SUMMARY
Obesity activates a complex systemic immune response that includes the recruitment of macrophages and other immune cells to key metabolic tissues. Current models postulate that obesity and excess lipids classically activate macrophages, polarizing them toward an M1 (inflammatory) state. Little is known about noninflammatory functions of adipose tissue macrophages (ATMs). Here, we show that the expansion of adipose tissue (AT) across models of obesity induces a program of lysosome biogenesis in ATMs and is associated with lipid catabolism but not a classic inflammatory phenotype. This program is induced by factors produced by AT and is tightly coupled to lipid accumulation by ATMs. Inhibition of ATM lysosome function impairs lipid metabolism and increases lipid content in ATMs and reduces whole AT lipolysis. These data argue that ATMs contribute quantitatively to the development of obesity-induced inflammation but also serve an important role in lipid trafficking independent of their inflammatory phenotype.
INTRODUCTION
Changes in metabolic state, including obesity, fasting, thermogenic challenges, weight loss, and caloric restriction, broadly activate the immune system (Feuerer et al., 2009; Kosteli et al., 2010; Nguyen et al., 2011; Obstfeld et al., 2010; Talukdar et al., 2012; Weisberg et al., 2003; Winer et al., 2011; Wu et al., 2011; Wu et al., 2007; Xu et al., 2003). In adipose tissue (AT), obesity leads to the accumulation of macrophages to the extent that, in the most obese individuals, as many as half of all cells in a fat depot are macrophages (Weisberg et al., 2003). Efforts to understand the role that AT macrophages (ATMs) play in metabolism have focused largely on characterizing the inflammatory phenotypes and functions of ATMs (Chawla et al., 2011). Indeed, several studies have suggested that, in addition to the quantitative increase in ATMs, obesity elicits a qualitative inflammatory switch in ATM phenotypes (Lumeng et al., 2007a, b).
These studies have described that, in lean animals, alternatively activated M2-like macrophages predominate, but, with the onset of obesity, there is recruitment and accumulation of classically activated macrophages that form multinucleated giant cells and express CD11c and markers of M1 polarization (Lumeng et al., 2007a, b). Most of these studies have focused on the expression of a few genes; e.g., Tnf, Il6, Arg1, or Nos2, or surface antigens (e.g., CD206) to categorize ATMs as M1 or M2 (Lumeng et al., 2007a, b). However, these findings are at odds with whole-tissue expression and other analysis of ATM populations. For example, in whole AT, the expression of Arg1 and CD206+ cells increases in obese individuals despite the fact that they are markers of M2 polarized cells (Bourlier et al., 2008; Shaul et al., 2010). Nonetheless, the observation that the treatment of non-ATMs, including bone-marrow-derived, peritoneal, and immortalized macrophage-like cells, with saturated fatty acids or conditioned medium of adipocyte cell lines induces an increase, albeit a modest increase in comparison to lipopolysaccharide (Lichtenstein et al., 2010; Shi et al., 2006; Suganami et al., 2007), in M1 gene expression has lead to a model in which excess lipids released from adipocytes during the development of obesity drives M1 polarization (Osborn and Olefsky, 2012).
Although macrophages share common immune and reparative roles and stereotypical inflammatory responses to many stimuli, they also possess distinct tissue-specific developmental programs, phenotypes, and functions that are regulated by their cellular context (Pollard, 2009). In AT, macrophages develop and differentiate in a lipid-rich environment, but the developmental program and tissue-specific functions of ATMs, including those related to lipid metabolism, have been largely unexplored. In contrast, the well-studied functions of osteoclasts (multinucleated bone macrophages) demonstrate that macrophages can play critical roles in local tissue-specific metabolic functions (Edwards and Mundy, 2011). We hypothesized that developmental signals produced by AT similarly drive the differentiation of ATMs and functions that are adapted to a lipid-rich environment.
Defining the tissue-specific functions of ATMs and how those functions are altered by obesity offers the possibility of identifying pathways that are fundamental to normal and pathologic function of AT. In an attempt to identify cellular functions of ATMs that are regulated by adiposity, we profiled AT and purified ATMs, finding that a program of lysosome biogenesis is activated by obesity. Surprisingly, we did not find that this program is associated with a classical inflammatory phenotype but, instead, is tightly coupled to lipid content and catabolism. Consistent with lysosomal-dependent lipid metabolism being a developmentally regulated process, AT induces the differentiation of bone marrow cells in a manner that recapitulates the same program and suggests a developmental role for ATMs in local lipid turnover.
RESULTS
Adiposity, Insulin Resistance, and Macrophage Content Are Associated with a Common Program of Lysosome Biogenesis in Whole Adipose Tissue
Macrophage content of AT is tightly correlated with systemic adiposity and insulin sensitivity. As a first step for identifying functions of ATMs that are activated by obesity, we asked whether there were any functional classes of genes whose expression in whole AT was correlated with adiposity, insulin sensitivity, and macrophage-specific gene expression. Using expression microarrays, we profiled the transcription of perigonadal AT (PGAT) from a population of 22- to 24-week-old mice that varied in adiposity and insulin sensitivity because of differences in diet (low- and high-fat diets), gender, segregation of alleles that cause obesity (obese, Ay), and treatment with insulin-sensitizing agents (thiazolidinedione and b-3 adrenergic agonist). We identified transcripts whose expression was correlated with body mass, fasting serum insulin concentration, or macrophage-specific gene expression (Pearson correlation coefficient > 0.6). For each phenotype—body mass, insulin sensitivity, and macrophage content—a functional class analysis was performed (DAVID) (Huang et al., 2009a, b), and classes of genes whose expression was correlated with each phenotype were identified (Table S1 available online).
We found 13 AT transcriptional programs coordinately and positively regulated by adiposity (Table S1), four programs regulated by insulin resistance (Table S1), and 22 associated with a macrophage transcriptional program (Table S1). Consistent with many previous analyses, inflammatory pathways (e.g., immune cell and inflammatory response) were over-represented among genes positively correlated with each phenotype. Among noninflammatory functions, we found one program, lysosomal biogenesis, that was also correlated with each phenotype.
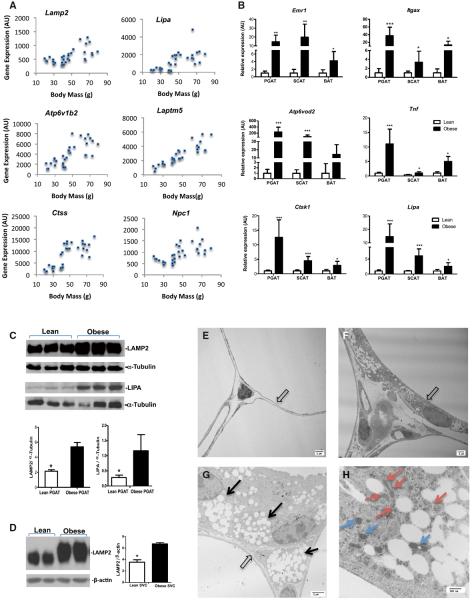
In a previous transcriptional profiling study, we noted in passing a preponderance of lysosomal-related transcripts among genes associated with body mass, but we had not rigorously analyzed the association (Weisberg et al., 2003). The statistical analysis of a larger, more varied data set here reveals that genes involved in all aspects of lysosome biogenesis and function are coordinately upregulated with insulin resistance and obesity: structural genes (Lamp2 and Laptm5), hydrolases (including acid lipases, phosphatases, and proteases [Lipa, Acp5, and Ctsk]), protein pumps required for lysosome acidification (Atp6v1b2), and transport proteins (Ncp1) (Figure 1A) (Saftig and Klumperman, 2009). The obesity-induced increase in lysosome gene transcription was not limited to PGAT and was seen in subcutaneous and brown AT (BAT) as well despite the fact that induction in BAT was less marked (Figure 1B). Obesity increases insulin resistance and macrophage content of AT in humans as well. Consistent with the obesity-dependent increase observed in mice, the expression of lysosomal genes in mesenteric AT from healthy obese human subjects was also elevated in comparison to expression in AT from lean subjects (Figure S1).
Figure 1. Obesity Activates a Program of Lysosome Biogenesis in Adipose Tissue.
(A) PGAT expression of genes encoding lysosome structural proteins (Lamp2 and Laptm5), hydrolases (Lipa and Ctss), and transport proteins (Npc and Atp6v0d2) correlate with body mass.
(B) The expression of macrophage (Emr1 and Itgax) inflammatory (Tnf) and lysosome (Adp6v0d2, Ctss, and Lipa) genes in multiple fat depots including PGAT, inguinal subcutaneous (SCAT), and intrascapular BAT from obese (C57BL/6J Lepob/ob) and lean (C57BL/6J Lep+/+) mice (n = 6).
(C) The expression of the lysosome protein LAMP2 and LIPA in whole PGAT.
(D and E) The expression of LAMP2 protein is strongly increased in SVCs of PGAT from obese (C57Bl/6J Lepob/ob) and lean (C57Bl/6J Lep+/+) mice; representative western blots and quantification (n = 2). Transmission electron micrographs of PGAT from a lean (C57BL/6J Lep+/+) mouse with thin adipocytes (E, open arrows) and few other interstitial cells or lysosomes.
(F and G) PGAT from obese (C57BL/6J Lepob/ob) mice revealing many interstitial nonadipocyte cells. At higher magnification, these nonadipocyte contain lipid-filled vesicles (G, black arrows).
(H) The lipid-filled vesicles are surrounded by lysosomal-like structures (red arrows) and mitochondria (blue arrows) in nonadipocyte cells (outlines) (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001). All values are means ± SD.
In whole PGAT, the obesity-induced increase in lysosomal transcriptional was also associated with increased expression of lysosomal proteins, including the structural lysosome protein LAMP2 and the acid lipase LIPA (Figure 1C); an increase in LAMP2 express was observed in subcutaneous tissue but not BAT (Figure S1). Although all nucleated mammalian cells contain lysosomes, the induction of lysosome protein was especially pronounced in the stromal vascular fraction of cells (Figure 1D). Lysosomes can be visualized as electron-dense bodies by electron microscopy (EM). Using transmission EM, we analyzed the ultrastructure of epididymal AT from lean (C57BL/6J Lep+/+) and obese (C57Bl/6J Lepob/ob) mice. Consistent with the transcriptional activation, lysosomes were abundant in cells from obese mice. Consistent with LAMP2 protein being induced in the stromal vascular faction, lysosomes were most abundant in cells that were distinct from adipocytes and contained what appeared to be filopodia and large lipid-filled vesicles (Figures 1E–1H). In addition, lipid vesicles and lysosomes were also associated with mitochondria (Figure 1H).
Obesity Activate Lysosome Biogenesis in ATMs
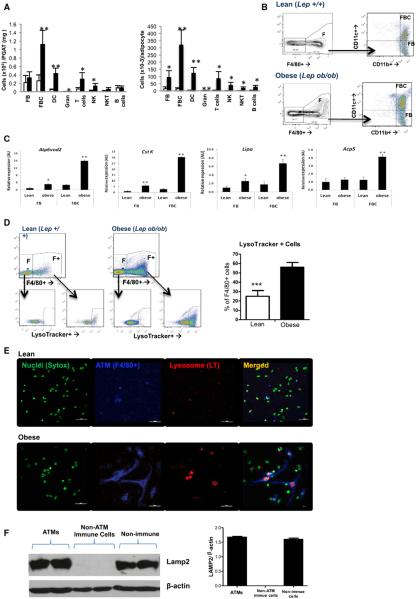
Our transcriptional profiling of whole AT suggested that the accumulation of ATMs in obesity and insulin-resistant states is coupled with lysosomal biogenesis. However, the positive associate of any transcriptional program with immune cells and macrophages in particular may not reflect a qualitative change in immune function but, rather, a quantitative increase in macrophage number. Therefore, to determine whether the lysosomal transcriptional program was activated in ATMs per se and whether it was associated with the M1 polarization state of ATMs, we isolated, quantified, and characterized AT immune cells from lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) mice. As reported previously, the number of macrophages per adipocyte increased more than 10-fold, and, although macrophages were the largest population of cells, all other immune cell populations analyzed increased substantially as well, including dendritic cells, T cells, B cells, granulocytes, NK cells, and NKT cells (Figure 2A).
Figure 2. Obesity Activities Lysosome Biogenesis in ATMs.
(A) Quantification of individual immune cell populations, including CD11c− (FB) and CD11c+ (FBC) macrophages, dendritic cells (DCs), granulocytes (Gran), T cells, NK cells (NK), NKT cells (NKT), and B cells in PGAT from lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) PGAT SVCs as measured by FACS (n = 5–6).
(B) FBC and FB macrophages were purified from lean and obese PGAT with FACS and profiled with expression microarrays.
(C) The expression of lysosome genes was increased in both CD11c− (FBs) and CD11c+ (FBCs) from obese mice in comparison to lean mice (n = 4).
(D) Lysosome content as measured by a fluorescent amine (LysoTracker) that accumulates in acidic cellular compartments is increased in F4/80+ ATMs (F+) from obese mice but not other immune cells (F−); representative FACS and quantification (n = 5).
(E) With the use of immunofluorescence staining of SVCs to identify nuclei (Sytox, green), macrophages (F4/80+, blue) and lysosomes (LysoTracker, red) revealed that most ATMs from obese mice contain visible lysosomes. Lysosomes were also visible in a population of non-ATM SVCs from obese mice (calibration mark = 40 mm).
(F) The expression of LAMP2 protein in ATMs (F4/80+), nonmacrophage immune cells (CD45+, F4/80−), and nonimmune SVCs from PGAT of obese (C57BL/6J Lepob/ob) mice (n = 4; *p < 0.05, **p < 0.01, ***p < 0.005). All values are means ± SD.
See also Figure S2.
In mice, ATMs express (as nearly all macrophage populations do) the antigens F4/80 (EMR1) and CD11b (ITGAM); however, two large subpopulations of ATMs have been identified one the basis of the expression of the integrin CD11c (ITGAX) (Lumeng et al., 2007a, 2008). CD11c− ATMs have been proposed to represent resident macrophages in an M2-polarized state. CD11c+ ATMs are thought to represent newly recruited M1-polarized cells. We found that both classes of ATMs, CD11c+ (F4/80+, CD11b+, CD11c+ [FBC]) and CD11c (F4/ 80+, CD11b+, CD11c− [FB]), increased in AT of obese (C57BL/6J Lepob/ob) mice in comparison to lean animals (C57BL/6J Lep+/+), although the increase in FBC was proportionally much greater (Figures 2A and 2B). Similar results were obtained when comparing low-fat-fed lean and high-fat-fed obese C57BL/6J mice (Figure S2A).
The large increase in ATM numbers may explain the increased expression of many inflammatory and lysosomal genes; i.e., an increased expression ~5- to 10-fold. However, some lysosomal genes (e.g., Atp6v0d2) were increased an order of magnitude disproportionate to the increase in cell number, suggesting a qualitative induction of a lysosomal program. In order to determine whether obesity activated a program of lysosome biogenesis coupled with inflammatory activation in either CD11c− (FB) or CD11c+ (FBC) macrophages, we used expression microarrays to generate transcription profiles of ATMs from lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) mice. Profiling purified FBs and FBCs, we identified 521 transcripts whose expression was differentially (nominal p < 0.01) expressed between FBs from lean and obese mice and 1,509 genes whose expression was differentially (nominal p < 0.01) expressed between FBCs from lean and obese mice (data not shown).
Using the same pathway analysis employed to analyze whole AT (DAVID), we found obesity activated four programs in both FBs and FBCs (Table S1). Most of the programs that were coordinately regulated by adiposity, by insulin resistance, and with macrophage content in whole AT were not upregulated by obesity in individual purified ATMs. Indeed, the only common program that was detectably increased in whole AT and both ATM populations of obese animals was the transcriptional program of lysosome biogenesis. The same individual lysosomal genes whose expression was correlated in whole AT with adiposity and insulin resistance were also upregulated in FBs and FBCs from obese mice in comparison to lean mice (Figure 2C).
To assess whether the transcriptional program of lysosome biogenesis program led to an increase lysosome content, ATMs were isolated from lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) mice and stained with a fluorescent weak amine that accumulates in late endosomes and lysosomes (LysoTracker, Invitrogen). Consistent with the transcriptional profile and our EM micrographs, the fraction of ATMs containing high levels of lysosomes as measured by LysoTracker accumulation doubled from 26% ± 6.2% to 57% ± 4.3% (p < 0.005) in obese mice (Figure 2D).
Lysosomes can also be visualized with the same fluorescent dye in stromal vascular cells (SVCs) isolated from AT. SVCs from lean (C57BL/6J Lep+/+) mice contained few ATMs and few identifiable lysosomes (Figure 2E). By comparison, many distinct lysosomal bodies were visible in SVCs from obese mice and, consistent with the fluorescence-activated cell sorting (FACS) analysis, the majority of lysosomes were contained within F4/80+-expressing ATMs. The LAMP2 protein, a structural component of lysosomes, was expressed in ATMs purified from AT of obese mice, but it was not detected in other immune cells. Consistent with recent findings that autophagy plays a role in adipocyte differentiation, nonimmune SVCs, which contain preadipocytes, also express measurable amounts of LAMP2 (Figure 2F).
Obesity-Induced Lysosomes Biogenesis Is Not Associated with M1 Polarization
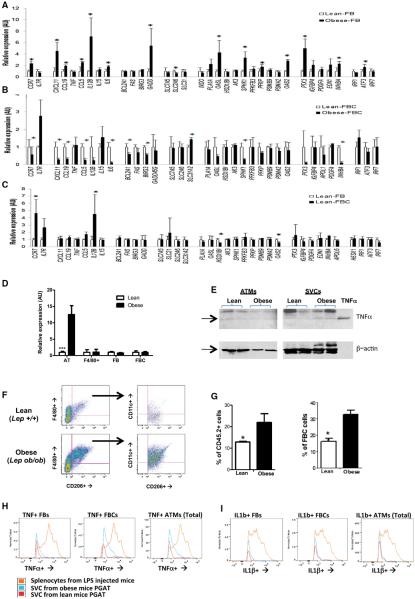
Lysosome biogenesis is not known to be induced during classical activation of macrophages or a common feature of M1 polarization (Martinez et al., 2006). Previous expression profiles had suggested that obesity polarized ATMs, and in particular FBCs, toward an M1 state (Lumeng et al., 2007b). However, our unbiased analysis did not find the enrichment of any immune or inflammatory pathways in FBCs or FBs of obese animals. Furthermore, comparing the expression of three dozen genes whose expression is upregulated in classically activated macrophages (Martinez et al., 2006) did not reveal a consistent trend; the expression of most inflammatory genes was unchanged between FBs and FBCs and between lean and obese ATMs (Figures 3A–3C). Indeed, although some of the M1 genes were actually increased in FBs by obesity, most of the M1 genes whose expression was altered in the “inflammatory” FBCs had reduced rather than increased expression. Similarly, the expression of genes activated in M2-polarized/alternatively activated macrophages was not different between ATM populations or ATMs in lean and obese mice (Figures S3A–S3C). This lack of classical activation was not specific to leptin deficiency; HFD-induced obesity also did not activate classic inflammatory gene expression in ATMs but did induce the lysosomal gene expression (Figures S3E and S3F).
Figure 3. Obesity Does Not Classically Activate or M1 Polarize ATMs.
The expression of genes activated by M1 polarization of macrophages in (A) lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) CD11c− (FB) or (B) CD11c+ (FBC) macrophages.
(C) Relative expression of M1 polarization genes in FBs from FBCs from lean mice (n = 5).
(D) Expression of Tnf, the prototypical M1 activation gene, in whole PGAT, ATMs (F4/80+), and subpopulations of FBs and FBCs from lean and obese mice.
(E) TNFa protein protein expression in whole SVCs and purified ATMs from obese mice in comparison to lean mice. The last lane contains purified TNFa (10 pg) as a positive control.
(F) Typical FACS plot of SVCs expressing the M2-marker CD206 and (G) increased numbers of CD206+ ATMs in PGAT from obese compared to lean mice as either the fraction of all immune CD45.2+ or fraction of ATMs.
(H) Typical FACS histograms of FBs, FBCs or all ATMs stained for TNFa. The beige curve represents data from splenocytes isolated from a mouse injected with LPS.
(I) Typical FACS histograms of FBs, FBCs or all ATMs stained for IL1b (n = 4). The beige curve represents data from splenocytes isolated from a mouse injected with LPS (*p < 0.05, **p < 0.01, ***p < 0.005). All values are means ± SD.
See also Figure S3.
TNFa production is a canonical marker of classically activated, M1-polarized macrophages, and its expression is increased in whole AT from obese animals (Figure 3D) (Gordon and Martinez, 2010). Though modest in comparison to classic activation induced in macrophages by LPS, we found, in addition to an increase in Tnf gene expression, an increase in expression of TNFa protein in total SVCs from obese (C57BL/6J Lepob/ob) mice. However, no measurable increase in TNFa among purified ATMs was detectable from obese mice in comparison to lean mice (C57BL/6J Lep+/+) (Figure 3E) by western blot. Using FACS analysis, we found that a small fraction of ATMs isolated from AT of obese mice (both FBs and FBCs) expresses detectable amounts of TNFa and Il-1b, but, in comparison to splenocytes isolated from LPS-injected mice, the expression of both classic inflammatory proteins as assessed by mean fluorescence intensity (MFI) was an order of magnitude lower in ATMs (Figure 3H). The only obesity-dependent effect we could detect was an increased shoulder in TNFa expression in FBCs associated with a large increase in total FBCs. Similarly, when we sought to use the surface antigen marker CD206 (MRC1) to identify M2-polarized/ alternatively activated ATMs, we found that obesity did not reduce, but actually increased, the fraction of CD206+ among immune cells, including FBCs (Figures 3F and 3G). Altogether, these data argue that obesity does not induce a simple classic binary switch in ATMs toward to an M1-polarized state. Instead, although some inflammatory genes are induced, others are reduced, and the M1/M2 paradigm does not neatly describe the effect of obesity on ATMs. These data also argue that the changes in the inflammatory profile of whole AT are attributable largely to quantitative changes in immune cells rather than a binary switch.
Lipid Droplet Accumulation Is Associated with ATM Lysosome Biogenesis
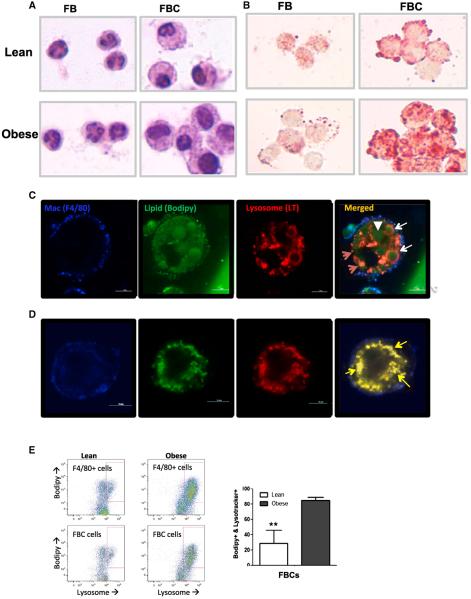
Lysosomes serve multiple functions in macrophage populations, including the processing of antigens for presentation, pathogen destruction, bone resorption, degradation of products of auto-phagy, and reverse cholesterol trafficking (Saftig and Klumperman, 2009). Recent studies suggest that lipid droplets are metabolized in a lysosomal-dependent autophagy pathway in nonadipocytes (Ouimet et al., 2011; Singh and Cuervo, 2012; Singh et al., 2009a,b; Zhang et al., 2009). Obesity and insulin resistance increase neutral lipid content in ATMs, though it is not known whether CD11c+ or CD11c− differentially accumulate lipid. Our expression, protein, and histological data suggest that both FB and FBC upregulate lysosome biogenesis; however, FBCs increase this program more than FBs (Figures 2C and S3). To investigate whether FBCs that more strongly upregu-late the lysosome biogenesis program also accumulate lipid, we isolated CD11c− (FB) and CD11c+ (FBC) ATMs from SVCs of lean (C57BL/6J Lep+/+) and obese leptin-deficient (C57BL/6J Lepob/ob) mice. If the accumulation of neutral lipid regulates lysosomes biogenesis in ATMs, then we predicted that FBCs from obese mice would contain more lipid than FBs. Indeed, FBCs from obese animals contain larger and more numerous lipid droplets than FBCs from lean animals or even FBs from obese mice (Figures 4A and 4B).
Figure 4. Lysosomes Accumulate in Lipid-Laden ATMs.
(A and B) Purified FB (CD11c−) and FBC (CD11c+) ATMs from lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) mice stained with hematoxylin (A) or Oil Red O (B) in order to reveal morphology and lipid content, respectively.
(C) Immunofluorescent confocal images of buoyant ATMs (copurify with adipocytes) isolated from the AT of obese mice and containing large lipid vesicles.
(D) Immunofluorescent confocal images of a live nonbuoyant ATM (with classic SVCs) containing smaller lipid vesicles. Lipid-laden ATMs expressing the macrophage specific antigen F4/80 (blue) and containing lipid (green) vesicles and lysosomes (red). White arrowheads identify large lipid-containing vesicles, red arrows indicate lysosomes, white arrows indicate lipid vesicles with acidic rings, and yellow arrows indicate colocalization of lysosome and lipid staining. Calibration mark = 10 μm.
(E) Lipid and lysosome content of macrophages (F/80+ cells) and FBCs from lean and obese mice were quantified by FACS (n = 5; **p < 0.01). All values are means ± SD.
See also Figure S4.
After the digestion of AT from obese mice, a large number of buoyant ATMs are found floating in the “adipocyte” fraction (Weisberg et al., 2003). We reasoned that ATMs in the adipocyte fraction would contain the largest amount of lipid and provide an opportunity to visualize the cellular relationship between lysosomes and lipid droplets. We digested and separated AT of obese (C57BL/6J Lepob/ob) and lean mice into buoyant and pelleted fractions. Using fluorescence confocal microscopy, we found virtually no ATMs in the buoyant samples from lean mice (data not shown) but many lipid-laden F4/80+ ATMs in the buoyant samples from obese animals. We found two classes of spherical acidic structures containing lipid: one class of these structures possessed a central core of lipid surrounded by an acidic ring, and in the second, typically smaller structures, the lipid and lysosome markers overlapped (Figures 4C and 4D).
To confirm and quantify that lipid droplets of ATMs from obese mice are associated with lysosome content, we used FACS to simultaneously quantify lipid and lysosome content among all ATMs. There was broad variation in neutral lipid and lysosome content, but, consistent with lipid droplet being tightly linked with lysosome biogenesis, the accumulation of the lysosome marker (LysoTracker) was linked with lipid accumulation (BODIPY) and most strongly with FBCs. The fraction of FBCs that were BODIPY+ and LysoTracker+ increased from 26% to more than 86% (Figure 4E).
Adipose Tissue Induces Lysosome Biogenesis, but Not Inflammatory Polarization, of Macrophages
Previous studies had found that saturated fatty acids and conditioned medium from adipocyte cell lines classically activate differentiated bone-marrow-derived and peritoneal macrophages, supporting a model of obesity-induced fatty acid activation of ATMs (Lichtenstein et al., 2010; Shi et al., 2006; Suganami et al., 2007). However, our data suggest that the developmental program of ATMs may elicit a different response to excess lipids, one in which lysosome biogenesis is a key phenotype. To test whether AT induces the development of macrophages that activate lysosome biogenesis, we differentiated bone marrow cells with a protocol similar to standard osteoclast differentiation methods but substituted AT for primary osteoblasts (Figure 5A).
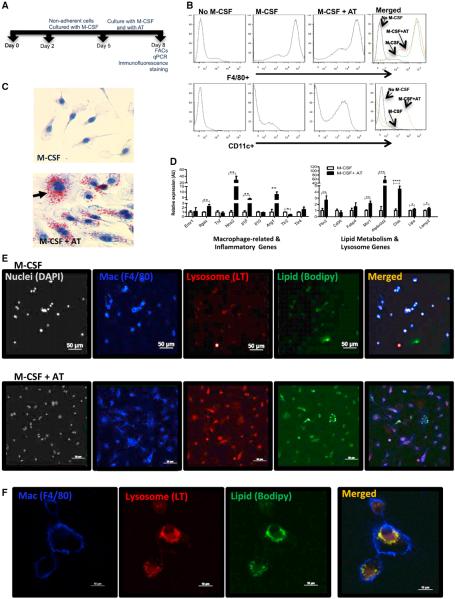
Figure 5. Adipose Tissue Induces an ATM Phenotype without Inflammation.
(A) Cartoon of ATM differentiation protocol with timeline of bone marrow cells cultured with M-CSF (CSF-1) in the absence or presence of AT.
(B) F4/80+ and CD11c+ populations identified by FACS among bone marrow cells differentiated in the absence of both M-CSF and AT in the presence of M-CSF only or in the presence of both M-CSF and AT.
(C) Hematoxylin and Oil Red O staining for lipid of bone marrow cells differentiated with M-CSF in the absence or presence of AT.
(D) Inconsistent induction of inflammatory gene expression when bone marrow cells are differentiated in the presence of AT; activation of lipid metabolism and lysosome genes by AT-induced differentiation (n = 5).
(E) Immunofluorescent staining of bone marrow cells differentiated in the absence or presence of AT to identify nuclei (DAPI, silver), macrophages (F4/80+, blue), lysosomes (LysoTracker [LT], red), and neutral lipid (Bodipy, green). Calibration mark = 50 μm.
(F) High magnification of immunofluorescent stained confocal images. Calibration mark = 10 μm (*p < 0.05, **p < 0.01, ***p < 0.005). All values are means ± SD.
See also Figure S5.
In the presence of M-CSF/CSF-1 alone, bone marrow cells efficiently differentiated into cells that possess characteristics typical of macrophages: adherent mononuclear cells with a high cytoplasm/nuclear ratio and high F4/80+ and low CD11c+ expression (Figure 5B). However, when cells were differentiated in the presence of AT, they took on characteristics observed in primary ATMs: intermediate F4/80 expression, high expression of CD11c (ITGAX), the appearance of multinucleated cells, the accumulation of neutral lipids (Figures 5B and 5C), and the induction of genes that are enriched in ATMs in comparison to other tissue myeloid cells (e.g., Cxcl1 and Tnfs9) (Figure S4). Consistent with our expression studies of primary ATMs, the differentiation of macrophages in the presence of AT did not induce the expression of a consistent M1 polarization program, no change occurring in the expression of Tnf, Tlr2, Tlr4, and Il10 (Figure 5D), and an increase in the expression of both M2-and M1-associated genes, including Arg1, Il1b, and Nos2 (Figure 5D).
Although differentiation in the presence of AT did not consistently induce inflammatory gene expression, it did strongly upregulate the expression of genes involved in lipid uptake and storage, including the scavenger receptor Msr1 and lipid-droplet-associated protein Plin2 (Figure 5D). AT also induced the expression of lysosome genes activated in ATMs by obesity, including Atp6v0d2, Lipa, and Ctsk. Consistent with these transcriptional phenotypes, macrophages that underwent terminal differentiation in our protocol accumulated neutral lipid and contained numerous lysosomes, whereas macrophages differentiated with only M-CSF/CSF-1 had scant lipid and few lysosomes (Figures 5E and 5F). These data suggest that secreted factors, including lipids, produced by AT do not classically activate ATMs per se but induce a differentiation program in which lipid uptake and lysosomal biogenesis are coordinately regulated.
Lysosomal-Dependent Metabolism of Lipids in ATMs and Adipose Tissue
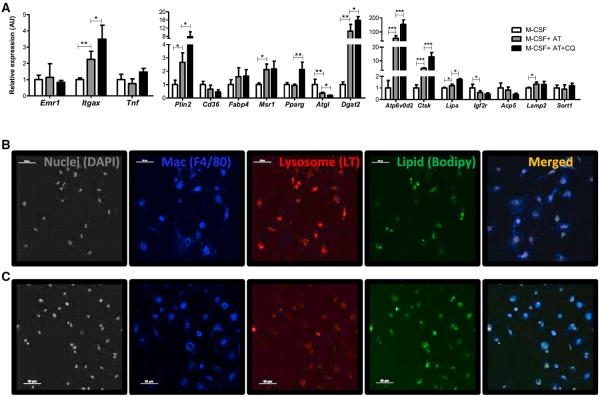
In both primary and in vitro differentiated ATMs lipid, droplets and lysosomes colocalize, suggesting that lysosomes may also play a role in lipid metabolism. A class of antimalarial drugs, including chloroquine, is lysosomotrophic and inhibits the acidification of lysosomes and acid-dependent lipase activity. If lysosomes function is necessary for lipid catabolism in ATMs, then inhibition by chloroquine should lead to the accumulation of lipid droplets. To test this directly, ATMs were differentiated in vitro with the protocol developed above and treated with chloroquine for 16 hr. As expected, treatment reduced detectable lysosomes in bone-marrow-derived ATMs (Figure 6C) and, as a compensatory response, increased the expression of lysosome genes, including Atp6v0d2, Lipa, and Ctsk (Figure 6A). Consistent with a role for lysosomes in lipid catabolism in ATMs, inhibiting lysosome function increased the lipid content markedly (Figures 6B and 6C). The accumulation of lipids in chloroquine treated ATMs without a clear inflammatory activation argue that lipids do not have an inherent inflammatory effect on ATMs (Figure 6A). A second inhibitor of lysosomal function had the same effect. Bafilomycin A1 prevents multiple steps in endocytic trafficking and lysosomal hydroplyses by inhibiting H+ATPases required for lysosomal acidification. Treating ATMs with bafilomycin A1 also reduced lysosomes and increased lipid content of ATMs (Figure S5).
Figure 6. Inhibition of Lysosome Function Increases Lipid Accumulation in ATMs.
(A) The expression of macrophage-related, lysosome and lipid metabolism genes in AT induced in ATM-like cells in the presence or absence of chloroquine. (n = 6; *p < 0.05, **p < 0.01, ***p < 0.005).
(B and C) Immunofluorescent confocal images of lysosomes and lipid among in vitro differentiated ATMs (B) and ATMs treated with chloroquine (C). Nuclei were identified with DAPI, macrophages (Mac) with an anti-F4/80, lysosomes with LT, neutral lipid with Bodipy. All values are means ± SD.
See also Figure S6.
Recent studies have found that lipid homeostasis in hepatocytes, adipocytes, and foam cells occurs in part through a lysosomal-dependent autophagy pathway, dubbed lipophagy (Ouimet et al., 2011; Singh and Cuervo, 2012; Singh et al., 2009a, b; Zhang et al., 2009). The inhibition of autophagy in hepatocytes and foam cells through target deletion of critical autophagy genes increased the lipid content of cells. However, when we generated ATMs that lacked the Atg7 necessary for normal autophagy, we found no increase in the lipid content of ATMs, arguing against autophagy being required for lysosomal degradation of lipids (Figure S5).
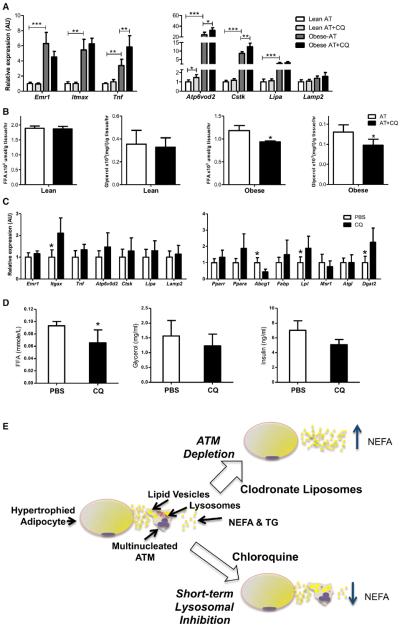
Reports dating back to the 1960s have demonstrated that chloroquine treatment acutely reduces AT lipolysis and improves systemic glucose and lipid homeostasis (Benoit, 1967; Powrie et al., 1993, 1991). It has been hypothesized that the systemic effect of chloroquine in reducing circulating lipids concentrations was secondary to increased circulating insulin concentrations due to reduced lysosomal-dependent hepatic clearance. To determine whether chloroquine could directly act on AT to reduce lipolysis in an ATM-dependent manner, we treated primary AT cultures with chloroquine. We compared the effect of chloroquine on lipolysis from AT of lean and obese mice. As would be predicted if chloroquine-induced reduction in lipolysis was ATM-dependent, treatment of ATM-poor AT from lean mice had no consistent effect on lipolysis, as measured by the release of nonesterified fatty acids or glycerol. In contrast, chloroquine treatment of AT from obese mice reduced lipolysis by ~20% (Figures 7A and 7B). ATMs are efficiently depleted from AT ex vivo by liposome-encapsulated clodronate. If ATM lysosomal function is critical for chloroquine's suppression of lipolysis by AT, then the depletion of ATMs should eliminate any chloroquine effects. Indeed, chloroquine did not reduce lipolysis in AT depleted of macrophages (Figure S6).
Figure 7. Lysosome Function Modulates Lipolysis in Macrophage-Rich Adipose Tissue.
(A) Expression of macrophage and lysosome genes from PGAT of lean (C57BL/6J Lep+/+) and obese (C57BL/6J Lepob/ob) mice treated ex vivo with chloroquine.
(B) The effect of lysosome inhibition by chloroquine on the release of free fatty acids and glycerol from PGAT of lean and obese mice.
(C) Macrophage, lysosome, and lipid metabolism genes in PGAT 24 hr after injection of either PBS or chloroquine.
(D) Circulating concentrations of free fatty acids, glycerol, and insulin 24 hr after injection of PBS or chloroquine into PGAT of obese mice (n = 5–6; *p < 0.05, **p < 0.01, ***p < 0.005). All values are means ± SD.
(E) During the development of obesity, adipocytes undergo hypertrophy and apoptosis, leading to increases in the local concentrations of lipids, including NEFAs and TGs. Excess lipids are recognized by the immune system and increase the accumulation of ATMs. By uptaking extracellular lipids, ATMs buffer surrounding cells from lipotoxic effects. Once in ATMs, the lipids are directed to lysosomal pathway of catabolism. In the absence of ATMs after clodronate treatment, the release of NEFAs by AT is increased. Conversely, short-term inhibition of lysosomal catabolism increases lipid content of ATMs and reduces net lipolysis of AT. Inhibition of lysosomes may reduce lipolysis through direct stoichiometric effects on lipid fluxes or via increased production of antilipolytic factors as lipid content increases in ATMs.
Although previous studies suggested that chloroquine's ability to improve metabolic phenotypes in patients with diabetes is secondary to increased plasma insulin concentration our data argue that there may also be direct effects on AT lipid trafficking. Using obese (C57BL/6J Lepob/ob) mice, we tested whether the direct delivery of chloroquine into AT would reduce lipolysis without increasing circulating insulin. Injection of chloroquine directly into visceral AT depots of obese mice lowered circulating nonesterified fatty acid concentrations within 24 hr. However, there was no effect of local chloroquine injection on serum insulin (Figures 7C and 7D). Altogether, the ex vivo and in vivo data argue that the acute effects of chloroquine are mediated, at least in part, by the local action of chloroquine in impairing lysosome function of ATMs in addition to any effects on increasing plasma insulin concentrations.
DISCUSSION
Studies aimed at understanding the role of the immune system in obesity and associated disorders have focused largely on quantitative changes in immune cell populations and classic inflammatory responses; the effect of adiposity on no-inflammatory functions has been less well studied. However, in other tissues, macrophages serve developmentally specific functions critical to organ function, including ovulation, steroid hormone production, bone homeostasis, and erythropoiesis (Pollard, 2009). Our efforts to identify noninflammatory functions of the immune system elicited by obesity have revealed that ATMs, the dominant immune cell in AT, respond to increases in adiposity by activating a program of lysosomal metabolism of lipids. This lysosomal program is partly under the developmental control of factors secreted from AT and closely coupled to lipid accumulation. The inhibition of lysosomal function increases lipid accumulation in ATMs and reduces the release of nonesterified fatty acids from AT. The net effect of ATMs is to buffer increased concentrations of lipids (Figure 7). Surprisingly, we found that obesity elicited no consistent polarization of ATMs toward a classically activated state, arguing that the increase in the inflammatory profile of AT associated with obesity derives primarily from quantitative increases in immune cell populations.
These observations suggest that the pathway of lipid release by AT is not simply a two-step process of hydrolysis of triglyceride (TG) in adipocytes and release of nonesterified fatty acid (NEFA) into the circulation, nor is the trafficking of lipids the same in lean and obese AT. Instead, the pathway appears to change with increasing adiposity; in AT with large adipocytes, released lipid is taken up by macrophages and targeted to lysosomes for metabolism. By either direct stoichiometric effects on lipid release or, more likely, through the indirect action of factors that reduce release of lipids by adipocytes, the inhibition of lysosomes and increased lipid accumulation in ATMs has the effect of reducing NEFA release by AT and buffering other cells from any toxic effects of excess lipids.
Because perturbations in the efficiency of lipid storage and trafficking by AT are central to the development of metabolic diseases, the role of ATM lysosome function may provide insights and potential therapeutic targets for understanding and treating obesity-related diabetes, hepatic steatosis, and cardiovascular disease. Any attempts to target ATM lysosome pathway to modulate AT metabolism, though, will require further elucidation of the uptake of lipid by ATMs, the signaling effects of lipid sensing and accumulation by ATMs, the ultimate fate of lysosomal products of lipid hydrolysis, and the anabolic factors elaborated by ATMs. Some clues may be gleaned from previous published observations and our data here.
In ATMs that contain the largest amount of lipid, there appear to be vesicles of at least two types. Both classes of lipid vesicles aggregate at the periphery of the cell near the plasma membrane. One population contains only neutral lipid, whereas the second has an acidic ring. One plausible explanation is that the neutral lipid vesicles fuse with primary lysosome forming an acidic-ringed secondary lysosome. In cells containing fewer lipids, there appears to be a single class of lipid containing vesicles that are acidic. If neutral lipid vesicles fuse with a lyso-some, then it would imply that the neutral lipid vesicles are not classic lipid droplet but rather membrane vesicles, raising the possibility that the lipid is derived through an endocyticphagocytic pathway. The phagocytosis of large lipid droplets may derive from dead or dying adipocytes or alternatively through some regulated process; e.g., the exocytosis of TG droplets or macrophage-adipocyte transfer. However, in addition to a pathway of TG uptake, we found transcriptional evidence for fatty acid uptake and esterification by ATMs. ATMs from obese animals upregulate the expression of fatty acid transporters, Msr1, Cd36, and esterification enzyme Dgat2 (data not shown). Given the ability of macrophages to broadly scavenge molecules through different pathways, it seems likely that multiple forms are taken up into lipid vesicles.
The fate of the lipid after it fuses with the primary lysosome is also not clear. Inhibition studies with chloroquine imply hydrolysis of neutral lipid into fatty acids and glycerol. If the ATMs function to buffer excess lipids, then it would make sense that a portion of the lipid is oxidized. Unlike the reduction in the expression of mitochondrial and peroxisomal-specific genes that occurs in adipocytes with the onset of obesity, the expression of several genes involved in medium- and long-chain fatty acid oxidation are upregulated in ATMs from obese mice. Alternatively, there may exist a complex recycling system in which FFAs are released by ATMs and either resterified by adipocytes or released into the circulation.
The reduction in AT lipolysis and plasma concentrations of nonesterified fatty acids induced by chloroquine and other lysosomotrophic agents has been known for several decades and attributed to enhanced insulin signaling. Our data suggest that, at least in the short-term, chloroquine reduces lipid metabolism via its effects on ATMs. These data argue that either a substantial fraction of the NEFA released from AT of obese individuals is released by ATMs or inhibition of lysosomes and lipid accumulation in ATMs secondarily suppresses adipocyte lipolysis via the release of antilipolytic factors. In the first model, long-term manipulation of ATMs' lysosomal function would have minimal effect and may prove detrimental. Alternatively, if ATMs sense lipid accumulation and activate lysosomes and secrete anabolic factors to reduce their lipid load, then modulating ATM lysosome function may afford indirect means of modulating adipocyte function. Unfortunately, the current studies do not distinguish between these actions but provide additional evidence that ATMs modulate lipid flux out of AT. Nonetheless, if the action of ATMs on lipid trafficking are mediated primarily via secreted factors, then our data would argue that increasing lipid content in ATMs favors the production of such anabolic and antilipolytic factors. Efforts are underway to identify such factors and define their regulation by lipid content.
Previous studies have argued that obesity polarizes the largest immune cell population, ATMs, toward a classically activated state. A model has emerged in which saturated free fatty acids released in proportion to adiposity activate pattern recognition receptors to recruit and activate a population of CD11c+ ATMs, increasing the local production of inflammatory molecules by increasing both the number of ATMs and the amount produced by individual cells. We did not observe an obesity-driven phenotypic switch of macrophages from an M2/anti-inflammatory to an M1/inflammatory state; there was no consistent obesity-induced increase in the expression of genes associated with M1 polarization in either CD11c+ or CD11c− ATMs, nor did we find that CD11c+ in comparison to CD11c− ATMs were simply more M1 polarized. These data are consistent with the observation that the obesity-induced increase in AT expression of prototypical inflammatory molecules including TNFα is commensurate with the increase in the number of macrophages and other immune cells; i.e., an order of magnitude increase in macrophage content occurs with a 10-fold increase in Tnf expression.
These findings do not imply that the expression of classical inflammatory molecules by ATMs is without metabolic consequences; a large amount of data suggest that, at least in rodents, genetic and pharmacologic manipulations that increase the expression of inflammatory molecules by ATMs impair the metabolic function of AT, and, conversely, reducing the inflammatory profile of ATMs reduces metabolic derangements associated with obesity. Instead, our data argue the degree to which macrophage-derived inflammatory cytokines increase the overall inflammatory profile and modulate AT metabolism is dominated by quantitative increases in cell numbers rather than qualitative changes in the classically defined inflammatory state. These data also do not preclude that obesity induces changes in the inflammatory state of subpopulations of macrophages, but they do argue that, in the aggregate, there is no polarization toward a traditionally defined M1 state or away from a traditional M2 state in obese rodents. In addition, although obesity did not induce changes in expression that recapitulated classical or alternative activation, there were consistent changes in a large number of immune regulatory genes, including obesity-induced reduction in Il6 and chemokines Cxcl11 and Ccl19. Thus, obesity alters the expression of immune regulatory genes but does so in a manner distinct from previously described classes of activation. How this distinct pattern of inflammatory gene expression affects metabolic function remains to be determined.
The unexpected lack of M1 polarization and the atypical transcriptional profile of ATMs from obese mice despite high local concentration of NEFA suggested that ATMs possess a developmental program and response to lipids distinct from other macrophage populations. Indeed, in vitro differentiation studies suggest that the response of ATMs to high concentrations of lipids and free fatty acids is distinct from typically differentiated bone marrow macrophages. The differentiation of cells in the presence of AT not only induced phenotypes characteristic of ATMs, including being multinucleated and expressing CD11c, and the coupled accumulation of lipids and lysosomes but also did not polarize the macrophages in a classical or M1 state. Perhaps, in retrospect, this should not be entirely unexpected; these findings reinforce the model of tissue macrophages as a group of cells that share common progenitors and immunologic characteristics but also possess distinct noninflammatory, trophic functions that are developmentally regulated by cellular context.
EXPERIMENTAL PROCEDURES
Animals and Animal Care
Mice were obtained from the Jackson Laboratory or from Masaaki Komatsu at Tokyo Metropolitan Institute of Medical Science (Komatsu et al., 2006). All procedures were approved by the Columbia University International Animal Care and Use Committee.
Isolation and Culturing of Primary Cells
AT or bone marrow were isolated with standard techniques from mice immediately after CO2 asphyxiation. Bone marrow cells were differentiated into ATM-like cells by coculturing nonadherent cells in the presence of whole PGAT.
Quantitative RT-PCR
Total RNA was extracted from tissues with either an acid-phenol- or silica-membrane-based methods. Complementary DNAs were generated with a commercially available kit, and quantitative PCR was carried out with fluores-cent double-stranded DNA dye. Data were normalized to Ppib with the ΔΔC(t) method and are presented as relative transcript levels. All primers used are listed in Table S2.
Immunophenotyping and Flow Cytometry
For standard FACS analysis, cells were collected in FACS buffer and incubated with blocking FcBlock (BD Pharmingen) antibody and with fluorophore-conjugated antibodies or fluorescent dyes.
Microarray Gene Expression
For whole AT samples, total RNA was extracted from the PGAT of individual mice. RNA was isolated from sorted FBC (F4/80+, CD11b+, and CD11c+) cells and FB (F4/80+, CD11b+, and CD11c−) cells with RNeasy micro kits (Qiagen) with a PicoPure RNA isolation kit and amplified for two rounds. Functional class analysis was performed with DAVID Suite software (NIAID, NIH http://david.abcc.ncifcrf.gov).
Western Blot
Purified SVCs, adipocyte-rich fractions, and whole AT samples were subjected to denaturing gel electrophoresis and standard western blotting techniques.
Macrophage Depletion and Lysosome Inhibition
Macrophage depletion was carried out with liposomes containing clodronate as previously described (Kosteli et al., 2010). Lysosomes function was inhibited in vitro with either chloroquine or bafilomycin a1 and in vivo with chloroquine.
Electron Microscopy
Transmission electron microscopy was performed on AT taken from mice perfused with fixative according to standard protocols.
Human Adipose Tissue
AT was collected from twenty obese (body mass index ranging from 34.8–61.3 kg/m2) Caucasian women with a mean age of 40.5 (26–50) years old who were scheduled for Roux-en-Y gastric bypass surgery were included. The study was approved by the Medical Ethical Committee of the Academic Medical Center, Amsterdam.
Statistics
Data are presented as mean ± SD. All p values were calculated with a two-tailed distribution, two-sample unequal variance Student's t test. All calculations were performed with Microsoft Excel and Statistica.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rudy Leibel, Ira Goldberg, and Domenico Accili for insightful comments and suggestions. We are grateful to Masaaki Komatsu for providing mice. These studies were supported by research and center grants from the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK; DK066525, DK063608, and DK026687) and the Russell Berrie Foundation. A.G. was support by a fellowship grant from the NIDDK (DK007647).
Footnotes
ACCESSION NUMBERS
RNA isolate data were deposited in NCBI Gene Expression Omnibus under accession number GSE8831.
SUPPLEMENTAL INFORMATION
Supplemental Information contains Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2013.11.001.
REFERENCES
- Benoit FL. The inhibitory effect of chloroquine on rat adipose tissue metabolism in vitro. Metabolism. 1967;16:557–561. doi: 10.1016/0026-0495(67)90086-8. [DOI] [PubMed] [Google Scholar]
- Bourlier V, Zakaroff-Girard A, Miranville A, De Barros S, Maumus M, Sengenes C, Galitzky J, Lafontan M, Karpe F, Frayn KN, Bouloumié A. Remodeling phenotype of human subcutaneous adipose tissue macrophages. Circulation. 2008;117:806–815. doi: 10.1161/CIRCULATIONAHA.107.724096. [DOI] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, Mundy GR. Advances in osteoclast biology: old findings and new insights from mouse models. Nat Rev Rheumatol. 2011;7:235–243. doi: 10.1038/nrrheum.2011.23. [DOI] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, Mathis D. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kosteli A, Sugaru E, Haemmerle G, Martin JF, Lei J, Zechner R, Ferrante AW., Jr. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Invest. 2010;120:3466–3479. doi: 10.1172/JCI42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein L, Mattijssen F, de Wit NJ, Georgiadi A, Hooiveld GJ, van der Meer R, He Y, Qi L, Köster A, Tamsma JT, et al. Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab. 2010;12:580–592. doi: 10.1016/j.cmet.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 2007a;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007b;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Nguyen KD, Qiu Y, Cui X, Goh YP, Mwangi J, David T, Mukundan L, Brombacher F, Locksley RM, Chawla A. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obstfeld AE, Sugaru E, Thearle M, Francisco AM, Gayet C, Ginsberg HN, Ables EV, Ferrante AW., Jr. C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes. 2010;59:916–925. doi: 10.2337/db09-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard JW. Trophic macrophages in development and disease. Nat. Rev. Immunol. 2009;9:259–270. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie JK, Smith GD, Shojaee-Moradie F, Sönksen PH, Jones RH. Mode of action of chloroquine in patients with non-insulin-dependent diabetes mellitus. Am. J. Physiol. 1991;260:E897–E904. doi: 10.1152/ajpendo.1991.260.6.E897. [DOI] [PubMed] [Google Scholar]
- Powrie JK, Shojaee-Moradie F, Watts GF, Smith GD, Sönksen PH, Jones RH. Effects of chloroquine on the dyslipidemia of non-insulin-dependent diabetes mellitus. Metabolism. 1993;42:415–419. doi: 10.1016/0026-0495(93)90096-7. [DOI] [PubMed] [Google Scholar]
- Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- Shaul ME, Bennett G, Strissel KJ, Greenberg AS, Obin MS. Dynamic, M2-like remodeling phenotypes of CD11c+ adipose tissue macrophages during high-fat diet—induced obesity in mice. Diabetes. 2010;59:1171–1181. doi: 10.2337/db09-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Invest. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009a;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 2009b;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganami T, Tanimoto-Koyama K, Nishida J, Itoh M, Yuan X, Mizuarai S, Kotani H, Yamaoka S, Miyake K, Aoe S, et al. Role of the Toll-like receptor 4/NF-kappaB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler. Thromb. Vasc. Biol. 2007;27:84–91. doi: 10.1161/01.ATV.0000251608.09329.9a. [DOI] [PubMed] [Google Scholar]
- Talukdar S, Oh Y, Bandyopadhyay G, Li D, Xu J, McNelis J, Lu M, Li P, Yan Q, Zhu Y, et al. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat. Med. 2012;18:1407–1412. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer DA, Winer S, Shen L, Wadia PP, Yantha J, Paltser G, Tsui H, Wu P, Davidson MG, Alonso MN, et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat. Med. 2011;17:610–617. doi: 10.1038/nm.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ghosh S, Perrard XD, Feng L, Garcia GE, Perrard JL, Sweeney JF, Peterson LE, Chan L, Smith CW, Ballantyne CM. T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation. 2007;115:1029–1038. doi: 10.1161/CIRCULATIONAHA.106.638379. [DOI] [PubMed] [Google Scholar]
- Wu D, Molofsky AB, Liang HE, Ricardo-Gonzalez RR, Jouihan HA, Bando JK, Chawla A, Locksley RM. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.