Abstract
In methicillin-resistant Staphylococcus aureus, β-lactam antibiotic resistance is mediated by the transmembrane protein BlaR1. The antibiotic-sensor domain BlaRS and the L2 loop of BlaR1 are on the membrane surface. We used NMR to investigate interactions between BlaRS and a water-soluble peptide from L2. This peptide binds BlaRS proximal to the antibiotic acylation site as an amphipathic helix. BlaRS acylation by penicillin G does not disrupt binding. These results suggest a signal transduction mechanism whereby the L2 helix, partially embedded in the membrane, propagates conformational changes caused by BlaRS acylation through the membrane via transmembrane segments, leading to antibiotic resistance.
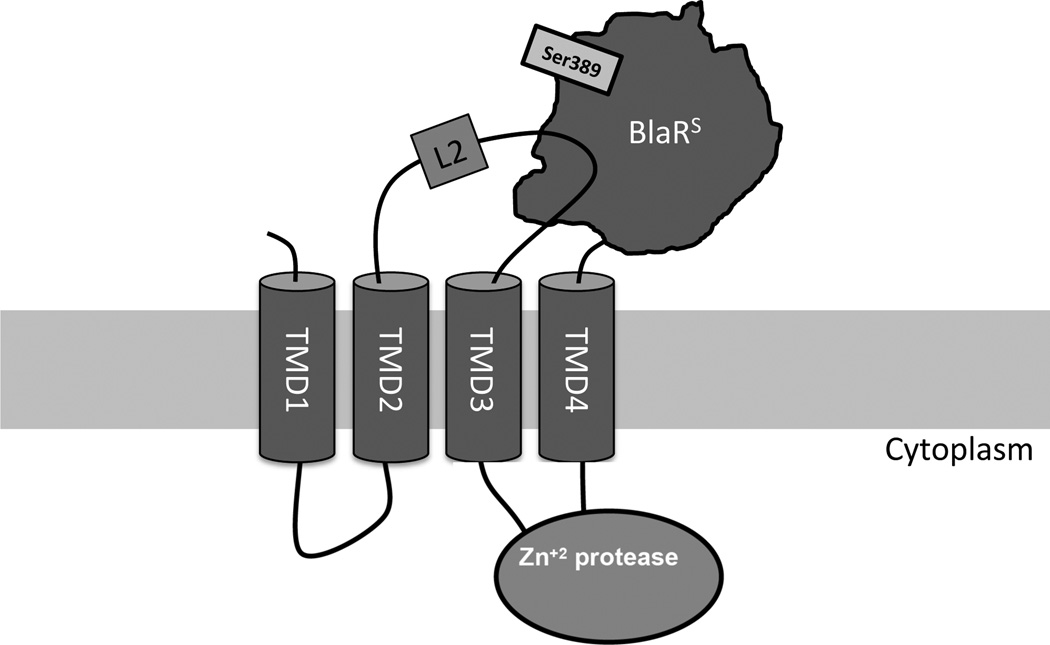
Methicillin-resistant Staphylococcus aureus (MRSA) is a global clinical scourge that has become resistant to virtually all β-lactam antibiotics. In many MRSA strains, the β-lactam resistance is induced by a transmembrane sensor/transducer protein, BlaR1 (Figure 1). The induction begins when the extracellular sensor domain of BlaR1, BlaRS, becomes acylated at Ser389 by a β-lactam antibiotic. Transduction of this signal to the BlaR1 cytoplasmic domain leads to transcription and expression of antibiotic resistance determinants (1–3).
Figure 1.
The arrangement of domains in the BlaR1 protein of methicillin-resistant S. aureus.
The initial events related to acylation are not fully understood. Previous studies of BlaR1 from Bacillus licheniformis (4) demonstrated an interaction between the aforementioned BlaRS and the extracellular transmembrane loop L2 (Figure 1). It is reasonable to hypothesize that this interaction plays a role in signal transduction. Herein, we describe studies that disclose the nature of the interactions between BlaRS and the L2 loop, and clarify the early events leading to transduction of the acylation signal through the membrane in Staphylococcus aureus.
We investigated the BlaRS/L2 interactions through solution NMR studies of the isolated sensor domain, BlaRS (residues 330–585) and a peptide corresponding to the C-terminal 33 amino acids of L2 (residues 73–105). The full-length L2 peptide (residues 39–105) was insoluble; therefore, we used the soluble truncated construct, L2short, for all NMR experiments.
Backbone amide 15N spin relaxation parameters (R1, R2, and heteronuclear NOE) of U-[15N] L2short in the absence versus presence of equimolar BlaRS (300 µM) indicated L2short undergoes rapid exchange between a major free state and a minor BlaRS-bound state (Supporting Information). From R1 decreases, we estimated an equilibrium dissociation constant (Kd) of 1.3 ± 0.4 mM. In intact BlaR1, co-localization of L2short and BlaRS on the membrane surface would impose a favorable entropic factor in their interactions that the millimolar Kd does not reflect. A reduced spectral density determination of L2short Jeff(0) values suggested the C-terminal residues 94–102 contact BlaRS directly (Figure S1).
Determining where L2short binds BlaRS was of key interest. Detecting intermolecular NOEs proved unsuccessful, most likely because of the weak binding affinity (5, 6). We therefore used paramagnetic relaxation enhancement (PRE) measurements, which are well suited for fast-exchange binding interactions (7–9). Our PRE measurements reported herein directly established the location and binding mode of the L2short/BlaRS interaction, which were inconclusive from chemical shift perturbations (CSPs), NOEs, and line broadening.
To do the PRE measurements, we generated a T92C variant of L2short, and attached paramagnetic and diamagnetic moieties consisting of MTSL and acetyl-MTSL, respectively (Figure S2). We chose T92 because of its proximity to the L2-binding interface residues revealed by Jeff(0) (Figure S1). We then measured the amide proton transverse relaxation rates R2(1HN) of [U-15N, 80% 2D] BlaRS in the presence of the paramagnetic and diamagnetic T92C variants. The PREs were the differences between the paramagnetic and diamagnetic R2(1HN) relaxation rates, namely, (8, 9). BlaRS amide protons with large Γ2 were sites that experienced greater electron-nuclear dipolar relaxation, indicating proximity to the L2short spin-label, and thus, involvement in the binding interface.
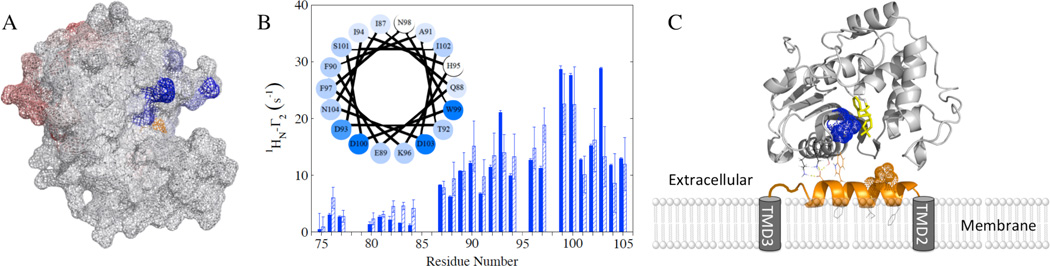
Two regions of BlaRS gave significant PREs, suggesting two binding sites (Figure 2A). The first region (blue) was proximal to the antibiotic-binding site (site of acylation), which includes residues in the β5–β6 turn. The second region (red) was distal from the antibiotic-binding site, and includes residues in the β6–β7 turn. The distal site PRE values were smaller, reflecting a different binding mode with larger intermolecular distances, lower binding affinity, or both. Addition of 10:1 L2short to 300 µM [U-15N, 80% 2D] BlaRS resulted in BlaRS CSPs that corroborate the PRE results (Figure S3).
Figure 2.
(A) BlaRS PRE Γ2 rates caused by spin-labeled L2short T92C mapped onto the BlaRS crystal structure (PDB 3Q7V, chain B). Blue (β5–β6 turn) and red (β6–β7 turn) indicate the two interaction regions. Darker shading indicates larger 1HN-Γ2. The antibiotic-binding-site residue S389 is in orange. (B) L2short PRE 1HN-Γ2 rates caused by IAP-labeled BlaRS I531C without (dark blue) and with (dashed) penicillin G. The helical wheel includes I87-N104 of L2short, with darker shading for larger 1HN Γ2 rates. (C) Model for L2 interaction with BlaRS, based on PREs from I531C (BlaRS) and T92C (L2short), and CSPs from the β5–β6 turn. Coloring: BlaRS (gray), L2short helix (orange helix), I531 (blue spheres), acylation site (penicillin G in yellow sticks).
We corroborated these results from the L2short perspective, by spin-labeling BlaRS and looking for amide protons PREs in [U-15N] L2short. We made two BlaRS variants for iodoacetamido-proxyl (IAP) (Figure S2) spin-labeling: I531C and N548C. I531 lies within the proximal L2short binding site (β5–β6 turn), while N548 is within the distal L2short binding site (β6–β7 turn).
The L2short PREs highlighted the same C-terminal region (residues 94–102) as the 15N Jeff(0) results; thus, these L2short residues clearly contribute to the binding interface. The L2short PREs caused by I531C (Figure 2B) were significantly greater than those caused by N548C; this is consistent with converse PRE experiments involving T92C. Figure S4 compares the two PRE profiles.
The L2short PREs from I531C showed a distinct undulation for residues 87–104, which indicated cyclic proximity of these residues to spin-labeled I531C. The pattern was consistent with an amphipathic α-helix (Figure 2B). 1H-1H NOESY spectra of L2short in the presence of BlaRS confirmed this α-helical model by giving the characteristic sequential 1HN-1HN NOEs (Figure S5) (10).
Figure 2C depicts our provisional model of the L2short/BlaRS binding mode at the proximal site; this was derived from HADDOCK (11) calculations using our PRE-derived intermolecular distances (12) and BlaRS CSPs. The L2short residues in the binding interface are mainly in the polar hydrophilic face of the α-helix, with putative interactions between D100 of L2short and K535/K562 of BlaRS. An exception is W99, which gave a very strong PRE. Hydrophobic interactions between W99 and BlaRS residues I531 and Y536 may enhance binding. Otherwise, the hydrophobic patches on the helix are opposite the BlaRS surface, allowing for partial embedding into the membrane (Figure 2C).
The binding mode for L2short at the BlaRS distal interaction site was unclear. The smaller L2short PREs precluded assessment of a similar amphipathic α-helix. The mainly polar residues at the distal site suggest binding is dominated by electrostatic interactions. PRE experiments at higher salt supported this hypothesis. The higher salt reduced the distal site PRE to near noise levels; by contrast, the proximal site PREs remained prominent, albeit, at a reduced level (Figure S3). HADDOCK modeling deems unlikely a scenario in which one L2short binds the proximal and distal sites simultaneously. Rather, L2short binds one or the other. The weaker PRE response and the lack of evidence for structured binding suggest the distal binding site reflects a non-specific interaction.
A natural question is whether the α-helicity of L2short is induced upon binding BlaRS. Far-UV circular dichroism (CD) measurements showed that the isolated L2short is disordered in solution (Figure S6). Yet, the same isolated L2short kept the α-helix NOE pattern (10) seen in the presence of BlaRS (Figure S5). These results are not contradictory: the r−6 distance dependence of the 1H-1H NOE is sensitive to sparsely populated conformers with short inter-proton distances (13, 14), such as that found within an α-helix. Together, the CD and NMR results suggest that the isolated L2short transiently samples the bound-state helix, which stabilizes upon binding BlaRS (conformational selection).
Finally, we investigated the effect of β-lactam acylation of BlaRS on the L2short interaction. For acylation, we added penicillin G (penG) to a five-fold molar excess over BlaRS. BlaRS 15N-1H CSPs monitored during a penG titration indicated that this excess was sufficient for acylation throughout the experiment. We then introduced [U-15N] L2short in a 1:1 ratio with the pre-acylated BlaRS. Both 15N relaxation (Figure S1) and PRE measurements for L2short (Figure S4) showed only minimal changes compared to the unacylated complex. Thus, acylation of BlaRS by the antibiotic does not disrupt its interaction with L2short, as suggested earlier (4). However, residue-specific changes remain possible and are likely contributors of the signal transduction mechanism. This lack of disruption of the complex on antibiotic binding is consistent with the L2short binding site being proximal to the acylation site, rather than occluding it.
In conclusion, our studies document the nature of the intramolecular interactions between L2 and the BlaRS sensor domain on the membrane surface, which are important for antibiotic recognition and signal transduction. BlaRS acylation by penG does not disrupt the interaction. The proximity of L2 to the membrane surface suggests that the amphipathic α-helix (Figure 2C) partially embeds itself into the membrane. Thus, we assert that L2/BlaRS interaction: (i) helps anchor the sensor domain to the membrane surface; and (ii) provides the means whereby conformational perturbations in the sensor domain BlaRS, resulting from its acylation, propagate through the membrane to initiate the events in the cytoplasm that lead to full-blown antibiotic resistance. This picture for the first time reveals the specific interactions and early events on the membrane surface of MRSA required for signaling from the cell exterior to the cytoplasm.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Leticia Llarrull, Mr. Michael Staude, Dr. Xingsheng Wang, Dr. Mandy Blackburn, and Dr. Gail Fanucci for valuable discussions.
Funding Sources
This work was supported by NIH grants AI104987 (to SM), and GM085109 (to JWP).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Materials and methods for all experiments and supporting figures S1–S6 can be accessed free of charge online at http://pubs.acs.org.
No competing financial interests have been declared.
References
- 1.Llarrull LI, Fisher JF, Mobashery S. Antimicrob Agents Chemother. 2009;53:4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llarrull LI, Toth M, Champion MM, Mobashery S. J Biol Chem. 2011;286:38148–38158. doi: 10.1074/jbc.M111.288985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llarrull LI, Mobashery S. Dissection of events in the resistance to beta-lactam antibiotics mediated by the protein BlaR1 from Staphylococcus aureus. Biochemistry. 2012;51:4642–4649. doi: 10.1021/bi300429p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanique S, Colombo ML, Goormaghtigh E, Soumillion P, Frere JM, Joris B. J Biol Chem. 2004;279:14264–14272. doi: 10.1074/jbc.M313488200. [DOI] [PubMed] [Google Scholar]
- 5.Zuiderweg ER. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 6.Peng JW, Moore JM, Abdul-Manan N. Prog NMR Spectr. 2004;44:225–256. [Google Scholar]
- 7.Jahnke W, Rudisser S, Zurini M. J Am Chem Soc. 2001;123:3149–3150. doi: 10.1021/ja005836g. [DOI] [PubMed] [Google Scholar]
- 8.Clore GM, Iwahara J. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwahara J, Clore GM. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 10.Wagner G. Prog NMR Spectr. 1990;22:101–139. [Google Scholar]
- 11.de Vries SJ, van Dijk M, Bonvin AM. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 12.Battiste JL, Wagner G. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 13.Bruschweiler R, Blackledge M, Ernst RR. J Biomol NMR. 1991;1:3–11. doi: 10.1007/BF01874565. [DOI] [PubMed] [Google Scholar]
- 14.Bonvin AM, Boelens R, Kaptein R. J Biomol NMR. 1994;4:143–149. doi: 10.1007/BF00178343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.