Abstract
Objectives
Cytokines and related molecules in immune-response pathways seem important in deciding the outcome of the host–pathogen interactions towards different polar forms in leprosy. We studied the role of significant and functionally important single-nucleotide polymorphisms (SNPs) in these genes, published independently from our research group, through combined interaction with an additional analysis of the in silico network outcome, to understand how these impact the susceptibility towards the disease, leprosy.
Design
The study was designed to assess an overall combined contribution of significantly associated individual SNPs to reflect on epistatic interactions and their outcome in the form of the disease, leprosy. Furthermore, in silico approach was adopted to carry out protein–protein interaction study between PARK2 and proinflammatory/anti-inflammatory cytokines.
Setting
Population-based case–control study involved the data of North India. Protein–protein interaction networks were constructed using cytoscape.
Participants
Study included the data available from 2305 Northern Indians samples (829 patients with leprosy; 1476 healthy controls), generated by our research group.
Primary and secondary outcome measures
For genotype interaction analysis, all possible genotype combinations between selected SNPs were used as an independent variable, using binary logistic regression with the forward likelihood ratio method, keeping the gender as a covariate.
Results
Interaction analysis between PARK2 and significant SNPs of anti-inflammatory/proinflammatory cytokine genes, including BAT1 to BTNL2-DR spanning the HLA (6p21.3) region in a case–control comparison, showed that the combined analysis of: (1) PARK2, tumour necrosis factor (TNF), BTNL2-DR, interleukin (IL)-10, IL-6 and TGFBR2 increased the risk towards leprosy (OR=2.54); (2) PARK2, BAT1, NFKBIL1, LTA, TNF-LTB, IL12B and IL10RB provided increased protection (OR=0.26) in comparison with their individual contribution.
Conclusions
Epistatic SNP–SNP interactions involving PARK2 and cytokine genes provide an additive risk towards leprosy susceptibility. Furthermore, in silico protein–protein interaction of PARK2 and important proinflammatory/anti-inflammatory molecules indicate that PARK2 is central to immune regulation, regulating the production of different cytokines on infection.
Keywords: PARK2, Pro-/Anti- Inflammatory Cytokine Genes, Leprosy, Gene Interaction
Strengths and limitations of this study.
Many of the genetic studies lack replication in different population groups, explaining the heterogeneity in associated genes and genomic regions. This may be due to the complex nature of a disease. The complexity, however, could partly be delineated at genetic level by assessing the quantum of the contribution of different loci to the disease in a combined manner instead of their individual role.
Our study highlights the importance of a combined effect of the important cytokine and other immune-regulatory genes, whose combined effect in diverse genotype combinations provides either increased risk or protection towards the complex genetic disease, leprosy.
Genetic interaction and an additional in silico pathway analysis provided an overall perspective on how PARK2 gene product, parkin, acts as a centrally placed molecule, playing an important role in regulating different pathways of the immune response and susceptibility to leprosy. This conclusion needs further support with future experiments in vitro or in vivo of T cell responses in different genetic backgrounds of the identified networks in this study.
Introduction
Leprosy caused by Mycobacterium leprae is a chronic infectious disease, characterised by clinically defined polar forms in which pathology and immunology are inextricably related, providing a critical model to explore the immunoregulatory mechanisms in humans. At one pole, tuberculoid form is associated with a strong cell-mediated immunity (CMI) and T helper 1 (Th1) cytokine profile, and, at the other end of the spectrum, the lepromatous form is associated with a strong humoral response and Th2 cytokine profile. Cytokines and other related molecules of the immunological pathways thus seem to be a part of significant group of candidates that are apparently critical for the host–pathogen interactions, where the outcome of the disease is majorly dependent on the host factors controlling the immune response, especially when M leprae possesses the lowest level of genetic diversity.1 This is supported by various studies of familial clustering,2 twin studies,3 complex segregation analysis,4 5 test of analysis with the HLA genes6 including recent genome-wide association studies,7 8 and studies of several genes that modulate CMI, with a role in either susceptibility to leprosy per se or to leprosy types.9 Various candidate gene studies and genome-wide approaches have implicated polymorphisms in cytokine genes, whose protein products are part of important immune modulatory molecules, playing a major role in influencing host–pathogen interactions and determine the outcome of many infectious and autoimmune diseases.10–16 However, only a few observations have been replicated unequivocally in different population groups, suggesting the polygenic nature of the disease with a high degree of heterogeneity among different populations.
We, recently, have studied various candidate genes of proinflammatory/anti-inflammatory cytokines in two independent population groups, North and East India-Orissa, and found a strong association with interleukin (IL)-10, IL-10RB, TGFBR2, IL-614 and IL-12B.17 Fine-mapping of a specific 6p (HLA) chromosomal region revealed a significant association of important candidates, BAT1, LTA, tumour necrosis factor (TNF) and BTNL2.16 A subsequent study of the 6q chromosomal region, involving the overlapping regulatory domain of PARK2-PACRG genes, revealed an involvement of significant single-nucleotide polymorphisms (SNPs) and presence of a differential LD structure in Indian populations as compared with Vietnamese.18 The latter observation and the functional role of PARK2, as a ubiquitin ligase, has recently been shown in providing resistance to intracellular pathogens19 through ubiquitin-mediated autophagy. Furthermore, the involvement of parkin in regulating production of cytokines upon infection,20 indeed, provides a strong hint for any functional variations in the gene having a profound effect in modulating the expression of the immune-regulatory genes. The importance of all the studied genes14–18 in the network of immune-response necessitated the analysis of an interaction between these genes as a whole to understand their contribution together towards the susceptibility of the complex disease, leprosy, where the outcome of the infection in all probabilities depends on the nature of gene interactions between the genes with the potential of contributing to the immune pathology.
Therefore, the aim of this study was to assess an overall interaction between the significant and functionally important SNPs studied in a case–control comparison of the samples from New Delhi, in Northern India, where most of these SNPs were replicated in an unrelated East Indian-Orissa population. These included an overall interaction of the PARK2 gene significant SNPs18 with the significant SNPs of anti-inflammatory cytokine genes (IL-10, IL-10RB, TGFBR2, IL-6),14 proinflammatory cytokine genes (TNFα, LT-α, IL-12B) and the genes spanning the HLA region of the chromosome 6p21.3, that is, BAT1 to BTNL2-DR16 17 to evaluate their combined contribution towards the outcome of the complex infectious disease, leprosy.
Methods
The study involved the revisit of our published work on individual candidate genes and regions, studied in North Indian population groups in case–control comparison, for a combined genotype interaction and for in silico protein–protein interaction (PPI) and network analysis. The data compiled were of 2305 samples from Northern India (including 829 patients with leprosy and 1476 unrelated healthy control participants from North India)14 16–18 with a complete coverage of genes belonging to proinflammatory, anti-inflammatory cytokines, selected HLA regions in 6p21.3 and common regulatory region of PARK2/PACRG genes located at 6q26 region.
The patients’ group was classified according to the WHO guidelines. An individual was regarded as having leprosy if he or she showed skin lesion consistent with leprosy and with definite sensory loss, with or without thickened nerves and positive skin smears test. Furthermore, patients were classified as paucibacillary (PB) or multibacillary (MB) according to the Ridley and Jopling criteria,21 including 421 patients with PB and 408 patients with MB, with a mean age of 32.30±3.2 years (range 6–80 years). All these patients were under treatment with multidrug therapy specific for MB and PB leprosy, as recommended by the WHO.
For genotype interaction analysis, all possible genotype combinations between selected SNPs (pairwise or multiple genes) were ascertained from a MassArray platform for the given genotypes of SNPs. However, only the combinations of significantly associated SNP genotypes were presented in the Ms for convenience. These interactions were tested using binary logistic regression with the forward likelihood ratio-based selection method, considering all variables independently and keeping gender as a covariate. In this selection method, entry testing based on the significance of the score statistics and removal testing based on the probability of a likelihood ratio statistics were applied. Furthermore, in multiple gene interaction analysis, all interactions with either risk or protection were combined against other interactions to observe the overall effect of all risk versus protective interactions. These analyses were performed using statistical software package SPSS V.17.0 (SPSS, Chicago, Illinois, USA) for Windows. p Value was considered significant at and below 0.05.
In silico approach to assess the network of the genes in a PPI of PARK2, using Agile Protein Interaction Database (APID), a comprehensive resource for protein interaction data, automatically accessed by cytoscape22 through the dedicated plugin APID2NET,23 was carried out to understand the involvement of the studied interactome. APID integrates in a single web-based tool all known experimentally validated PPI from BIND,24 BioGRID,25 DIP,26 HPRD,27 IntAct28 and MINT29 databases.
Results
The interaction analysis carried out between PARK2 gene regulatory region SNPs (rs9365492 and rs9355403)18 and SNPs of the anti-inflammatory cytokines14 provided a significant risk towards the leprosy susceptibility, combining individually with SNPs of IL-10 (OR=1.99), IL-6 (OR=1.33) and TGFBR2 (OR=1.29) cytokine genes. However, with IL10RB (receptor β), the result showed a significant protection towards the disease (OR=0.61). Similar analysis between PARK2 SNPs with proinflammatory cytokine genes TNFα and BTNL2-DRA interval (showing strong LD with the BTNL2 promoter SNPs)16 provided a significant risk towards leprosy susceptibility with OR=2.10 and 5.40, respectively. However, the SNPs of BAT-1, NFKABIL1, LTA, TNF-LTB and IL12B16 17 provided a significant protection towards leprosy with OR=0.65, 0.58, 0.61, 0.54 and 0.71, respectively (table 1 and see online supplementary figure S1).
Table 1.
Genotype interaction analysis of PARK2 SNPs with proinflammatory /anti-inflammatory cytokines gene SNPs providing risk/protection towards leprosy susceptibility
| PARK2 (SNPs are within 63.8 kb upstream gene region) | Samples (n) |
95% CI for EXP(B) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNPs providing risk | Patients | Controls | Significance | OR | Lower | Upper | ||||
| rs9365492; minor, risk allele-C | rs9355403; minor, risk allele-A | IL-10 | rs1800871 (−819); minor, risk allele-T | rs1554286 (intron 3 boundary); minor, risk allele-T | rs1800872 (−592); minor, risk allele-A | ||||||
| TC+CC | GA+AA | CT+TT | TT | CA+AA | 82 | 84 | 3.22E-05 | 1.997 | 1.441 | 2.767 | |
| TGFBR2 | rs2228048 (3' UTR downstream); minor, risk allele-T | rs744751 (3′ UTR downstream); minor, risk allele-G | |||||||||
| CT+CC | GG | 287 | 400 | 1.04E-02 | 1.293 | 1.062 | 1.575 | ||||
| IL-6 | rs1800797 (−718); minor, risk allele-G | ||||||||||
| GG | 320 | 432 | 2.90E-03 | 1.333 | 1.103 | 1.611 | |||||
| TNF | rs1800629 (−308); minor, risk allele-G | rs1800610 (Intron-1); minor, risk allele-G | |||||||||
| GG | GG | 311 | 420 | 2.06E-09 | 2.103 | 1.649 | 2.682 | ||||
| BTNL2-DRA interval | rs3135365; minor, risk allele-C | rs7773756; minor, risk allele-T | |||||||||
| CA+CC | CT+TT | 272 | 269 | 1.22E-21 | 5.4 | 3.821 | 7.631 | ||||
| SNPs providing protection | |||||||||||
| TT | GG | LTA | rs13192469 (13 kb upstream); minor, risk allele-C | rs36221459 (−1409); minor, risk allele-DEL | |||||||
| TT | GTTT | 240 | 571 | 3.56E-07 | 0.616 | 0.512 | 0.743 | ||||
| IL-10RB2 | rs3171425 (3’ UTR); major, risk allele-G | rs7281762 (3′ UTR downstream); minor, risk allele-A | |||||||||
| GA+AA | GA+GG | 164 | 391 | 1.10E-05 | 0.61 | 0.489 | 0.76 | ||||
| BAT1 | rs2523504 (−603); minor, risk allele-T | ||||||||||
| CC | 192 | 475 | 4.15E-05 | 0.645 | 0.523 | 0.795 | |||||
| NFKBIL | rs2230365 (exon-3); minor, risk allele-T | ||||||||||
| CC | 195 | 486 | 1.01E-07 | 0.589 | 0.484 | 0.715 | |||||
| TNF-LTB | rs769178 (gene downstream); major, risk allele-G | ||||||||||
| GT+TT | 66 | 211 | 8.93E-05 | 0.546 | 0.404 | 0.739 | |||||
| IL12B | rs2853694; major, risk allele-A | ||||||||||
| CA+CC | 233 | 519 | 5.03E-04 | 0.705 | 0.579 | 0.858 | |||||
IL, interleukin; SNP, single-nucleotide polymorphism; TNF, tumour necrosis factor.
In the second step of combined interaction analysis with all the genes, providing either protection or risk towards leprosy, showed that the combined genotypic interaction analysis of the SNP loci PARK2, TNF, BTNL2-DR, IL10, IL-6 and TGFBR2 further increased the risk of leprosy (OR=2.54), and a similar combined analysis for loci PARK2, BAT1, NFKBIL1, LTA, TNF-LTB, IL12B and IL10RB increased the protection towards leprosy (OR=0.26) in comparison with their individual contribution (table 2A,B). Dividing the patients into PB and MB subtypes of leprosy revealed PB subtype to carry a higher risk (OR=3.02) and protection (OR=0.11) towards leprosy in comparison with MB subtype, for respective combinations.
Table 2.
Combined interaction analysis of all the SNPs providing either protection or risk towards leprosy susceptibility
| (A) Analysis of SNPs providing protection | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples (n) |
PARK2 | PARK2 | BAT1 promoter | NFKBIL | LTA 13 kb upstream | LTA promoter | TNF-LTB | IL10RB | IL10RB | IL12B | Significance | OR | 95% CI for EXP(B) | |||
| Pat | Cont | rs9365492 | rs9355403 | rs2523504 | rs2230365 | rs13192469 | rs36221459 | rs769178 | rs3171425 | rs7281762 | rs2853694 | Lower | Upper | |||
| Alleles | T/C | G/A | C/T | C/T | T/C | GTTT/DEL | G/T | G/A | G/A | A/C | ||||||
| Risk allele | C | A | T | T | C | DEL | G | G | A | A | ||||||
| TOTAL | 10 | 59 | TT | GG | CC | CC | TT | GTTT | GT+TT | GA+AA | GA+GG | CA+CC | 1.15E-04 | 0.263 | 0.133 | 0.518 |
| PB/HC | 2 | 59 | TT | GG | CC | CC | TT | GTTT | GT+TT | GA+AA | GA+GG | CA+CC | 2.36E-03 | 0.111 | 0.027 | 0.458 |
| MB/HC | NA | |||||||||||||||
| (B) Analysis of SNPs providing risk | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples (n) |
PARK2 | PARK2 | IL-10 | IL-10 | IL-10 | IL-6 | TGFBR2 | TGFBR2 | TNF (-308) promoter | TNF intron 1 | BTNL2-DRA interval | BTNL2-DRA interval | Significance | OR | 95% CI for EXP(B) |
|||
| Pat | Cont | rs9365492 | rs9355403 | rs1800871 | rs1554286 | rs1800872 | rs1800797 | rs2228048 | rs744751 | rs1800629 | rs1800610 | rs3135365 | rs7773756 | Lower | Upper | |||
| Alleles | T/C | G/A | C/T | C/T | C/A | G/A | C/T | G/A | G/A | G/A | A/C | C/T | ||||||
| Risk allele | C | A | T | T | A | G | T | G | G | G | C | T | ||||||
| Total | 57 | 44 | TC+CC | GA+AA | CT+TT | CT+CC | CA+AA | GG | CT+CC | GA+AA | GG | GG | CA+CC | CT+TT | 5.77E-06 | 2.543 | 1.699 | 3.806 |
| PB/HC | 32 | 44 | TC+CC | GA+AA | CT+TT | CT+CC | CA+AA | GG | CT+CC | GG | GG | GG | CA+CC | CT+TT | 3.73E-06 | 3.028 | 1.894 | 4.843 |
| MB/HC | 25 | 44 | TC+CC | GA+AA | CT+TT | CT+CC | CA+AA | GG | CT+CC | GG | GG | GG | CA+CC | CT+TT | 3.16E-03 | 2.133 | 1.29 | 3.528 |
IL, interleukin; MB, multibacillary; PB, paucibacillary; SNP, single-nucleotide polymorphism; TNF, tumour necrosis factor.
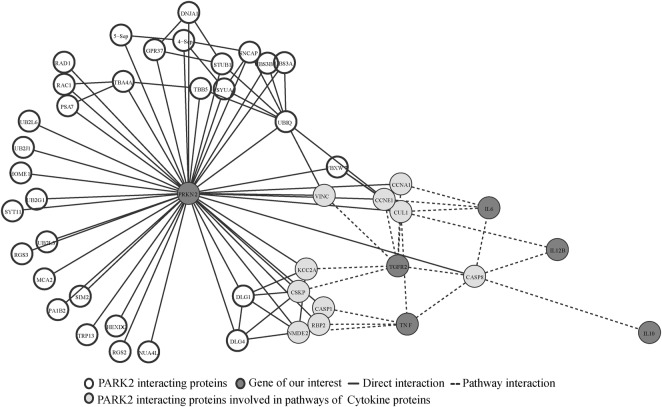
We further performed in silico analysis to identify the PPI of PARK2. We used APID2NET and cytoscape tools for PARK2 interaction Data retrieval, providing a total of 43 PARK2 interacting proteins. However, the result did not provide any direct interaction of the PARK2 with the cytokines studied by us in North Indian population.14 16–18 Furthermore, we considered 43 PARK2 interacting proteins for pathways analysis by using KEGG, BioCarta, Nci-Nature and Reactome tools, which confirmed these 43 proteins to be involved in 253 different pathways (without removing overlapping pathways). Similarly, in the second step of pathway analysis, we considered 11 cytokine proteins studied by us in North Indian population,14 16–18 and the results revealed the involvement of five cytokine proteins; IL12B, IL6, TNF, TGFR2 and IL10 in 94 pathways, not involving BTNL2, BAT1, NFKBIL, LTA, IL10RB2 and BTNL2-DR in any pathways. Comparing both pathways, 253 PARK2 interacting proteins pathways and 94 cytokine proteins pathways revealed 27 commonly involved pathways, via CASP8, CUL1, CCNE1 and CCNA proteins, involving only 5 (IL12B, IL6, TNF, TGFR2 and IL10) of 11 cytokine proteins studied in North Indian population (figure 1), connecting majorly through Toll-like receptor (TLR) signalling pathways (figure 1, see online supplementary table S1).
Figure 1.
PARK2 interaction analysis: unfilled circles showing the PARK2 interacting proteins. Light grey circles showing protein links between the PARK2 interacting protein and five cytokines protein (dark grey circles) study by us in North India population.
Discussion
Leprosy, an ideal model of a chronic human complex infectious disease, provides an opportunity to dissect the components of the host-dependent polygenic susceptibility to this disease. Many loci have been shown to be individually associated and providing the risk towards the disease; justifying to find out interesting gene–gene interactions at different risk loci which may prove to provide a strong association towards the disease susceptibility. In order to understand the role of multiple genes together, an interaction analysis was carried out between the genotype status of functionally different variants of different genetic loci involved in immune response, with an expected combined effect on the outcome of the disease in different polar forms of the disease.
Considering the above facts, we first carried out pairwise interaction analysis of PARK2 gene with proinflammatory/anti-inflammatory cytokine genes (table 1), followed by multiple gene interaction analysis (table 2). Analysis of PARK2 with TNF, BTNL2-DR, IL10, IL-6 and TGFBR2 showed an increased risk towards leprosy (OR=2.54 (1.69 to 3.80), p=5.77e-06), while the combined analysis of PARK2 with BAT1, NFKBIL1, LTA, TNF-LTB, IL12B and IL10RB showed protection towards the disease (OR=0.26 (0.13 to 0.51), p=1.15e-04). PARK2, encoding E3 ubiquitin ligase protein-parkin, has been shown to be involved in the cellular ubiquitination metabolism,30 providing resistance to intracellular pathogen via ubiquitin-mediated autophagy,19 essentially shown to be involved in the host responses to M leprae31 and for pathogenesis of the disease.8 32 Recently, parkin protein has shown to be involved to respond to infection in a regulated way by producing important cytokines,20 suppressing molecules that limit proinflammatory-IL-2,33 TNFα cytokine production and enhancing the production of anti-inflammatory cytokines, IL-4, IL-10 and IL-13.34–39 All these observations indicate the in vivo importance of PARK2 gene product, parkin, to be centrally involved in regulating the production of critical cytokines during immune response against the invading mycobacterium and justifying our study, where combination of risk genotype at different loci of important immune response gene with PARK2 provides increased and significant risk towards this complex disease. These interesting results of gene–gene interaction analysis suggest the in vivo effect of the invading mycobacterium in future, where immune response to specific antigens is assessed in cells with different background of important variations in the PARK2 promoter region followed by the effect on the expression levels of proinflammatory/anti-inflammatory cytokines.
An in silico approach was used to understand the role of immune-regulatory PPI between PARK2 and other cytokine genes, and an indirect interaction was observed between PARK2 and IL12B, IL6, TNF, TGFR2 and IL10 genes. All these interactions were found to be connected with TLR signalling pathway (see online supplementary table S1). As already known, the polymorphisms in different TLRs, important molecules of innate immune response, are associated with leprosy and its subtypes,7 40–44 influencing recognition of M leprae. A simultaneous involvement of PARK2, a ubiqutin ligase protein involved in innate immunity by modulating the production of important cytokines, including IL6,20 hints at the involvement of all these important molecules to be interconnected through a TLR receptor signalling pathway to fight against the invading mycobacterium.
The above interaction and pathway analysis allows us to propose that the complex genetic background is the predominant factor for the outcome of the disease, where the combined effect of the variant risk alleles of the PARK2 gene, responsible for affecting transcription binding site and lowering the expression of the reporter gene by in vitro experiment,18 along with the risk alleles of the anti-inflammatory cytokine genes—IL-10, IL-6, TGFBR2, responsible for lowering the CMI response towards the invading bacteria and proinflammatory cytokines—TNFα, is responsible in providing highly significant risk towards leprosy. The study opens a way for future in vivo work of immune-response readouts in complex variant genomic backgrounds to understand the wide gap in understanding the balance in the network of all the immune regulatory molecules operational in providing either susceptibility or resistance towards disease.
Supplementary Material
Acknowledgments
The authors would like to thank all who participated in the study. They also thank Dr Shobit Caroli and Dr Sudhanshu, Dr Shalu the attending physicians at LNJP Hospital and Professor Jason P Sinnwell for his invaluable support in providing Haplo.Stats package.
Footnotes
Contributors: RNKB and RC contributed in planning, designing and execution of work and wrote the article; RC, SA, PK, SA and AKS contributed in biostatistics and in silico analysis; VKG and SNB contributed in patient evaluation, clinical categorisation and discussion. All authors critically reviewed the manuscript. RNKB led the research effort.
Funding: Financial and Infrastructural support for the production of manuscript was provided by Shri Mata Vaishno Devi University, University Grants Commission; and high-throughput project from DBT to the National Center of Applied Human Genetics.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: Approved by Jawaharlal Nehru University ethics committee, New Delhi, India.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.Monot M, Honore N, Garnier T, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 2009;41:1282–9 [DOI] [PubMed] [Google Scholar]
- 2.Shields ED, Russell DA, Pericak-Vance MA. Genetic epidemiology of the susceptibility to leprosy. J Clin Invest 1987;79:1139–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravartti M, Vogel F. A twin study on leprosy. Topics in human genetics. Vol. 1 Stuttgart, Germany: Georg Thieme Verlag, 1973:1–123 [Google Scholar]
- 4.Abel L, Demenais F. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Desirade island. Am J Hum Genet 1988;42:256–66 [PMC free article] [PubMed] [Google Scholar]
- 5.Abel L, Vu DL, Oberti J, et al. Complex segregation analysis of leprosy in southern Vietnam. Genet Epidemiol 1995;12:63–82 [DOI] [PubMed] [Google Scholar]
- 6.Todd JR, West BC, McDonald JC. Human leukocyte antigen and leprosy: study in northern Louisiana and review. Rev Infect Dis 1990;12:63–74 [DOI] [PubMed] [Google Scholar]
- 7.Wong SH, Gochhait S, Malhotra D, et al. Leprosy and the adaptation of human toll-like receptor 1. PLoS Pathog 2010;6:e1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med 2009;361:2609–18 [DOI] [PubMed] [Google Scholar]
- 9.Fitness J, Tosh K, Hill AV. Genetics of susceptibility to leprosy. Genes Immunity 2002;3:441–53 [DOI] [PubMed] [Google Scholar]
- 10.Gomolka M, Menninger H, Saal JE, et al. Immunoprinting: various genes are associated with increased risk to develop rheumatoid arthritis in different groups of adult patients. J Mol Med 1995;73:19–29 [DOI] [PubMed] [Google Scholar]
- 11.Mok CC, Lanchbury JS, Chan DW, et al. Interleukin-10 promoter polymorphisms in Southern Chinese patients with systemic lupus erythematosus. Arthritis Rheum 1998;41:1090–5 [DOI] [PubMed] [Google Scholar]
- 12.Loughrey BV, Maxwell AP, Fogarty DG, et al. An interluekin 1B allele, which correlates with a high secretor phenotype, is associated with diabetic nephropathy. Cytokine 1998;10:984–8 [DOI] [PubMed] [Google Scholar]
- 13.Morahan G, Huang D, Wu M, et al. Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet 2002;360:455–9 [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal S, Ali S, Chopra R, et al. Genetic variations and interactions in anti-inflammatory cytokine pathway genes in the outcome of leprosy: a study conducted on a MassARRAY platform. J Infect Dis 2011;204:1264–73 [DOI] [PubMed] [Google Scholar]
- 15.Alcais A, Alter A, Antoni G, et al. Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat Genet 2007;39:517–22 [DOI] [PubMed] [Google Scholar]
- 16.Ali S, Chopra R, Aggarwal S, et al. Association of variants in BAT1-LTA-TNF-BTNL2 genes within 6p21.3 region show graded risk to leprosy in unrelated cohorts of Indian population. Hum Genet 2012;131:703–16 [DOI] [PubMed] [Google Scholar]
- 17.Ali S, Srivastava AK, Chopra R, et al. IL12B SNPs and copy number variation in IL23R gene associated with susceptibility to leprosy. J Med Genet 2013;50:34–42 [DOI] [PubMed] [Google Scholar]
- 18.Chopra R, Ali S, Srivastava AK, et al. Mapping of PARK2 and PACRG overlapping regulatory region reveals LD structure and functional variants in association with leprosy in unrelated Indian population groups. PLoS Genet 2013;9:e1003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manzanillo PS, Ayres JS, Watson RO, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013;501:512–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Leseleuc L, Orlova M, Cobat A, et al. PARK2 mediates interleukin 6 and monocyte chemoattractant protein 1 production by human macrophages. PLoS Negl Trop Dis 2013;7:e2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danielssen DC, Boeck W, Losting JL. Om Spedalskhed [on leprosy]. Christiana Chr Grondahl 1847
- 22.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Toro J, Prieto C, De las Rivas J. APID2NET: unified interactome graphic analyzer. Bioinformatics 2007;23:2495–7 [DOI] [PubMed] [Google Scholar]
- 24.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res 2003;31:248–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res 2013;41(Database issue):D816–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salwinski L, Miller CS, Smith AJ, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res 2004;32(Database issue):D449–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human protein reference database—2009 update. Nucleic Acids Research 2009;37(Database issue):D767–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerrien S, Aranda B, Breuza L, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res 2012;40(Database issue):D841–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Licata L, Briganti L, Peluso D, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 2012;40(Database issue):D857–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 2000;25:302–5 [DOI] [PubMed] [Google Scholar]
- 31.Mira MT, Alcais A, Nguyen VT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 2004;427:636–40 [DOI] [PubMed] [Google Scholar]
- 32.Schurr E, Alcais A, de Leseleuc L, et al. Genetic predisposition to leprosy: a major gene reveals novel pathways of immunity to Mycobacterium leprae. Semin Immunol 2006;18:404–10 [DOI] [PubMed] [Google Scholar]
- 33.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat Immunol 2004;5:883–90 [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Covarrubias L, Manning EW, III, Sorell LT, et al. Ubiquitin enhances the Th2 cytokine response and attenuates ischemia-reperfusion injury in the lung. Crit Care Med 2008;36:979–82 [DOI] [PubMed] [Google Scholar]
- 35.Majetschak M. Extracellular ubiquitin: immune modulator and endogenous opponent of damage-associated molecular pattern molecules. J Leukoc Biol 2011;89:205–19 [DOI] [PubMed] [Google Scholar]
- 36.Majetschak M, Krehmeier U, Bardenheuer M, et al. Extracellular ubiquitin inhibits the TNF-alpha response to endotoxin in peripheral blood mononuclear cells and regulates endotoxin hyporesponsiveness in critical illness. Blood 2003;101:1882–90 [DOI] [PubMed] [Google Scholar]
- 37.Patel MB, Majetschak M. Distribution and interrelationship of ubiquitin proteasome pathway component activities and ubiquitin pools in various porcine tissues. Physiol Res 2007;56:341–50 [DOI] [PubMed] [Google Scholar]
- 38.Saini V, Romero J, Marchese A, et al. Ubiquitin receptor binding and signaling in primary human leukocytes. Commun Integr Biol 2010;3:608–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh M, Roginskaya M, Dalal S, et al. Extracellular ubiquitin inhibits beta-AR-stimulated apoptosis in cardiac myocytes: role of GSK-3beta and mitochondrial pathways. Cardiovasc Res 2010;86:20–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bochud PY, Hawn TR, Siddiqui MR, et al. Toll-like receptor 2 (TLR2) polymorphisms are associated with reversal reaction in leprosy. J Infect Dis 2008;197:253–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bochud PY, Sinsimer D, Aderem A, et al. Polymorphisms in Toll-like receptor 4 (TLR4) are associated with protection against leprosy. Eur J Clin Microbiol 2009;28:1055–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson CM, Lyle EA, Omueti KO, et al. Cutting edge: a common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol 2007;178:7520–4 [DOI] [PubMed] [Google Scholar]
- 43.Misch EA, Macdonald M, Ranjit C, et al. Human TLR1 deficiency is associated with impaired mycobacterial signaling and protection from leprosy reversal reaction. PLoS Negl Trop Dis 2008;2:e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuring RP, Hamann L, Faber WR, et al. Polymorphism N248S in the human Toll-like receptor 1 gene is related to leprosy and leprosy reactions. J Infect Dis 2009;199:1816–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.