Abstract
Image-guided radiotherapy (IGRT) requires fast and accurate localization of prostate in treatment CTs, which is challenging due to low tissue contrast and large anatomical variations across patients. On the other hand, in IGRT workflow, a series of CT images is acquired from the same patient under treatment, which contains valuable patient-specific information yet is often neglected by previous works. In this paper, we propose a novel learning framework, namely incremental learning with selective memory (ILSM), to effectively learn the patient-specific appearance characteristics from these patient-specific images. Specifically, starting with a population-based discriminative appearance model, ILSM aims to “personalize” the model to fit patient-specific appearance characteristics. Particularly, the model is personalized with two steps, backward pruning that discards obsolete population-based knowledge, and forward learning that incorporates patient-specific characteristics. By effectively combining the patient-specific characteristics with the general population statistics, the incrementally learned appearance model can localize the prostate of the specific patient much more accurately. Validated on a large dataset (349 CT scans), our method achieved high localization accuracy (DSC ~ 0.87) in 4 seconds.
1 Introduction
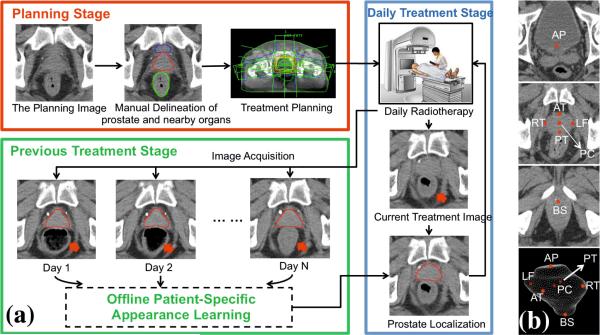
Image Guided Radiation Therapy (IGRT) is a newly developed technology for prostate cancer treatment. It consists of planning and treatment stages (Fig. 1(a)). In the planning stage, a planning CT scan is acquired and radiation oncologists manually delineate the prostate for treatment planning. In the treatment stage, to account for daily prostate motions, a treatment CT scan is acquired at each treatment day right before the radiation therapy. Guided by the treatment CTs, radiation oncologist adapt the treatment plan to precisely target radiation dose to the current positions of tumors and avoid neighboring healthy tissues. Consequently, IGRT increases the probability of tumor control and typically shortens radiation therapy schedules. In order to effectively adapt the treatment plan, it is critical to localize the prostate in the daily treatment images fast and accurately. Thus, an automatic localization algorithm becomes very desirable.
Fig. 1.
(a) Illustration of Image Guided Radiation Therapy. (b) Seven prostate anatomical landmarks used in our study.
However, prostate localization in treatment CT is quite challenging due to the low tissue contrast (Fig. 1(a)), unpredicted prostate motion [1], and potential large intra-patient anatomical variations that might happen across treatment days (pointed by red arrows in Fig. 1(a)). On the other hand, however, IGRT workflow provides a remarkable opportunity to the algorithm - at each treatment day, a few CT scans of the same patient have already been acquired in the planning and previous treatment stages. In addition, prostate boundaries have also been delineated in these images. If the prostate appearance characteristics of this specific patient can be learned from these patient-specific images, the algorithm can exploit this information to localize the prostate much more effectively.
To effectively exploit the patient-specific appearance characteristics, we propose to localize prostate in daily treatment CTs using a novel learning scheme, namely incremental learning with selective memory (ILSM), which leverages both the large number of population data (CT scans of other patients) and the very limited number of patient-specific data. Our learning framework starts with learning a population-based discriminative appearance model. This model is then “personalized” according to the appearance information from CTs of the specific patient under treatment. Instead of either preserving or discarding all knowledge learned from population, our method selectively inherits part of population-based knowledge that is in accordance with the current patient, and at the same time incrementally learns the patient-specific characteristics. This is where the name “incremental learning with selective memory” comes from. Once the population-based discriminative appearance model is personalized, it can be used to detect distinctive anatomical landmarks in new treatment images of the same patient for fast prostate localization.
Related Work
Costa et al. [2] proposed coupled deformable models to localize the prostate by considering the non-overlapping constraint from bladder. Chen et al. [3] used a Bayesian framework that integrates anatomical constraints from pelvic bones for prostate localization. Foskey et al. [4] proposed to use large deformable registration to localize the prostate by warping the planning image to daily treatment images. While these methods have exhibited certain effectiveness in CT prostate localization, without fully exploiting the patient-specific information, their performance is still limited. Recently, Liao et al. [5] proposed a feature-based registration method by exploiting the patient-specific information and achieved accurate prostate localization. However, it takes 3.8 minutes to localize the prostate. If the prostate unexpectedly moves during the long localization procedure, the localization result might become meaningless for IGRT.
2 Methodology
Overview
To localize the prostate in daily treatment images, we learn a set of local discriminative appearance models. Specifically, these models are used as anatomy detectors to detect distinctive prostate anatomical landmarks as shown in Fig. 1(b). The detected anatomical landmarks are further pruned by RANSAC [9]. Finally, based on the pruned landmarks, the prostate is localized by transforming the prostate surface delineated in the planning image (Fig. 1 (a)) onto the treatment image space. Fig. 2 shows the flowchart of our method.
Fig. 2.
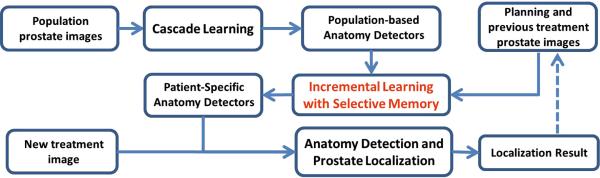
The flowchart of our CT prostate localization method
2.1 Cascade Learning for Anatomy Detection
Our prostate localization method relies on several prostate anatomical landmarks. Inspired by Viola's face detection work [6], we adopt a learning-based detection method, which formulates landmark detection as a classification problem. Specifically, for each image, the voxel of the specific landmark is positive and all others are negatives. In the training stage, we employ a cascade learning framework that aims to learn a sequence of classifiers to gradually separate negatives from positives (Fig. 3). Compared to learning a single classifier, cascade learning has shown better classification accuracy and runtime efficiency [6]. Mathematically, cascade learning can be formulated as:
Input: Positive voxel set , negative voxel set , and label set
Classifier: , denotes the appearance features of a voxel x.
Initial Set:
- Objective: Optimize Ck, k = 1, 2, · · · , K, such that
where Xk = {x|x ∈ Xk–1 and Ck(x) = +1}.
Fig. 3.
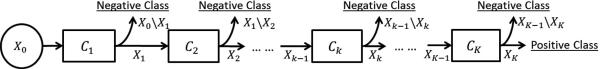
Illustration of cascade learning
The cascade classifiers Ck, k = 1, 2, · · · , K, are optimized sequentially. As shown in Eq. 1, Ck is optimized to minimize the false positives left over by the previous k – 1 classifiers.
| (1) |
where ∥.∥ denotes the cardinality of a set. It is worth noting that the constraint in Eq. (1) can be simply satisfied by adjusting the perceptron threshold of Ck [6] to make sure that all positive training samples are correctly classified. This cascade learning framework is general to any image feature and classifier. Extended Haar wavelets [7][8] and Adaboost classifier are employed in our study.
Once the cascade classifiers {Ck(x)} are learned, they have incorporated the appearance characteristics of the specific landmark. Given a testing image, the learned cascade is applied to each voxel. The voxel with the highest classification score after going through the cascade is considered as the detected landmark.
2.2 Incremental Learning with Selective Memory (ILSM)
Motivation
Using cascade learning, one can learn anatomy detectors from training images of different patients (population-based learning). However, since intra-patient anatomy variations are much less than inter-patient variations (Fig. 4), patient-specific appearance information available in IGRT workflow should be exploited to improve the detection accuracy on the specific patient. Unfortunately, the number of patient-specific images is often very limited, especially in the beginning of IGRT. As a result, cascade learning using only patient-specific data (pure patient-specific learning) often suffers from overfitting. One can also mix population and patient-specific images for training (mixture learning). However, since patient-specific images are the “minority” in the training samples, detectors trained by mixed samples might not capture patient-specific characteristics well. To address this problem, we propose a new learning scheme, ILSM, to combine the general information in the population images with the personal information in the patient-specific images. Specifically, population-based anatomy detector is served as an initial appearance model and is then “personalized” by the limited patient-specific data. Specifically, ILSM consists of backward pruning for discarding obsolete population appearance information and forward learning for incorporating the online-learned patient-specific appearance characteristics.
Fig. 4.
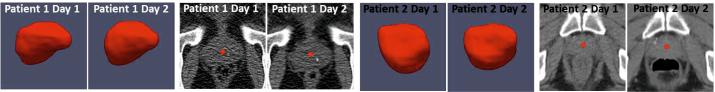
Inter- and intra-patient prostate shape and appearance variations
Notations
is the population-based anatomy detector (learned Sec. 2.1), which contains a cascade of classifiers. and are positives and negatives from the patient-specific training images, respectively. D(x) denotes the class label of voxel x predicted by detector D.
Backward Pruning
The general appearance model learned from population is not necessarily applicable to the specific patient. Specifically, the anatomy landmarks in the patient-specific images (positives) may be classified as negatives by the population-based anatomy detectors, i.e., , . In order to discard parts of population appearance model that do not fit the patient-specific characteristics, we propose backward pruning to tailor the population-based detector. As shown in Alg. 1, in backward pruning, the cascade is pruned from the last level until all patient-specific positives can pass through the cascade. It is equivalent to searching for the maximum number of cascade levels that could be preserved from the population-based anatomy detector (Eq. 2).
| (2) |
Forward Learning
Once the population cascade has been tailored, the remaining cascade of classifiers encodes the population appearance information that is consistent with the patient-specific characteristics. Yet till now no real patient-specific information has been incorporated into the cascade. More specifically, false positives might exist in the patient-specific samples, i.e., , , . In the forward learning stage, we will use the remaining cascade as an initialization, and adopt forward learning to eliminate the patient-specific false positives left over by the previously inherited population classifiers. As shown in Alg. 2, a greedy strategy is adopted to sequentially optimize a set of additional patient-specific classifiers .
After backward pruning and forward learning, the personalized anatomy detector includes two groups of classifiers. While encode patient-specific characteristics, contain population information applicable to this specific patient. This information effectively remedies the limited variability from small number of patient-specific training images.
2.3 Robust Surface Transformation
Once the population-based anatomy detectors are “personalized” by ILSM, they are used to detect the corresponding prostate anatomical landmarks (Fig. 1(b)) in new treatment images. To account for those erroneously detected landmarks (outliers), the relative positions of landmarks in the planning image are served as a reference geometric model, and RANSAC [9] algorithm is adopted to fit this model onto the detected landmarks for outlier removal and re-prediction. Finally, based on the refined landmarks, the prostate surface delineated in the planning image is transformed onto the treatment image space for fast localization. Considering the fact that prostate shape changes slightly under radiotherapy as well as computational efficiency, rigid transformation is used in our work.
3 Experimental Results
Data Description
Our experiment dataset consists of 349 prostate CTs. The prostate in each image is manually delineated by an expert to serve as ground-truth. Each patient has 14 CT scans on average. The typical image size of each CT scan is 512 × 512 × 60 with voxel size 1mm × 1mm × 3mm. Five-fold cross validation is used to evaluate the algorithm performance. For each fold, about 250 CT images are used to train the population-based detectors. τ in Alg. 2 is set to allow at most 1 false positive in each patient-specific training image.
Learning Approaches for Comparison
To illustrate the effectiveness of our learning framework, we first compare ILSM with other four learning-based approaches. All of these methods localize the prostate through learning-based anatomy detection with the same features, classifiers and cascade framework (as described in section 2.1). Their differences lie at the training images and learning strategies, which are shown in Table. 1. Note that for all patient-specific training images, artificial transformations are applied to increase the variability. To emulate the real clinical setting, for prostate localization in treatment day N + 1, we use previous N treatment images as patient-specific training data (Fig. 1).
Table 1.
Quantitative comparisons between ILSM and four learning-based methods. (POP: population-based learning; PPAT: pure patient-specific learning; MIX: population and patient-specific mixture learning; IL: incremental learning without backward pruning; ILSM: proposed incremental learning with selective memory.)
| POP | PPAT | MIX | IL | ILSM | ||
|---|---|---|---|---|---|---|
| Training images | Population | ✓ | ✓ | ✓ | ✓ | |
| Patient-specific | ✓ | ✓ | ✓ | ✓ | ||
| Learning strategies | Cascade Learning | ✓ | ✓ | ✓ | ✓ | ✓ |
| Backward Pruning | ✓ | |||||
| Forward Learning | ✓ | ✓ | ||||
| Evaluation | Mean DSC | 0.81 ± 0.10 | 0.84 ± 0.15 | 0.83 ± 0.09 | 0.83 ± 0.09 | 0.87 ± 0.06 |
| Acceptance Rate | 66% | 85% | 74% | 77% | 90% | |
| Landmark Error | 7.52mm | 6.25mm | 6.34mm | 6.50mm | 4.52mm |
Table 1 compared the four learning-based approaches with ILSM on our dataset. Here, “Acceptance Rate” denotes the ratio of images where an algorithm performs more accurate than inter-operator variability (DSC = 0.81) [4]. These results can be accepted without manual editing. We can see that ILSM achieves the best localization accuracy among them. Besides, not surprisingly, by utilizing patient-specific information, all three methods (i.e., PPAT, MIX and IL) outperform POP. However, their performances are still inferior to ILSM, which shows the effectiveness of ILSM.
Comparison with other CT Prostate Localization Methods
Our method can achieve localization accuracy at average DSC 0.87 and average surface distance 2.02 mm. Table 2 shows the comparison of our method with other four state-of-the-art methods, which employ deformable model (DM) [2][3] and registration [4][5]. Since neither their data nor their executables are publicly available, we only cited the numbers reported in their publications. Based on the reported performances, we achieve better localization accuracy than Costa [2], Chen [3] and Foskey [4]. While Liao [5] is slightly more accurate than ours, it takes much longer (228s vs. 4s) to localize the prostate. If the prostate unexpectedly moves during localization, that algorithm has to be performed again. Since the localization error of our method (DSC = 0.87 ± 0.06) is less than the inter-operator variability (DSC = 0.81 ± 0.06) [4], our method in fact well satisfies both accuracy and speed requirements of IGRT. For other applications requiring higher accuracy, our method can be combined with sophisticated segmentation methods (e.g., deformable model [10]) for better accuracy. The typical runtime for our method to localize the prostate is around 4 seconds (on an Intel Q6600 2.4GHz desktop with 4 GB memory). The offline training time for incremental learning is about 30 minutes per landmark detector.
Table 2.
Quantitative comparison with other prostate localization methods based on DSC, sensitivity (Sen.), positive predictive value (PPV.) and speed (seconds)
4 Conclusion
In this paper, we propose a novel learning scheme, namely incremental learning with selective memory (ILSM), which can take both specificity and generalization into account by leveraging population and patient-specific data. It is applied to extract the patient-specific appearance information in IGRT for accurate prostate localization. Validated on 349 CT images, our method can localize prostate accurately (mean DSC ~ 0.87) and fast (4 seconds).
Algorithm 1.
Backward pruning algorithm
| Input: - the population-based detector |
| - positive samples from the patient-specific training images |
| Output: Dbk - the tailored population-based detector |
| Init: k = Kpop, Dbk = Dpop. |
| while do |
| k = k – 1 |
| end while |
| Kbk = k |
| return |
Algorithm 2.
Forward learning algorithm
| Input: - the tailored population-based detector |
| - positive samples from the patient-specific training images |
| - negative samples from the patient-specific training images |
| τ - the parameter controlling the tolerance ratio of false positives |
| Output: Dpat - the patient-specific detector |
| Init: k = 1, Dpat = Dbk, |
| while do |
| Train the classifier by minimizing the equation below |
| k = k + 1 |
| end while |
| Kpat = k – 1 |
| return |
References
- 1.Liu W, Qian J, Hancock JL, Xing L, Luxton G. Clinical development of a failure detection-based online repositioning strategy for prostate IMRT— Experiments, simulation, and dosimetry study. Med. Phys. 2010;37(10):5287–5297. doi: 10.1118/1.3488887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa MJ, Delingette H, Novellas S, Ayache N. Automatic segmentation of bladder and prostate using coupled 3D deformable models. In: Ayache N, Ourselin S, Maeder A, editors. MICCAI 2007, Part I. LNCS. Vol. 4791. Springer; Heidelberg: 2007. pp. 252–260. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Lovelock DM, Radke RJ. Segmenting the prostate and rectum in CT imagery using anatomical constraints. Medical Image Analysis. 2011;15(1):1–11. doi: 10.1016/j.media.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Foskey M, Brad D, Lav G, Ed C, Nathalie S, Sandrine T, Julian R, Sarang J. Large deformation 3D image registration in image-guided radiation therapy. Physics in Medicine and Biology. 2005;50(24):5869–5892. doi: 10.1088/0031-9155/50/24/008. [DOI] [PubMed] [Google Scholar]
- 5.Liao S, Shen D. A Feature-Based Learning Framework for Accurate Prostate Localization in CT Images. IEEE TIP. 2012;21(8):3546–3559. doi: 10.1109/TIP.2012.2194296. [DOI] [PubMed] [Google Scholar]
- 6.Viola P, Jones M. Robust Real-Time Face Detection. IJCV. 2004;57(2):137–154. [Google Scholar]
- 7.Zhan Y, Dewan M, Harder M, Krishnan A, Zhou XS. Robust Automatic Knee MR Slice Positioning Through Redundant and Hierarchical Anatomy Detection. IEEE TMI. 2011;30(12):2087–2100. doi: 10.1109/TMI.2011.2162634. [DOI] [PubMed] [Google Scholar]
- 8.Zhan Y, Zhou XS, Peng Z, Krishnan A. Active Scheduling of Organ Detection and Segmentation in Whole-Body Medical Images. In: Metaxas D, Axel L, Fichtinger G, Székely G, editors. MICCAI 2008, Part I. LNCS. Vol. 5241. Springer; Heidelberg: 2008. pp. 313–321. [DOI] [PubMed] [Google Scholar]
- 9.Fischler MA, Bolles RC. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM. 1981;24(6):381–395. [Google Scholar]
- 10.Zhang S, Zhan Y, Dewan M, Huang J, Metaxas DN, Zhou XS. Deformable Segmentation via Sparse Shape Representation. In: Fichtinger G, Martel A, Peters T, editors. MICCAI 2011, Part II. LNCS. Vol. 6892. Springer; Heidelberg: 2011. pp. 451–458. [DOI] [PubMed] [Google Scholar]