovascular Wellness at Bastyr University in Seattle, WA from 2005-2013. In addition to ten years in practice, Dr. Bradley is an accomplished medical researcher, and has published his research in leading peer-reviewed medical journals. His research focuses on many areas including the chemical properties of natural products and their application in the reduction of diet-induced oxidative stress and vascular dysfunction.
Thalia Frasco is a junior at Clarkson University majoring in chemical engineering with a minor in law studies. She started college at Clarkson University in 2011 as part of the Clarkson School, an early college entrance program that allowed her to skip her senior year of high school. She has been working under Dr. Silvana Andreescu in the Department of Chemistry and Biomolecular Science from May 2012 to present.
Dilhani I. Jayathilake received her Bachelor of Science in Chemistry and Biology from the University of Peradeniya, Sri Lanka in 2010. She gained research experience in the fields of Chemistry and Civil Engineering at Clarkson University in 2012 and 2013.
Amanda Marsh is a senior chemistry major with a biochemistry concentration and creative writing minor, at Utica College in Utica, NY. She has participated in both independent research at her home institution and as a NSF-REU fellow through two Research Experience for Undergraduate (REU) programs funded by the National Science Foundation at Clarkson University and Duke University. In the summer of 2012, Amanda worked under Dr. Silvana Andreescu and Erica Sharpe at Clarkson on electrochemical and colorimetric sensors to enhance their effectiveness for the detection of antioxidants.
Dr. Silvana Andreescu is a Professor of Chemistry and Biomolecular science at Clarkson University. She received her PhD in 2002 from the University of Bucharest, Romania and University of Perpignan, France and completed postdoctoral training at State University of New York at Binghamton before joining the Department of Chemistry and Biomolecular Science at Clarkson University in 2005. Her research interests are in bionanotechnology, biointerfaces, bioanalytical chemistry and biosensors development and their applications for environmental, food and clinical monitoring. She is the recipient of a French government graduate fellowship, a NATO-NSF postdoctoral fellowship, the NSF-CAREER award and the Clarkson University's John W. Graham Research Award. She has published more than 70 peer-reviewed journal articles and 15 book and encyclopedia chapters, has co-edited two books, and has delivered some 100 presentations at professional and academic conferences throughout the world.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available:
The supporting information for this paper details: the effect of pH on sensor response; the comprehensive database of polyphenol Pantone© ID codes for four metal oxide sensor types (CeO2, Ti(IV)oxo, ZnO and Fe2O3); and the analytical parameters necessary for complete use of this database (linear equations for polyphenol calibrations, antioxidant activities, correlation coefficients, and linear ranges for each compound, determined using all metal oxide sensor types). This information can be used for rapid field sample analysis.
References
- 1.Spencer J, Kuhnle G. Intracellular metabolism and bioactivity of quercetin and its in vivo metabolites. Biochem. J. 2003;372:173–181. doi: 10.1042/BJ20021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacic P, Somanathan R. Multifaceted approach to resveratrol bioactivity. Oxidative Medicine and Cellular Longevity. 2010;3:86–100. doi: 10.4161/oxim.3.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Yin JJ, Shen SR. Growth Inhibition of Prostate Cancer Cells by Epigallocatechin Gallate in the Presence of Cu2+ Journal of Agricultural and Food Chemistry. 2004;52:462–466. doi: 10.1021/jf035057u. [DOI] [PubMed] [Google Scholar]
- 4.Zafra-Stone S, Yasmin T, Bagchi M. Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol. Nutr. Food Res. 2007;51:675–683. doi: 10.1002/mnfr.200700002. [DOI] [PubMed] [Google Scholar]
- 5.Zheng W WS. Oxygen radical absorbing capacity of phenolics in blueberries, cranberries, chokeberries, and lingonberries. Journal of agricultural and food chemistry. 2003;51:502–509. doi: 10.1021/jf020728u. [DOI] [PubMed] [Google Scholar]
- 6.Wollgast J, Anklam E. Review on polyphenols in Theobroma cacao: changes in composition during the manufacture of chocolate and methodology for identification and quantification. Food Research International. 2000;33:423–447. [Google Scholar]
- 7.Kang J, Xie C, Li Z, Nagarajan S, Schauss AG, Wu T, Wu X. Flavonoids from acai (euterpe oleracea mart.) Pulp and their antioxidant and anti-inflammatory activities. Journal of Food Chemistry. 2011;128:152–157. doi: 10.1016/j.foodchem.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Scholz, E EZ, Katus HA, Karle CA. Cardiovascular ion channels as a molecular target of flavonoids. Cardiovasc. Ther. 2010;28:46–52. doi: 10.1111/j.1755-5922.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 9.Cook JA VD, Dasgupta A, Mounkaila G, Glew RS, Blackwell W, Glew RH. Use of the Trolox assay to estimate the antioxidant content of seventeen edible wild plants of Niger. Life Sciences. 1998;63:105–110. doi: 10.1016/s0024-3205(98)00245-8. [DOI] [PubMed] [Google Scholar]
- 10.Sharpe E FT, Andreescu D, Andreescu S. Portable ceria nanoparticle-based assay for rapid detection of food antioxidants (NanoCerac) Analyst. 2013;138:249–262. doi: 10.1039/c2an36205h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao G AH, Cutler RG. Oxygen-radical absorbance capacity assay for antioxidants. Free Radical Biol. Med. 1993;14:303–311. doi: 10.1016/0891-5849(93)90027-r. [DOI] [PubMed] [Google Scholar]
- 12.Benzie IFF SJ. The Ferric Reducing Ability of Plasma (FRAP) as a measure of “Antioxidant Power”: The FRAP Assay. Analytical Biochemistry. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- 13.Apak R GK, Oxyurek M, Karademir SE. Novel total antioxidant capacity index for dietary polyphenols and vitamins C and E, using their cupric ion reducing capability in the presence of neocuproine: CUPRAC method. J. Agric. Food Chem. 2004;52:7970–7981. doi: 10.1021/jf048741x. [DOI] [PubMed] [Google Scholar]
- 14.Vasilescu, E. AS, Andreescu S. Nanoparticle-Based Technologies for the Detection of Food Antioxidants. Current Analytical Chemistry. 2012;8 [Google Scholar]
- 15.Tsao RD, Zeyuan Separation procedures for naturally occurring antioxidant phytochemicals. Journal of Chromatography B. 2004;812:85–99. doi: 10.1016/j.jchromb.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 16.Ornatska M, Sharpe E, Andreescu D, Andreescu S. Paper Bioassay Based on Ceria Nanoparticles as Colorimetric Probes. Analytical Chemistry. 2011;83:4273–4280. doi: 10.1021/ac200697y. [DOI] [PubMed] [Google Scholar]
- 17.Xu M, Bunes BR, Zang L. Paper-based vapor detection of hydrogen peroxide: colorimetric sensing with tunable interface. ACS applied materials & interfaces. 2011;3:642–647. doi: 10.1021/am1012535. [DOI] [PubMed] [Google Scholar]
- 18.Henning S, Fajardo-Lira C, Lee HW, Youssefian AA, Go VLW, Heber D. Catechin Content of 18 Teas and a Green Tea Extract Supplement Correlates With the Antioxidant Capacity. Nutrition and Cancer. 2003;45:226–235. doi: 10.1207/S15327914NC4502_13. [DOI] [PubMed] [Google Scholar]
- 19.Tsao R. Chemistry and Biochemistry of Dietary Polyphenols. Nutrients. 2010;2:1231–1246. doi: 10.3390/nu2121231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berman H, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doreleijers, S. JM, Maziuk D, et al. BioMagResBank database with sets of experimental NMR constraints corresponding to the structures of over 1400 biomolecules deposited in the Protein Data Bank. Journal of Biomolecular NMR. 2003;26:139–146. doi: 10.1023/a:1023514106644. [DOI] [PubMed] [Google Scholar]
- 22.Singleton V, Orthofer R, Lamuela-Raventos R. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods in Enzymology. 1999;299:152–178. [Google Scholar]
- 23.Graham H. Stabilization of the Prussian blue color in the determination of polyphenols. J. Agric. Food Chem. 1992;40:801–805. [Google Scholar]
- 24.Gornall A, Bardawill CJ, David MM. Determination of serum proteins by means of the biuret reaction. The Journal of Biological Chemistry. 1949;177:751–766. [PubMed] [Google Scholar]
- 25.Bradford M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 26.Sharpe E, Andreescu D, Andreescu S. Artificial Nanoparticle Antioxidants. In: Andreescu S, Hepel M, editors. Oxidative Stress: Diagnostics, Prevention, and Therapy. American Chemical Society; 2011. pp. 235–253. [Google Scholar]
- 27.Xiao-Ling L.-X.J. Cheng, Teng Qing-Feng, Chang Jin, Yao Xiao-Jun, Dai Fang, Qian Yi-Ping, Tang Jiang-Jiang, Li Xiu-Zhuang, Zhou Bo. Antioxidant activity of a-pyridoin and its derivatives: possible mechanism. Organic & Biomolecular Chemistry. 2010;8:1058–1063. doi: 10.1039/b922673g. [DOI] [PubMed] [Google Scholar]
- 28.Apak R, Guclu K, Demirata B, Ozyurek M, Celik SE, Bektasoglu B, Berker KI, Ozyurt D. Comparative evaluation of various total antioxidant capacity assays applied to phenolic compounds with the CUPRAC assay. Molecules. 2007;12:1496–1547. doi: 10.3390/12071496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yashin YYA, Nemzer B. Determination of Antioxidant Activity in Tea Extracts, and Their Total Antioxidant Content. Am. J. Biomed. Sci. 2011;3:322–335. [Google Scholar]
- 30.Carlsen BLHH, Holte K, Bohn SK, Dragland S, Sampson L, Willey C, Senoo H, Umenzono Y, Sanada C, Barikmo I, Berhe N, Willett WC, Phillips KM, Jacobs DR, Blomhoff R. The total antioxidant content of more than 1300 foods, beverages, spices, herbs and supplements used worldwide. Nutr. J. 2010 doi: 10.1186/1475-2891-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo M, Zhang LL, Ji HP, Kou Y, Ou BX. Bioavailability and Antioxidant Effects of a Xanthone-Rich Mangosteen (Garcinia mangostana) Product in Humans. Journal of Agricultural and Food Chemistry. 2009;57:8788–8792. doi: 10.1021/jf901012f. [DOI] [PubMed] [Google Scholar]
- 32.Speisky H, Lopez-Alarcon C, Gomez M, Fuentes J, Sandoval-Acuna C. First Web-Based Database on Total Phenolics and Oxygen Radical Absorbance Capacity (ORAC) of Fruits Produced and Consumed within the South Andes Region of South America. Journal of Agricultural and Food Chemistry. 2012;60:8851–8859. doi: 10.1021/jf205167k. [DOI] [PubMed] [Google Scholar]
- 33.Boligon RBFAA, Brum TF, Piana M, Belke BV, Rocha JBT, Athayde ML. Phytochemical constituents and in vitro antioxidant capacity of Tabernaemontana catharinensis A. DC. Free Radicals and Antioxidants. 2013;3:77–80. [Google Scholar]
- 34.Ananth TSDA, Rameshkumar A, Jeyadevi R, Aseervatham SB. Chemical constituents, in vitro antioxidant and antimicrobial potential of Caryota urens L. Free Radicals and Antioxidants. 2013;3:107–112. [Google Scholar]
- 35.Kim YJ, Lee HJ, Shin Y. Optimization and Validation of High-Performance Liquid Chromatography Method for Individual Curcuminoids in Turmeric by Heat-Refluxed Extraction. J Agric Food Chem. 2013 doi: 10.1021/jf402483c. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Centeno MR, Jourdes M, Femenia A, Simal S, Rossello C, Teissedre PL. Characterization of Polyphenols and Antioxidant Potential of White Grape Pomace Byproducts (Vitis vinifera L.) J Agric Food Chem. 2013 doi: 10.1021/jf403168k. [DOI] [PubMed] [Google Scholar]
- 37.United States Department of Agriculture Agricultural Research Service Nutrient Data Products and Services: Oxygen Radical Absorbance Capacity (ORAC) of Selected Foods. Release. 2010 May 16;2 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.