Abstract
Image representation plays an important role in medical image analysis. The key to the success of different medical image analysis algorithms is heavily dependent on how we represent the input data, namely features used to characterize the input image. In the literature, feature engineering remains as an active research topic, and many novel hand-crafted features are designed such as Haar wavelet, histogram of oriented gradient, and local binary patterns. However, such features are not designed with the guidance of the underlying dataset at hand. To this end, we argue that the most effective features should be designed in a learning based manner, namely representation learning, which can be adapted to different patient datasets at hand. In this paper, we introduce a deep learning framework to achieve this goal. Specifically, a stacked independent subspace analysis (ISA) network is adopted to learn the most effective features in a hierarchical and unsupervised manner. The learnt features are adapted to the dataset at hand and encode high level semantic anatomical information. The proposed method is evaluated on the application of automatic prostate MR segmentation. Experimental results show that significant segmentation accuracy improvement can be achieved by the proposed deep learning method compared to other state-of-the-art segmentation approaches.
1 Introduction
The key to the success of many medical image analysis methods is mainly dependent on the choice of data representation (or features) used to characterize the input images. Over the past decades, feature engineering has been an active research topic in medical image analysis. For instances, Shen et al. [1] proposed the geometric moment invariant based features for feature-guided non-rigid image registration, and Guo et al. [2] proposed the discriminative local binary patterns feature for protein cellular classification. A common and important finding in these studies is the performance of the same registration, segmentation or classification framework with different types of image features adopted can be significantly different, which reveals the importance of image representation in medical image analysis. Other widely used image features in medical image analysis include, but not limited to Haar wavelet [3] and histogram of oriented gradient (HOG) [4].
A major limitation of the features widely used for medical image analysis discussed in the previous paragraph is their inability to extract and organize salient information from the data at hand. In other words, these are hand-crafted features, and their representation power can be varied across different patient datasets. In order to resolve this limitation, it is essential to develop a framework which can extract specialized features adapted to different datasets at hand.
Therefore, we are motivated to propose a deep learning framework for unsupervised automatic feature extraction. Deep learning is an active research topic which learns data adaptive representations. Representative deep learning methods include stacked auto-encoders [5] and deep belief nets [6]. In this paper, a stacked independent subspace analysis (ISA) network [7] is adopted. In the low level, basic image microscopic features such as spots, edges, and curves are learnt and captured from the input data. The learnt low level features are served as input to the high level of the ISA network, which encodes more abstract and higher level semantic image context information of the input data.
We evaluate the proposed method on the application of automatic prostate MR segmentation for prostate cancer diagnosis. Our method is also compared with other state-of-the-art segmentation methods and hand-crafted features, and experimental results show that significant improvement of the segmentation accuracy can be achieved by the proposed method.
2 Methodology
In this section, we will first introduce the basic independent subspace analysis (ISA) network, which is an unsupervised learning algorithm for feature extraction. Then, the stacked ISA network is introduced to construct the deep learning framework. Finally, details will be given for integrating the learnt features with a sparse label propagation framework for automatic prostate MR segmentation.
2.1 The Basic Independent Subspace Analysis (ISA) Network
The basic ISA network [8] can be characterized as a two-layer network to learn features in an unsupervised manner from input images. The inputs of the basic ISA network are image patches sampled from training images, as illustrated in Figure 1. The first layer of the basic ISA network consists of atomic learning units called simple units. The simple units capture the square nonlinearity relationships among the input patches. The atomic learning units which form the second layer of the basic ISA network are called pooling units, which group and integrate the responses from different simple units in the first layer.
Fig. 1.

Schematic illustration of the basic ISA network, where inputs of the basic ISA network are image patches sampled from the training images. The basic ISA network contains two layers. The first layer is consisted of simple units (green circles) to capture the square nonlinearity among the input patches. The second layer is consisted of pooling units (red circles) to group and integrate the responses from the simple units in the first layer to capture the square root nonlinearity relationships. R1,R2,#x2026;,Rm denote the responses from the second layer.
The basic ISA network can be mathematically formulated as follows: Given N input patches xi (i.e., each input patch is reshaped into a column vector), ISA aims to estimate the optimal parameter matrix W ∈ Rk×d corresponding to the first layer, and the optimal parameter matrix V ∈ Rm×k corresponding to the second layer by minimizing the energy function expressed by Equation 1:
| (1) |
where and I is the identity matrix. d, k and m denotes the dimension of each input xi, number of simple units in the first layer, and the number of pooling units in the second layer, respectively. In this paper, V is set to constant similar to [7].
It should be noted that each row of W in Equation 1 is corresponding to one simple unit in the first layer of the ISA network shown in Figure 1, and if we reshape it to the original resolution of input patches, it can be viewed as a filter learnt from input patches to extract features. Figure 2 shows typical filters learnt from 30 prostate T2 MR images with patch size 16 × 16 × 2.
Fig. 2.

Typical filters learnt by the ISA network from 30 prostate T2 MR images with patch size 16 × 16 × 2. Each patch is normalized to have zero mean and unit variance before the training process.
2.2 Deep Learning Framework with Stacked ISA Networks
In order to capture different levels of anatomical information of input training images, it is essential to build the ISA network in a hierarchical manner.
The stacked ISA networks [7] are used to construct the deep learning architecture. Specifically, we first train the ISA network with small input patch sizes. Then, the learnt ISA network is used to convolve with larger regions of input training images. This idea can be illustrated by Figure 3. The PCA operation aims to obtain more compact representation from low-level ISA network.
Fig. 3.
Schematic illustration of the stacked ISA deep learning architecture. We first learn the lower level ISA network with small input patches. Then, for each larger patch, we can represent it as s overlapping small patches, and we can obtain the pooling unit responses of each overlapping small patch based on the previously learnt lower level ISA network. The pooling unit responses of each overlapping small patch are then put through a PCA dimensionality reduction procedure to serve as input to train the higher level ISA network.
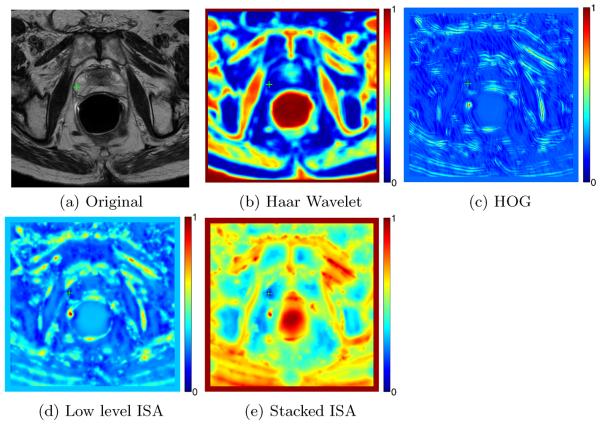
Figure 4 shows the color coded difference maps obtained by different features in a prostate T2 MR image shown in Figure 4 (a). As shown in Figures 4 (b) and (c), hand-crafted features such as Haar wavelet [3] and HOG [4] cannot satisfactorily discriminate the reference voxel highlighted by the green cross with other voxels. On the other hand, using features learnt by the lower level ISA network alone still cannot effectively separate the reference voxel from other voxels. By integrating features learnt by both the lower level ISA and higher level ISA networks, the reference voxel can be satisfactorily separated from other voxels as it is only similar to neighboring voxels with similar anatomical properties.
Fig. 4.
(a) A prostate T2 MR image, with the reference voxel highlighted by the green cross. (b), (c), (d), and (e) denote the color coded difference map obtained by computing the Euclidean distance between the reference voxel and all the other voxels in (a) by using features extracted by Haar wavelet [3], HOG [4], low level ISA network, and the stacked ISA network, respectively.
2.3 Feature Based Sparse Label Propagation
Label propagation is a popular approach [9,10] for multi-atlases based image segmentation. It can be described as follows: Given T atlas images It and their segmentation groundtruths Lt (t = 1, …, T) registered to the target image Inew, the label at each voxel position x ∈ Inew can be computed by Equation 2:
| (2) |
where denotes the local neighborhood of x in It. wt(x, y) is the graph weight reflecting the contribution of voxel y ∈ It to estimate the anatomical label of x. Lnew is the estimated label probability map of Inew.
A popular method to estimate the graph weight wt(x, y) in Equation 2 is sparse coding [11]. Specifically, for each voxel x ∈ Inew, its feature signature is denoted as fx. For each voxel , we also denote its feature signature as and organize them into a matrix A. Then, the sparse coefficient vector βx is estimated by minimizing Equation 3:
| (3) |
where ∥·∥1 denotes the L1 norm, and wt(x, y) is set to the corresponding element in the optimal sparse coefficient vector with respect to y ∈ It.
In this paper, features learnt by the stacked ISA network are used as signature for each voxel in the sparse label propagation framework for automatic prostate T2 MR segmentation. The registration procedure in [12] was adopted in our method before label propagation.
3 Experimental Results
We evaluate the discriminant power of features learnt by the stacked ISA network by prostate T2 MR segmentation. 30 prostate T2 MR images from different patients were obtained from the University of Chicago Hospital. The 30 images were taken from different MR scanners and the segmentation groundtruth of each image is also provided by a clinical expert. The multi-resolution fast free-form deformation (FFD) algorithm similar in [12] was used to perform registration for sparse label propagation. The segmentation accuracy of different methods is evaluated in a leave-one-out cross validation manner.
The following parameter settings were adopted for our method: The patch size is set to 16 × 16 × 2 and 20 × 20 × 3 in the low level and high level ISA network, respectively. Each patch with size 20 × 20 × 3 trained in the higher level ISA network is decomposed into s = 8 overlapping patches with size 16 × 16 × 2 in the lower level ISA network. λ is set to 10−4 in Equation 3.
Our method was compared with three widely used hand-crafted features: Haar wavelet [3], HOG [4], and local binary patterns (LBP) [13]. It was also compared with three multi-atlases based segmentation methods proposed by Klein et al. [12], Coupe et al. [10] and Liao et al. [14]. Three evaluation metrics were used: Dice ratio, average surface distance (ASD), and Hausdorff distance. Table 1 lists the average values and standard deviations (SD) of different metrics obtained by different methods.
Table 1.
Segmentation accuracies obtained by different methods associated with different evaluation metrics. Here SL denotes the sparse label propagation framework introduced in Section 2.3. The last two rows show the segmentation accuracies obtained by our method using only the single lower level ISA network and the stacked ISA network, respectively. The best results are bolded.
| Method | Mean Dice + SD (in %) | Mean Hausdorff + SD (in mm) | Mean ASD + SD (in mm) |
|---|---|---|---|
|
| |||
| Klein's Method [12] | 83.4 ± 3.1 | 10.2 ± 2.6 | 2.5 ± 1.4 |
| Coupe's Method [10] | 81.7 ± 2.7 | 12.4 ± 2.8 | 3.6 ± 1.6 |
| Liao's Method [14] | 84.4 ± 1.6 | 11.5 ± 2.2 | 2.3 ± 1.5 |
| Haar + SL | 84.2 ± 3.4 | 9.7 ± 3.2 | 2.3 ± 1.7 |
| HOG + SL | 80.5 ± 2.7 | 12.2 ± 4.5 | 3.8 ± 2.2 |
| LBP + SL | 82.6 ± 3.2 | 10.4 ± 2.2 | 3.0 ± 1.9 |
| Haar + HOG + LBP + SL | 84.9 ± 3.6 | 9.8 ± 3.3 | 2.5 ± 1.8 |
|
| |||
| Single ISA + SL | 84.8 ± 2.5 | 9.5 ± 2.4 | 2.2 ± 1.8 |
| Stacked ISA + SL | 86.7 ± 2.2 | 8.2 ± 2.5 | 1.9 ± 1.6 |
It can be observed from Table 1 that features learnt by the stacked ISA network consistently outperform other hand-crafted features such as Haar, HOG, and LBP. Figure 5 shows some typical segmentation results by our method. It can be observed that the estimated prostate boundaries of the two patients corresponding to the first two rows are very close to the groundtruths, while the estimated prostate boundary of the patient in the third row is not very accurate in the apex region, which needs further improvement in future work. Our method takes around 1.4 mins to segment a 3D prostate MR image with image size around 300 × 300 × 120, and the computation time without image registration is around 0.08 min. All the experiments were conducted on an Intel Xeon 2.66-GHz CPU computer with parallel computing implementation.
Fig. 5.
Typical segmentation results obtained by the proposed method. Each row shows the segmentation results of a patient. The estimated prostate boundary is highlighted in yellow, and the groundtruth prostate boundary is highlighted in red. The third row shows a failure case.
4 Conclusion
In this paper, we present a representation learning method for automatic feature extraction in medical image analysis. Specifically, a stacked independent subspace analysis (ISA) deep learning framework is proposed to automatically learn the most informative features from the input data. The major difference between the proposed feature extraction method and conventional hand-crafted features such as Haar, HOG, and LBP is that it can dynamically learn the most informative features adapted to the input data at hand, instead of blindly fixing the feature extraction kernel. The stacked ISA network simultaneously learns the low level basic microscopic image structure information and the high level image abstract and semantic information. Its discriminant power is evaluated on the application of prostate T2 MR image segmentation and compared with other types of image features and state-of-the-art segmentation algorithms. Experimental results show that the proposed method consistently achieves the highest segmentation accuracies than other methods under comparison.
References
- 1.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE TMI. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y, Zhao G, Pietikainen M. Discriminative features for texture description. Pattern Recognition. 2012;45:3834–3843. [Google Scholar]
- 3.Mallat G. A theory for multiresolution signal decomposition: the wavelet representation. IEEE PAMI. 1989;11:674–693. [Google Scholar]
- 4.Dalal N, Triggs B. CVPR. 2005. Histograms of oriented gradients for human detection; pp. 886–893. [Google Scholar]
- 5.Bengio Y, Lamblin P, Popovici D, Larochelle H. NIPS. 2006. Greedy layerwise training of deep networks; pp. 153–160. [Google Scholar]
- 6.Hinton G, Osindero S, Teh Y. A fast learning algorithm for deep belief nets. Neu. Comp. 2006:1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 7.Le Q, Zou W, Yeung S, Ng A. CVPR. 2011. Learning hierarchical invariant spatio-temporal features for action recognition with independent subspace analysis; pp. 3361–3368. [Google Scholar]
- 8.Hyvarinen A, Hurri J, Hoyer P. Natural Image Statistics. Springer; 2009. [Google Scholar]
- 9.Rousseau F, Habas P, Studholme C. A supervised patch-based approach for human brain labeling. IEEE TMI. 2011;30:1852–1862. doi: 10.1109/TMI.2011.2156806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coupe P, Manjon J, Fonov V, Pruessner J, Robles M, Collins D. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, Zhan Y, Dewan M, Huang J, Metaxas D, Zhou X. Towards robust and effective shape modeling: sparse shape composition. MedIA. 2012;16:265–277. doi: 10.1016/j.media.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Klein S, Heide U, Lips I, Vulpen M, Staring M, Pluim J. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Medical Physics. 2008;35:1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]
- 13.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE PAMI. 2002;24:971–987. [Google Scholar]
- 14.Liao S, Gao Y, Shen D. Sparse patch based prostate segmentation in CT images. In: Ayache N, Delingette H, Golland P, Mori K, editors. MICCAI 2012, Part III. LNCS. vol. 7512. Springer; Heidelberg: 2012. pp. 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]