Significance
Despite intensive efforts, the structure of the native HIV-1 envelope trimer—the sole relevant target for vaccine design—has remained elusive. Our work identifies a key structural constraint that stabilizes the native envelope conformation and modulates its sensitivity to neutralization. We show that this constraint is established by previously unrecognized sulfated tyrosines within the second variable loop (V2) of the envelope glycoprotein subunit gp120, which mediate intramolecular interaction with the base of the third variable loop, V3. Strikingly, the V2 sulfotyrosines functionally mimic those present in the N terminus of the CCR5 coreceptor, which bind to the same V3 region. Our results shed light on the mechanisms adopted by HIV-1 to elude immunologic control and open new perspectives for vaccine design.
Abstract
Elicitation of broadly neutralizing antibodies is essential for the development of a protective vaccine against HIV-1. However, the native HIV-1 envelope adopts a protected conformation that conceals highly conserved sites of vulnerability from antibody recognition. Although high-definition structures of the monomeric core of the envelope glycoprotein subunit gp120 and, more recently, of a stabilized soluble gp140 trimer have been solved, fundamental aspects related to the conformation and function of the native envelope remain unresolved. Here, we show that the conserved central region of the second variable loop (V2) of gp120 contains sulfated tyrosines (Tys173 and Tys177) that in the CD4-unbound prefusion state mediate intramolecular interaction between V2 and the conserved base of the third variable loop (V3), functionally mimicking sulfated tyrosines in CCR5 and anti–coreceptor-binding-site antibodies such as 412d. Recombinant gp120 expressed in continuous cell lines displays low constitutive levels of V2 tyrosine sulfation, which can be enhanced markedly by overexpression of the tyrosyl sulfotransferase TPST2. In contrast, virion-associated gp120 produced by primary CD4+ T cells is inherently highly sulfated. Consistent with a functional role of the V2 sulfotyrosines, enhancement of tyrosine sulfation decreased binding and neutralization of HIV-1 BaL by monomeric soluble CD4, 412d, and anti-V3 antibodies and increased recognition by the trimer-preferring antibodies PG9, PG16, CH01, and PGT145. Conversely, inhibition of tyrosine sulfation increased sensitivity to soluble CD4, 412d, and anti-V3 antibodies and diminished recognition by trimer-preferring antibodies. These results identify the sulfotyrosine-mediated V2–V3 interaction as a critical constraint that stabilizes the native HIV-1 envelope trimer and modulates its sensitivity to neutralization.
The development of a protective vaccine remains a high priority for the global control of the HIV/AIDS epidemic (1). However, the unique biological features of HIV-1 make this task extremely challenging. The main obstacles include the ability of the virus to integrate into the host chromosomes, a remarkable degree of genetic variability, and the cryptic, antibody-shielded conformation adopted by the viral envelope in the native spikes that protrude from the virion surface (2). These spikes are composed of homotrimers of heterodimers of the envelope glycoprotein subunits gp120 and gp41 maintained in an energetically unfavorable, metastable conformation (3, 4). Upon binding to CD4 and a coreceptor such as CCR5 or CXCR4, gp120 undergoes dramatic conformational changes that lead to a low-energy state, creating permissive conditions for activation of the gp41 fusogenic mechanism (3). In the prefusion conformation, gp120 effectively conceals its highly conserved receptor- and coreceptor-binding sites from antibody recognition, imposing a high-entropy penalty for interaction with CD4 or antibodies to the coreceptor-binding site such as 17b; in contrast, in the open, low-energy conformation, gp120 interacts with CD4 and 17b with minimal thermodynamic changes (4, 5). This conformational masking of the vulnerable receptor- and coreceptor-binding sites is believed to be a primary mechanism of immune evasion by HIV-1 (4).
The inherent conformational flexibility of gp120, along with the extensive N-linked glycosylation that covers most of the exposed surface of the glycoprotein, has severely hampered attempts to elucidate the native structure of the HIV-1 envelope spike. As a consequence, most of the available high-definition structures of gp120 have been obtained with deglycosylated, variable loop-truncated core monomers in complex with stabilizing ligands such as soluble CD4 (sCD4) (6–10). Important information regarding the overall conformation and ligand interactions of the trimeric spike at intermediate resolution has emerged from the use of increasingly refined cryo-electron microscopy (cryo-EM) technologies (11–17). Moreover, the crystal structure of a stabilized, soluble, cleaved gp140 trimer (BG505 SOSIP.664) at 4.7-Å resolution was reported recently (18). However, despite these advances, many critical aspects related to the structural mechanisms of HIV-1 immune vulnerability and evasion remain unresolved. In particular, the fine molecular details of the interaction between the second and third variable loops (V2 and V3, respectively) of gp120, which are believed to play a critical role in stabilizing the prefusion envelope structure (19–21), are elucidated only partially. Functionally, V2 and V3 cooperate in the formation of quaternary epitopes targeted by some of the most potent and broadly neutralizing mAbs hitherto identified (22, 23), and V2 effectively masks neutralization epitopes in V3 (24–27). Cryo-EM studies have provided evidence that in the prefusion conformation V2 and V3 are spatially contiguous and account for most of the density at the apex of the trimeric envelope spike (12–17). Although various fragments of V2 and V3 were crystallized separately using antibody-complexed synthetic peptides (28, 29), scaffolded chimeric constructs of the first and second variable loops (V1V2) (30, 31), or a V3-containing gp120 core monomer (8), the only study in which the two loops were visualized simultaneously is the recent report of the BG505 SOSIP.664 trimer crystal structure (18). In this artificially stabilized trimer, which displays several antigenic features of the native envelope (32), V2 and V3 appear to interact directly at the trimer apex with the V3 β-hairpin extensively buried under the V1V2 four-stranded Greek-key β-sheet (18).
In the present study, we provide evidence that the conserved central region of the gp120 V2 loop contains previously unrecognized sulfated tyrosines that, in the CD4-unbound prefusion state, mediate intramolecular interaction between V2 and the CCR5-binding site at the base of V3. Our results suggest that the sulfotyrosine-bolstered interaction between V2 and V3 is a key structural constraint that stabilizes the native conformation of the HIV-1 envelope trimer.
Results
The Conserved Central Region of the V2 Loop of HIV-1 gp120 Contains Sulfated Tyrosines.
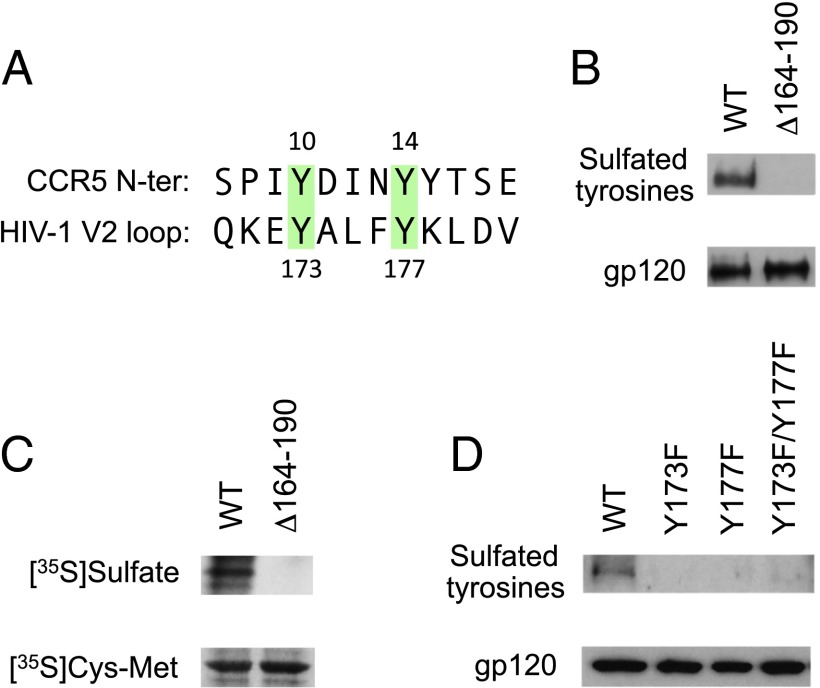
Despite their definition as variable loops, the V2 and V3 regions of gp120 contain highly conserved domains (33). Because several lines of evidence suggest that V3 establishes direct interaction with V2 (12–21), and the conserved base of V3 is the binding site for the N-terminal region of the CCR5 coreceptor (34), we looked for potential structural homology between V2 and CCR5. We recognized that the conserved central region of V2 contains two tyrosine residues (Tyr173 and Tyr177) with spacing identical to that of two tyrosines (Tyr10 and Tyr14) present in the N-terminal region of CCR5 (Fig. 1A). Tyr177 is highly conserved across all HIV-1 subtypes (>99%), whereas Tyr173 is conserved in subtypes A, B, and C but often is replaced by histidine in subtypes D, E, and F (∼66% cross-subtype conservation). Because Tyr10 and Tyr14 in CCR5 were shown to be posttranslationally modified by O-sulfation (35) and to play a critical role in the interaction of CCR5 with V3 (36), we investigated whether the V2 tyrosines also can be sulfated. Western blot analysis using a highly specific anti-sulfotyrosine (Tys) mAb (Fig. S1) documented the presence of sulfated tyrosines in gp120 immunoprecipitated from the surface of HeLa cells expressing full-length cleavable gp160 from the subtype-B HIV-1 isolate BaL (Fig. 1B). Because gp120 contains additional tyrosine residues both within and outside V2, we produced a partial V2-deletion mutant that selectively excludes Tyr173 and Tyr177 (BaL Δ164–190). Despite comparable levels of gp120 expression, the Tys signal was abrogated in the deletion mutant (Fig. 1B), indicating that gp120 sulfation is specific and occurs selectively at Tyr173 and Tyr177.
Fig. 1.
The V2 domain of HIV-1 gp120 contains sulfated tyrosines. (A) Sequence alignment of the CCR5 N-terminal domain and the conserved central region of the V2 domain of HIV-1 gp120 (consensus sequence for subtype B). Two colinear conserved tyrosine residues present in CCR5 and gp120 V2 are highlighted in green. (B) Detection of sulfated tyrosines by Western blot in gp120 purified from HeLa cells expressing WT HIV-1 BaL gp160 or a partially V2-deleted mutant (Δ164–190) lacking Tyr173 and Tyr177 by vaccinia technology. gp120 was immunoprecipitated from the cell surface and analyzed by Western blot using a specific mAb for sulfated tyrosines (1C-A2); a murine anti-gp120 mAb (b13) was tested in parallel as a loading control (gp120). (C) Detection of sulfated tyrosines by autoradiography in metabolically labeled HEK293 cells expressing HIV-1 BaL WT or Δ164–190 gp160 by vaccinia technology. The cells were labeled with free [35S]sulfate or [35S]cysteine/[35S]methionine by overnight culture in sulfate-free or cysteine/methionine-free medium, respectively. The films were exposed for 48 h for [35S]sulfate labeling and for 18 h for [35S]cysteine/[35S]methionine labeling. (D) Detection of sulfated tyrosines by Western blot in HEK293 cells transfected with a plasmid encoding WT HIV-1 BaL gp160 or the phenylalanine-substituted mutants BaL Y173F, BaL Y177F, and BaL Y173F/Y177F. Western blot analysis was performed as in B.
To confirm the presence of sulfated tyrosines in gp120 and their selective localization in the V2 loop using a different methodology, we performed metabolic labeling with free [35S]sulfate in cells expressing HIV-1 BaL WT or Δ164–190. Gp120 was immunoprecipitated from metabolically labeled cells, deglycosylated to exclude [35S]sulfate incorporated into glycans, and analyzed by autoradiography. Although both WT and Δ164–190 gp120 were labeled with comparable amounts of [35S]cysteine and [35S]methionine, only WT gp120 incorporated free [35S]sulfate (Fig. 1C), confirming the presence of sulfated tyrosines in gp120 and their specific localization within the central region of V2.
The identity of the gp120 sulfotyrosines was investigated further by mutating Tyr173 and Tyr177 to phenylalanine either individually (Y173F, Y177F) or in combination (Y173F/177F). All three mutants showed a loss of Tys signal by Western blot (Fig. 1D), confirming that the central V2 tyrosines selectively account for gp120 sulfation. The absence of signal in both positions with individual phenylalanine mutants is in line with observations made with single phenylalanine mutants of the CCR5 N terminus (35), suggesting that tyrosine sulfation is hampered by the presence of neighboring phenylalanines.
Overexpression of Tyrosyl Protein Sulfotransferase 2 Enhances the Low Constitutive Levels of gp120 Tyrosine Sulfation in Continuous Cell Lines.
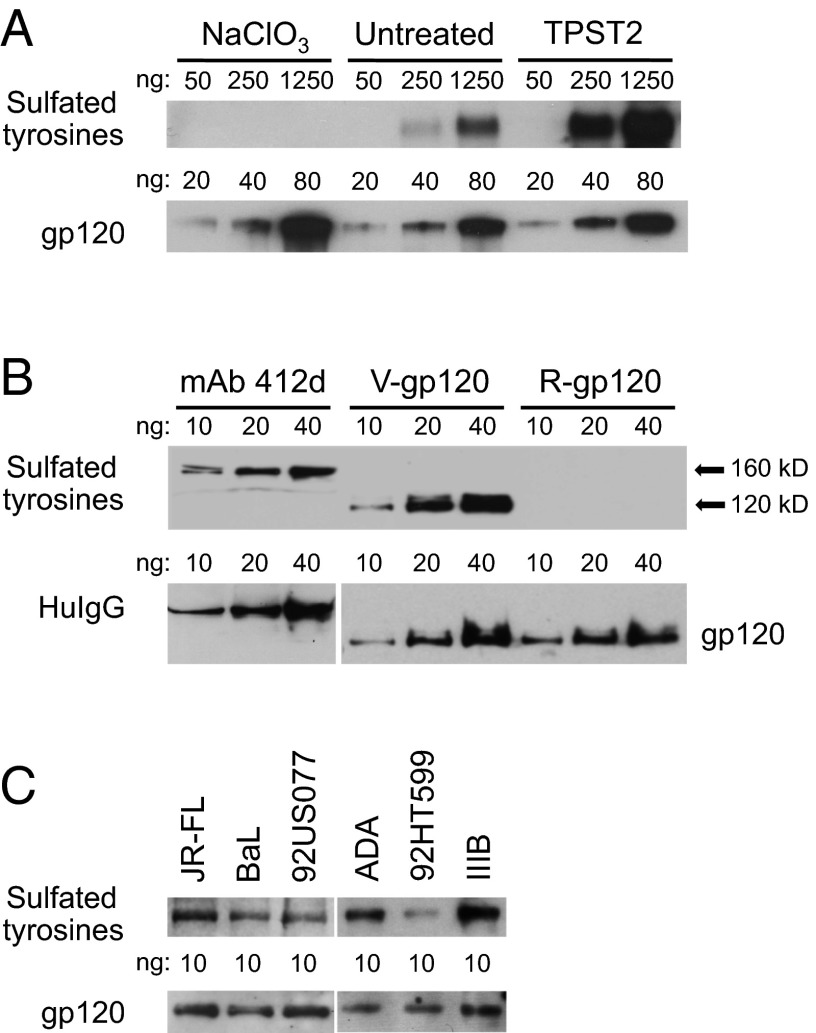
Because continuous cell lines are generally inefficient in posttranslationally modifying tyrosines by O-sulfation (37), we evaluated by Western blot the constitutive levels of gp120 tyrosine sulfation in CHO cells and the effects of overexpression of tyrosyl protein sulfotransferase 2 (TPST2) or treatment with the sulfotransferase inhibitor sodium chlorate (NaClO3) (35). To visualize low levels of tyrosine sulfation, high amounts of gp120 were loaded (fivefold dilutions starting at 1,250 ng per lane). Fig. 2A shows that the constitutive levels of tyrosine sulfation in CHO-expressed recombinant gp120 (BaL) were low, but they could be enhanced markedly by overexpression of TPST2, whereas treatment with 30 mM NaClO3 totally abrogated the signal (Fig. 2A), even though it had no adverse effects on cell viability (Fig. S2).
Fig. 2.
Efficiency of V2 tyrosine sulfation in recombinant gp120 produced in continuous cell lines and virion-associated gp120 produced in primary CD4+ T cells. (A) Detection of sulfated tyrosines in recombinant gp120 produced in CHO cells. The effects of overexpression of the sulfotransferase TPST2 or treatment with the sulfotransferase inhibitor NaClO3 (30 mM) were tested in parallel. Gp120 was expressed by transfection, purified from the cell culture supernatants by lectin affinity, and quantified. Identical amounts of serially diluted gp120s were loaded onto the gel, as indicated over each lane. The presence of sulfated tyrosines was evaluated by Western blot with the specific mAb 1C-A2; loading controls were revealed with a murine anti-gp120 mAb (b24). To visualize low levels of tyrosine sulfation, high amounts of gp120 were used (fivefold dilutions starting from 1,250 ng per lane). (B) Direct side-by-side comparison of tyrosine sulfation levels in a reference sulfated mAb (412d) versus virion-associated gp120 (BaL) produced by infected primary human CD4+ T cells (V-gp120) and recombinant gp120 (BaL) produced in HEK293 cells (R-gp120). Identical amounts of serially diluted mAb 412d and purified gp120s were loaded onto the gels under nondenaturing conditions, as indicated over each lane. For V-gp120, the protein was immunoprecipitated directly from the infectious viral stock (BaL), quantified, and analyzed by Western blot as described in A. The loading control for mAb 412d was revealed using a murine mAb specific for human IgG. (C) Detection of sulfated tyrosines in virion-associated gp120 purified from different HIV-1 isolates grown in primary human CD4+ T cells. Infectious viral stocks from six primary and laboratory isolates with different coreceptor-use phenotypes were used: Three were CCR5-tropic (BaL, JR-FL, ADA), two were dual-tropic (92US077, 92HT599), and one was CXCR4-tropic (IIIB). The sequences of the tyrosine-sulfated segments of V2 (amino acids 170–181) from the tested isolates are as follows: JR-FL: QKEYALFYKLDV; BaL: QKEYALFYELDI; 92US077: QKEDAFFYKSDV; ADA: KKDYALFYRLDV; 92HT599: QKEYALFSKLDV; IIIB: QKEYAFFYKLDI. Although BaL contains an unusual acidic residue (Glu) at position 178, which may favor tyrosine sulfation at position 177, all the other isolates contained a basic residue (Lys or Arg) at position 178. The gp120 proteins were immunoprecipitated directly from the viral stocks, quantified, and analyzed by Western blot as in A.
Virion-Associated gp120 Produced by Infected Primary CD4+ T Cells Is Highly Sulfated.
Next, we tested the levels of V2 tyrosine sulfation in gp120 purified from HIV-1 virions produced by primary human CD4+ T lymphocytes, which are physiologically relevant target cells for HIV-1 infection. Relative quantification of sulfation levels was obtained by direct side-by-side comparison of purified gp120 with a reference sulfated human mAb, 412d, which contains two sulfotyrosines in its CDRH3 domain (37). When identical amounts of serially diluted virion-associated gp120 from HIV-1 BaL (V-gp120) and mAb 412d were loaded onto the same gel, both proteins displayed high levels of tyrosine sulfation, as indicated by the presence of visible bands at protein concentrations as low as 10 ng per lane, which is close to the lower detection limit of the method (Fig. 2B). In contrast, no signal was detected with recombinant homologous gp120 (BaL) produced in HEK293 cells (R-gp120). These results indicate that a major fraction of virion-associated gp120 produced by primary CD4+ T cells contains sulfated tyrosines.
To confirm the presence and efficiency of tyrosine sulfation in different HIV-1 envelopes, we tested gp120 purified from whole virions of six unrelated viral isolates produced in primary CD4+ T cells, including both laboratory-expanded and primary isolates displaying different coreceptor-use phenotypes: three isolates were CCR5-tropic (BaL, JR-FL, ADA); two were dual-tropic (92US077, 92HT599); and one was CXCR4-tropic (IIIB). Fig. 2C shows that sulfotyrosine signals were detected in all cases with 10 ng of gp120, indicating a high degree of sulfation efficiency. Of note, 92HT599, a rare isolate that lacks the highly conserved Tyr177, displayed a reduced sulfotyrosine signal. Taken together, these data demonstrate that in different HIV-1 isolates grown in physiologically relevant target cells the V2 tyrosines are sulfated efficiently, in sharp contrast with the inefficiency documented in continuous cell lines.
A Tyrosine-Sulfated V2 Peptide Mimetic Interacts with the CCR5-Binding Site at the Base of V3.
Having established that the V2 loop of gp120 contains sulfated tyrosines with spacing identical to that in the CCR5 N terminus, we hypothesized that these modified tyrosines could functionally mimic the CCR5 sulfotyrosines and mediate intramolecular interaction of V2 with the base of V3 in the CD4-unbound, prefusion gp120 state. This interaction is compatible with the recently published crystal structure of a soluble SOSIP gp140 trimer (18), in which the 173–177 segment is directly juxtaposed to the CCR5-binding region at the base of V3, even though sulfotyrosines were not detected in this structure (most likely because the trimer was produced in HEK293 cells). Because the sulfated human mAb 412d interacts with the CCR5-binding site in V3 (34), we tested the ability of a tyrosine-sulfated 18-amino acid peptide mimetic derived from the central region of V2 (pV2α-Tys; amino acids 168–185, bearing sulfations on both Tyr173 and Tyr177) to compete with 412d binding to gp120 (BaL) by surface plasmon resonance. Because the 412d-binding site becomes accessible only after binding to CD4 (37), gp120 was precomplexed with sCD4. Fig. 3A shows that peptide pV2α-Tys potently inhibited binding of surface-bound mAb 412d to CD4-activated gp120, whereas its unsulfated counterpart (pV2α) had a limited effect. No binding was detected in the absence of sCD4. As an additional control, we tested a tyrosine-sulfated peptide derived from the CCR5 N terminus, pCCR5-Tys (amino acids 1–22, bearing sulfations on both Tyr10 and Tyr14), which, in line with previous observations, markedly reduced gp120-sCD4 binding to 412d (37), whereas the corresponding unsulfated peptide (pCCR5) had no effect. Specificity was further confirmed by the lack of inhibition of gp120 binding to a control mAb, F105 (Fig. 3A).
Fig. 3.
Competition of a tyrosine-sulfated V2 mimetic peptide with mAb 412d binding to gp120. (A) Effect of a tyrosine-sulfated V2-loop mimetic peptide (pV2α-Tys; amino acids 168–185) on mAb 412d binding to gp120 as assessed by surface plasmon resonance. A tyrosine-sulfated peptide derived from the CCR5 N terminus (pCCR5-Tys; amino acids 1–22) and unsulfated peptides from V2 (pV2α) and CCR5 (pCCR5) were tested in parallel as controls. MAb 412d was immobilized on the sensor surface and tested for binding to gp120 (BaL) pretreated with two-domain sCD4 in the presence or absence of the peptides used at 166 μM. A control antibody (F105; no sCD4 pretreatment) was tested in parallel as a control. The data are from a representative experiment of three that were performed with similar results. (B) Effect of tyrosine-sulfated and unsulfated V2-loop and CCR5 N terminus mimetic peptides on HIV-1 virion capture by mAb 412d. Infectious viral stocks from HIV-1 BaL were pretreated with sCD4 (5 μg/mL) in the presence or absence of the indicated peptides (each at 100 μM) and then were mixed with immunomagnetic beads prearmed with mAb 412d. A control antibody (2G12; no sCD4 pretreatment) was tested in parallel as a further specificity control. The data presented are mean values (± SD) from three independent experiments. Asterisks denote significant differences from the virion capture in the peptide-untreated control (P < 0.01 by unpaired Student t test).
The interaction of the V2 peptide mimetic with the 412d-binding site was confirmed using a virion-capture assay in which the HIV-1 envelope is displayed on the surface of intact virions. To reveal the 412d-binding site, the viral stock (BaL) was treated with sCD4 before capture by antibody-coated magnetic beads. Fig. 3B shows that peptide pV2α-Tys dramatically reduced virion capture by 412d, whereas the unsulfated peptide (pV2α) had a limited effect; likewise, 412d-mediated virion capture was significantly reduced by the sulfated CCR5 peptide but not by its unsulfated counterpart. As an additional proof of specificity, none the peptides affected virion capture by a control mAb, 2G12 (Fig. 3B). Taken together, these results support the concept that in unliganded gp120 the V2 sulfotyrosines interact with the CCR5- and 412d-binding site at the base of V3.
Tyrosine Sulfation in the V2 Loop Specifically Modulates gp120 Epitope Exposure and Neutralization Sensitivity.
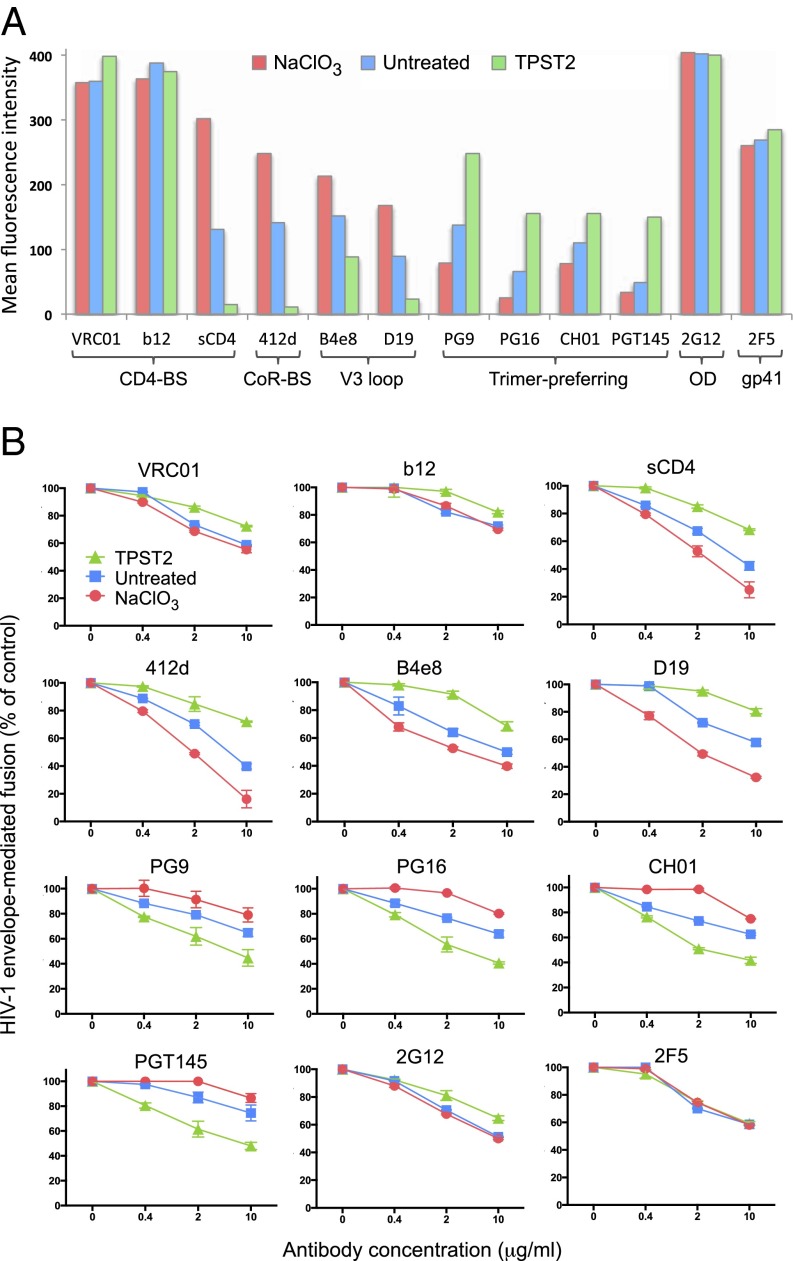
To investigate the functional role of the V2 sulfotyrosines, we tested the effects of the modulation of tyrosine sulfation on gp120 epitope accessibility and neutralization using a panel of antibodies to the major neutralization regions of gp120 and monomeric sCD4. The experiments were performed on full-length, cleavable gp160 derived from two unrelated HIV-1 isolates, BaL and YU2, expressed on the surface of HeLa cells. A striking dichotomous effect was observed on the antigenic profile of gp120 from both isolates (Fig. 4A and Fig. S3). Enhancement of tyrosine sulfation by TPST2 overexpression dramatically reduced binding of monomeric sCD4 (receptor-binding site), mAb 412d (coreceptor-binding site) (37), and anti-V3 loop mAbs B4e8 (38) and D19 (39), all ligands with a restricted binding capacity to the native trimeric spike. In contrast, TPST2 overexpression markedly increased recognition by the trimer-preferring antibodies PG9, PG16, CH01, and PGT145, which are directed to quaternary, glycan-dependent V2 epitopes that are stabilized on the native trimer (22, 23, 40). Opposite effects were observed when tyrosine sulfation was inhibited by NaClO3 treatment, with a marked increase of sCD4, 412d, and anti-V3 antibody binding and a reciprocal decrease in recognition by trimer-preferring antibodies. Binding of three broadly neutralizing antibodies which readily recognize the native trimer, b12 and VRC01 (CD4-binding site) (9, 41), and 2G12 (outer domain) (42), was not affected significantly. Of note, neither TPST2 overexpression nor NaClO3 treatment had adverse effects on the levels of gp160 expression, as evaluated by reactivity with a control antibody, 2F5, directed against the gp41 membrane-proximal external region (Fig. 4A and Fig. S3).
Fig. 4.
Effect of V2 tyrosine sulfation on gp120 (BaL) epitope exposure and neutralization. (A) Modulation of V2 tyrosine sulfation alters gp120 (BaL) epitope accessibility. Flow cytometry was performed on HeLa cells expressing full-length, cleavable HIV-1 BaL gp160 on their surface. V2 tyrosine sulfation was enhanced by overexpression of the sulfotransferase TPST2 or inhibited by treatment with the sulfotransferase inhibitor NaClO3 (30 mM). A panel of human mAbs directed to the indicated domains of gp120 and gp41 or four-domain soluble CD4 (sCD4) (each at 5 μg/mL) was used. BS, binding site; CoR, coreceptor; OD, outer domain. The sequence of the tyrosine-sulfated segment of V2 (amino acids 170–181) of HIV-1 BaL is QKEYALFYELDI. The data represent mean fluorescence intensity values after subtraction of background fluorescence levels. The signals obtained with the reference antibody 2F5 were used to verify that cell-surface envelope expression levels were comparable in the three cultures. The data presented are from a representative experiment of three that were performed with similar results. (B) Modulation of V2 tyrosine sulfation alters HIV-1 BaL neutralization sensitivity. Neutralization experiments were performed using an HIV-1 envelope-mediated fusion assay with the same panel of anti-gp120/gp41 antibodies and sCD4 as in A, which were tested at the indicated concentrations. The fusion assay was performed using vaccinia technology as described (39) with HeLa cells expressing full-length cleavable HIV-1 BaL gp160 as effector cells and NIH 3T3 cells expressing human CD4 and CCR5 as target cells. Tyrosine sulfation was enhanced and inhibited as described in A. The fusion values were normalized to the value obtained with untreated controls for each culture condition. The mean absolute levels of envelope-mediated fusion for the experiment shown, after background subtraction, were the following: untreated, 1.136 optical density units; NaClO3-treated, 1.192 optical density units; TPST2 overexpression, 0.891 optical density units. The data presented are mean values (± SE) of duplicate wells from a representative experiment of three that were performed with similar results.
The pattern of epitope exposure observed by flow cytometry was mirrored precisely by the results of neutralization studies. As illustrated in Fig. 4B, TPST2 overexpression increased the resistance of HIV-1 BaL to neutralization by sCD4, 412d, and anti-V3 antibodies but at the same time made the virus more sensitive to neutralization by PG9, PG16, CH01, and PDT145. On the other hand, inhibition of V2 tyrosine sulfation by NaClO3 treatment increased neutralization by sCD4, 412d, and anti-V3 antibodies and decreased sensitivity to trimer-preferring antibodies. Overexpression of TPST2 had only a slight inhibitory effect on neutralization by 2G12, b12, and VRC01, in accordance with the ability of these antibodies to bind with high affinity to the native trimeric spike. Neutralization by the control anti-gp41 antibody 2F5 was totally unaffected. Consistent with a diminished interaction with CD4, the efficiency of the fusion reaction was reduced slightly in TPST2-expressing cells compared with untreated and NaClO3-treated cells (see the legend for Fig. 4B). As a further proof of specificity, NaClO3 treatment did not affect the neutralization sensitivity of the gp160 Δ164–190 mutant (Fig. S4), which lacks sulfated tyrosines (Fig. 1 B and C). Although these results await confirmation on a larger panel of HIV-1 isolates of different genetic subtypes, they suggest that the sulfotyrosine-mediated V2–V3 interaction plays a critical role in stabilizing the trimeric envelope conformation and specifically modulates the sensitivity of HIV-1 to antibody-mediated neutralization.
Discussion
In this study, we provide evidence for the presence of sulfated tyrosines within the V2 loop of the external HIV-1 envelope glycoprotein subunit gp120 and show that these modified tyrosines play an important role in the functional structuring of the native envelope by stabilizing the intramolecular interaction between V2 and V3. Tyrosine sulfation is a posttranslational modification that is estimated to occur in ∼7% of mammalian proteins and is increasingly recognized as an important modulator of protein–protein interaction (43). Tyrosine sulfation of functional relevance was identified previously in several host proteins that bind to gp120, including coreceptors (35), antibodies to the coreceptor-binding site such as 412d (37), and, more recently, antibodies to glycan-dependent quaternary epitopes such as PG9 and PG16 (44). Unlike these previous examples, however, our data identify tyrosine sulfation within a viral protein, as reported for varicella-zoster virus envelope glycoproteins (45). Furthermore, it is remarkable that HIV-1 appears to have modeled the tyrosine-sulfated region of the V2 loop after the N-terminal domain of the CCR5 coreceptor, which interacts with the same conserved region at the base of V3. Our results shed light on the mechanisms whereby V2 occludes the coreceptor-binding site in the prefusion envelope conformation, preventing access to 412d and other sulfated antibodies. Upon binding to CD4, however, V2 unclamps from V3, unraveling the coreceptor-binding site and allowing the sulfated coreceptor N terminus to replace V2 at the base of V3. These events promote subsequent conformational changes that eventually lead to the fusion event.
The direct V2–V3 interaction described herein is supported by biological and structural evidence. Functional and antigenic interactions between V2 and V3 have long been recognized (21–27), and spatial proximity between these two loops has been documented by both cryo-EM studies (12–17) and the recently solved crystal structure of a stabilized soluble SOSIP trimer at 4.7-Å resolution (18). Despite the lack of sulfated tyrosines (presumably because the soluble trimer was produced in HEK293 cells), the latter structure is consistent with our model because it shows the 173–177 segment of V2 directly juxtaposed to the CCR5-binding region at the base of V3, with the most conserved of the two sulfotyrosines, Tyr177, establishing van der Waals contacts with Ile420, Leu154, and Leu175, as well as an H-bond with Asn302, all residues that are proximal to the predicted binding pocket for the homologous sulfotyrosine (Tys14) of CCR5 (34). It is likely that the addition of sulfate groups would markedly affect the local conformations and interactions, including the establishment of additional H-bonds between Tys177 and Arg298, Asn300, and the backbone of Arg327, resulting in a significant increase in the total binding energy. Thus, the absence of sulfated tyrosines may be one of the reasons why the SOSIP.664 trimer, albeit reportedly in a “near-native” conformation (32), maintains some phenotypic properties of the open gp120 structure, such as reactivity with anti-V3 mAbs that do not neutralize the homologous virus, intermediate-affinity binding to monomeric sCD4, and an unfavorable ratio between the concentrations of trimer-preferring antibodies required for binding versus neutralization of the homologous virus (32).
The divergent effects on gp120 epitope exposure and neutralization that we observed upon modulation of sulfation attest to the biological relevance of tyrosine sulfation in the V2 loop. Although the present results were obtained with only two HIV-1 envelopes and need to be validated on a larger panel of genotypically and phenotypically diverse isolates, they suggest that the sulfotyrosine-bolstered V2–V3 interaction is a critical mechanism whereby HIV-1 constrains the envelope trimer in its metastable native conformation. The native trimer was shown to shield specific vulnerable sites—most notably the receptor- and coreceptor-binding sites (1–4)—effectively from antibody recognition, but at the same time it stabilizes quaternary epitopes recognized by a unique group of trimer-preferring antibodies, such as PG9, CH01, and PGT145, that make direct contact with high-mannose glycan moieties at the trimer apex (22, 23, 40). Thus, HIV-1 must cope with an immunologic oxymoron by which protection of key vulnerable sites is associated with exposure of other vulnerable sites. However, the generation of trimer-preferring antibodies is a challenging process for the host immune system that requires selection for long CDRH3 domains and extensive somatic hypermutation (2, 40).
Several considerations point to Tys177, analogous to Tys14 in CCR5, as the most important V2 sulfotyrosine for stabilizing the interaction with V3, whereas Tys173, corresponding to Tys10 in CCR5, may play a less critical role. In agreement with this model, Tys177 is extremely conserved across all HIV-1 subtypes, whereas Tys173 shows greater variability, being conserved in subtypes A, B, and C but often replaced by histidine in subtypes D, E, and F. Although we cannot exclude that different viral genotypes may have devised slightly different solutions to stabilize the V2–V3 interaction, genetic and phenotypic diversity is a true hallmark of HIV-1, as reflected by a broad range of neutralization sensitivities among primary isolates. Thus, variation at position 173 or neighboring amino acids that may influence the efficiency of V2 tyrosine sulfation (43) might be exploited by different HIV-1 isolates to fine-tune the tightness of their antibody shield, balancing the need to maintain a high replication fitness and the need to elude immunologic recognition of the receptor- and coreceptor-binding sites.
As seen with other posttranslational modifications such as N-linked glycosylation, tyrosine sulfation varies remarkably in different cell lineages. Indeed, we found that gp120 sulfation in certain continuous cell lines is inefficient, presumably because of limited endogenous expression of tyrosyl sulfotransferases. This observation has important practical implications, because cell lines such as CHO and HEK293 are widely used for the production of recombinant gp120 for structural, biological, and clinical studies, including atomic-level structure determination (6–10, 18), standardized neutralization assays (46), and experimental vaccine trials (47, 48). These considerations emphasize the need to use in vitro expression systems that implement physiologically relevant posttranslational modifications. Importantly, we found that in primary human CD4+ T lymphocytes, which are the main physiological target cells for HIV-1 infection, gp120 from different viral isolates was sulfated efficiently, corroborating the biological relevance of this modification.
Finally, our findings provide a further rationale for considering the conserved central region of V2 as a potential target for HIV-1 vaccine development, emphasizing tyrosine sulfation in this region not only as a component of the immunogenic epitopes but also as a key modulator of their 3D structure. Thus, the presence and extent of V2 tyrosine sulfation may be relevant criteria for the evaluation of candidate vaccine immunogens, because a high rate of gp120 sulfation may favor the adoption of a more stable prefusion conformation. Moreover, the stabilizing effect of V2 tyrosine sulfation on the HIV-1 envelope structure may be exploited to promote crystallization of gp120 in both monomeric and trimeric forms.
Materials and Methods
The presence and localization of sulfated tyrosines in HIV-1 gp120 were investigated by Western blot using a specific anti-sulfotyrosine antibody and by metabolic labeling. Details of materials and methods used in this work are given in SI Materials and Methods. This section describes methods for the detection of sulfated tyrosines, modulation of tyrosine sulfation, peptide synthesis, surface plasmon resonance, HIV-1 virion capture, flow cytometry, neutralization of HIV-1 envelope-mediated fusion, and statistical analysis.
Supplementary Material
Acknowledgments
We thank Edward A. Berger and Anthony S. Fauci for helpful suggestions and critical reading of the manuscript; John R. Mascola, George K. Lewis, and James E. Robinson for anti-gp120 mAbs; Michael Farzan for the TPST2-expressing vector; Jocelyn Ray for help in producing recombinant gp120; the IAVI consortium for mAbs PG9, PG16, CH01, and PGT145; and the National Institutes of Health (NIH) AIDS Reagent Program for cell lines and reagents. This work was supported by the National Institute for Allergy and Infectious Diseases, NIH, Intramural Research Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314718111/-/DCSupplemental.
References
- 1.Burton DR, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12(4):396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: Understanding nature’s pathways. Immunol Rev. 2013;254(1):225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 4.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 5.Myszka DG, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97(16):9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong PD, et al. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8(12):1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 8.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326(5956):1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107(3):1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: Strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6(12):e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu G, Liu J, Taylor KA, Roux KH. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J Virol. 2011;85(6):2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci USA. 2011;108(28):11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci USA. 2013;110(11):4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20(12):1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan N, Thali M, Furman C, Ho DD, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67(6):3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang SH, et al. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84(7):3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci USA. 2012;109(15):5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker LM, et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao J, et al. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71(12):9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinter A, et al. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Cimbro R, Lusso P, Berger EA. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci USA. 2011;108(50):20148–20153. doi: 10.1073/pnas.1104840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusert P, et al. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J Exp Med. 2011;208(7):1419–1433. doi: 10.1084/jem.20110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghiara JB, Stura EA, Stanfield RL, Profy AT, Wilson IA. Crystal structure of the principal neutralization site of HIV-1. Science. 1994;264(5155):82–85. doi: 10.1126/science.7511253. [DOI] [PubMed] [Google Scholar]
- 29.Liao HX, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38(1):176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancera M, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20(7):804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol. 2010;10(7):527–535. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846):1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 36.Farzan M, et al. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275(43):33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- 37.Choe H, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114(2):161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 38.Cavacini LA, et al. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS. 2003;17(5):685–689. doi: 10.1097/00002030-200303280-00006. [DOI] [PubMed] [Google Scholar]
- 39.Lusso P, et al. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J Virol. 2005;79(11):6957–6968. doi: 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem. 2003;278(27):24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 44.Pejchal R, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107(25):11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edson CM. Tyrosine sulfation of varicella-zoster virus envelope glycoprotein gpl. Virology. 1993;197(1):159–165. doi: 10.1006/viro.1993.1576. [DOI] [PubMed] [Google Scholar]
- 46.Mascola JR, et al. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79(16):10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitisuttithum P, et al. Bangkok Vaccine Evaluation Group Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194(12):1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 48.Rerks-Ngarm S, et al. MOPH-TAVEG Investigators Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.